a9cc945610e63825758231ba07354053.ppt
- Количество слайдов: 40
 QSAR/QSPR: the Universal Approach to the Prediction of Properties of Chemical Compounds and Materials V. A. Palyulin, I. I. Baskin, N. S. Zefirov Department of Chemistry Moscow State University
QSAR/QSPR: the Universal Approach to the Prediction of Properties of Chemical Compounds and Materials V. A. Palyulin, I. I. Baskin, N. S. Zefirov Department of Chemistry Moscow State University
 "Every attempt to employ mathematical methods in the study of chemical questions must be considered profoundly irrational and contrary to the spirit of chemistry. If mathematical analysis should ever hold a prominent place in chemistry an aberration which is happily almost impossible - it would occasion a rapid and widespread degeneration of that science. " A. Compte, 1798 -1857
"Every attempt to employ mathematical methods in the study of chemical questions must be considered profoundly irrational and contrary to the spirit of chemistry. If mathematical analysis should ever hold a prominent place in chemistry an aberration which is happily almost impossible - it would occasion a rapid and widespread degeneration of that science. " A. Compte, 1798 -1857
 Fundamental Problem in Chemistry: Evaluation of relationships between the structures of chemical compounds and their properties or biological activity
Fundamental Problem in Chemistry: Evaluation of relationships between the structures of chemical compounds and their properties or biological activity
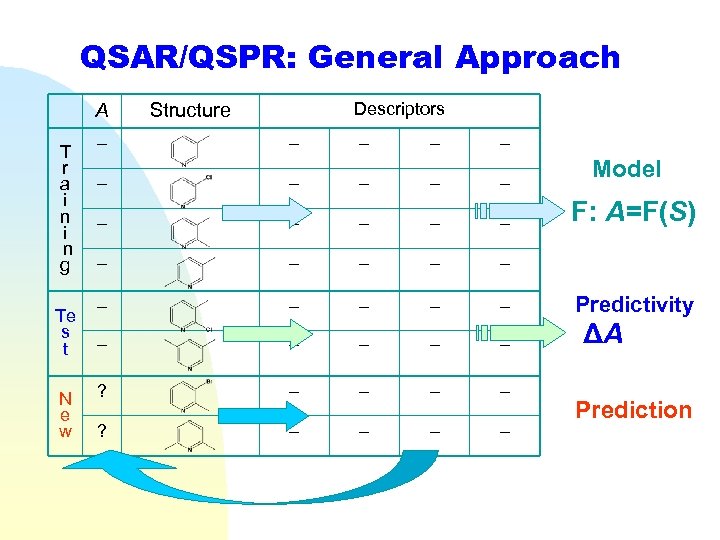 QSAR/QSPR: General Approach A T r a i n g Te s t N e w Descriptors Structure – – – – – – – – ? – – – – Model F: A=F(S) Predictivity ΔA Prediction
QSAR/QSPR: General Approach A T r a i n g Te s t N e w Descriptors Structure – – – – – – – – ? – – – – Model F: A=F(S) Predictivity ΔA Prediction
 PROPERTIES Physico-chemical properties: Boiling points, melting points, density, viscosity, surface tension, solubility in various solvents, lipophilicity, magnetic susceptibility, retention indices, dipole moments, enthalpy of formation, etc. Biological activity: IC 50, EC 50, LD 50, MEC, ILS, etc.
PROPERTIES Physico-chemical properties: Boiling points, melting points, density, viscosity, surface tension, solubility in various solvents, lipophilicity, magnetic susceptibility, retention indices, dipole moments, enthalpy of formation, etc. Biological activity: IC 50, EC 50, LD 50, MEC, ILS, etc.
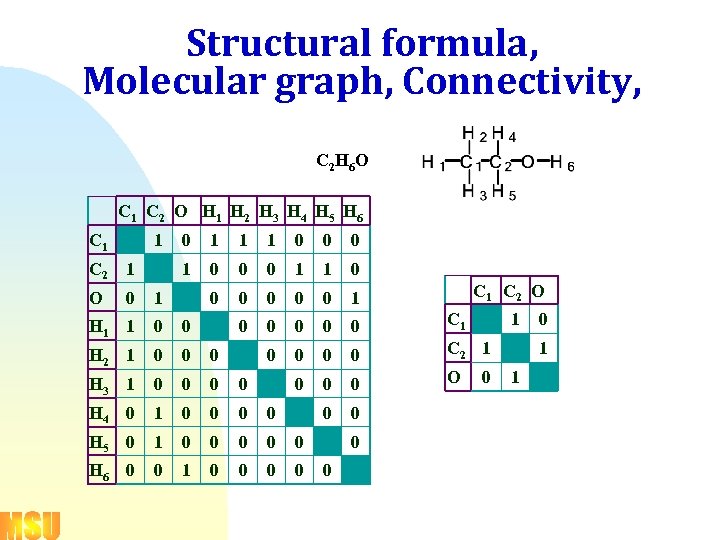 Structural formula, Molecular graph, Connectivity, C 2 H 6 O C 1 C 2 O H 1 H 2 H 3 H 4 H 5 H 6 C 1 1 O 1 1 1 0 0 0 1 C 2 1 0 0 1 1 0 0 0 C 1 0 0 C 2 1 0 0 0 O 0 0 0 1 H 1 1 0 0 H 2 1 0 0 0 H 3 1 0 0 H 4 0 1 0 0 H 5 0 1 0 0 0 H 6 0 0 1 0 0 0 C 1 C 2 O 1 0 0 1 1
Structural formula, Molecular graph, Connectivity, C 2 H 6 O C 1 C 2 O H 1 H 2 H 3 H 4 H 5 H 6 C 1 1 O 1 1 1 0 0 0 1 C 2 1 0 0 1 1 0 0 0 C 1 0 0 C 2 1 0 0 0 O 0 0 0 1 H 1 1 0 0 H 2 1 0 0 0 H 3 1 0 0 H 4 0 1 0 0 H 5 0 1 0 0 0 H 6 0 0 1 0 0 0 C 1 C 2 O 1 0 0 1 1
 DESCRIPTORS Topological indices: Connectivity indices (Randic, c; Kier-Hall, mcv, solvation indices mcs), Wiener W and expanded Wiener, Balaban J, Gutman indices, Hosoya, Merrifield-Simmons indices, indices based on local invariants, informational indices, … Fragmental descriptors: The number of fragments of various size (chains, cycles, branched fragments) in a molecule with several levels of classification of atoms Physico-chemical descriptors: Indices based on atomic charges and electronegativities, atomic inductive constants, Vd. W volume and surface, H-bond descriptors, Lipophilicity (Log P), … Quantum-mechanical 3 D Usp. Khim. (Russ. Chem. Rev. ), 57 (3), 337 -366 (1988)
DESCRIPTORS Topological indices: Connectivity indices (Randic, c; Kier-Hall, mcv, solvation indices mcs), Wiener W and expanded Wiener, Balaban J, Gutman indices, Hosoya, Merrifield-Simmons indices, indices based on local invariants, informational indices, … Fragmental descriptors: The number of fragments of various size (chains, cycles, branched fragments) in a molecule with several levels of classification of atoms Physico-chemical descriptors: Indices based on atomic charges and electronegativities, atomic inductive constants, Vd. W volume and surface, H-bond descriptors, Lipophilicity (Log P), … Quantum-mechanical 3 D Usp. Khim. (Russ. Chem. Rev. ), 57 (3), 337 -366 (1988)
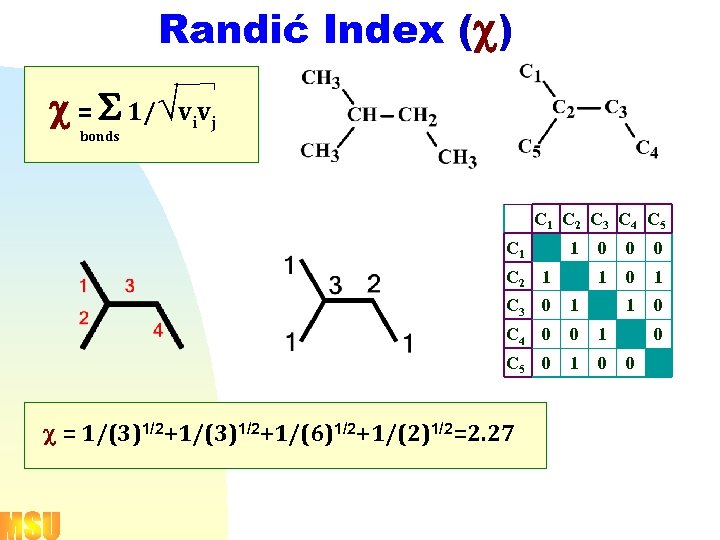 Randić Index (c) ¾Ø c = S 1/Övivj bonds C 1 C 2 C 3 C 4 C 5 C 1 1 0 0 1 C 2 1 0 0 1 1 0 C 3 0 1 C 4 0 0 1 C 5 0 1 0 c = 1/(3)1/2+1/(6)1/2+1/(2)1/2=2. 27 0 0
Randić Index (c) ¾Ø c = S 1/Övivj bonds C 1 C 2 C 3 C 4 C 5 C 1 1 0 0 1 C 2 1 0 0 1 1 0 C 3 0 1 C 4 0 0 1 C 5 0 1 0 c = 1/(3)1/2+1/(6)1/2+1/(2)1/2=2. 27 0 0
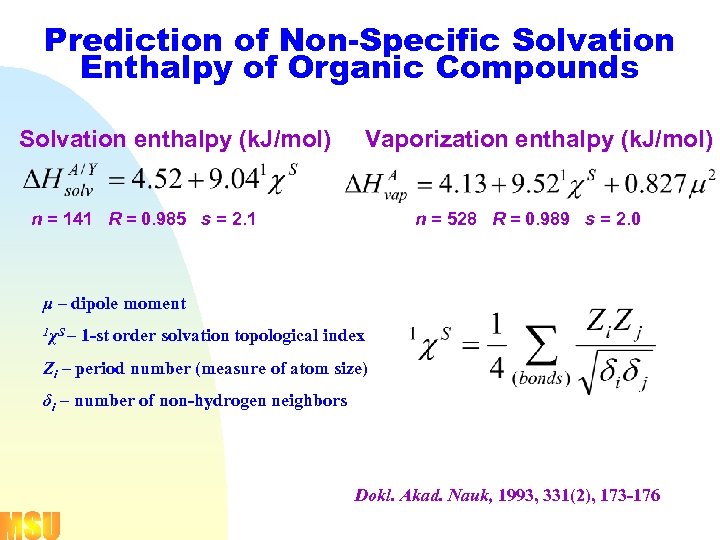 Prediction of Non-Specific Solvation Enthalpy of Organic Compounds Solvation enthalpy (k. J/mol) Vaporization enthalpy (k. J/mol) n = 141 R = 0. 985 s = 2. 1 n = 528 R = 0. 989 s = 2. 0 μ – dipole moment 1 χS – 1 -st order solvation topological index Zi – period number (measure of atom size) δi – number of non-hydrogen neighbors Dokl. Akad. Nauk, 1993, 331(2), 173 -176
Prediction of Non-Specific Solvation Enthalpy of Organic Compounds Solvation enthalpy (k. J/mol) Vaporization enthalpy (k. J/mol) n = 141 R = 0. 985 s = 2. 1 n = 528 R = 0. 989 s = 2. 0 μ – dipole moment 1 χS – 1 -st order solvation topological index Zi – period number (measure of atom size) δi – number of non-hydrogen neighbors Dokl. Akad. Nauk, 1993, 331(2), 173 -176
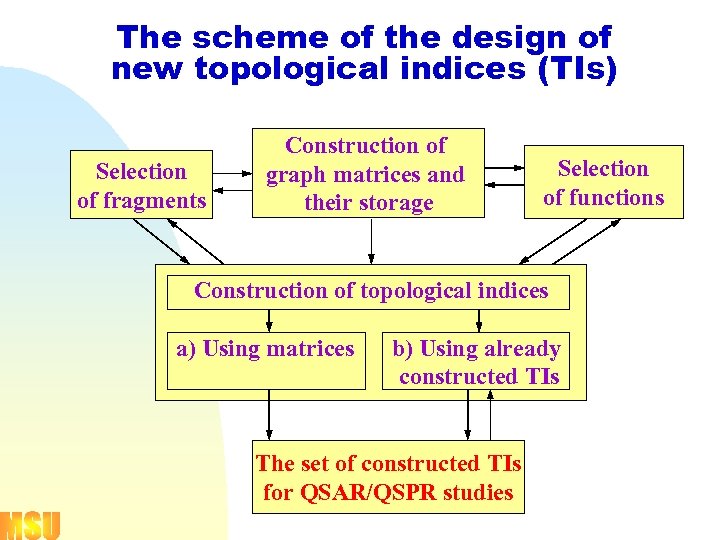 The scheme of the design of new topological indices (TIs) Selection of fragments Construction of a graph matrices and their storage Selection of functions Construction of topological indices a) Using matrices b) Using already constructed TIs The set of constructed TIs for QSAR/QSPR studies
The scheme of the design of new topological indices (TIs) Selection of fragments Construction of a graph matrices and their storage Selection of functions Construction of topological indices a) Using matrices b) Using already constructed TIs The set of constructed TIs for QSAR/QSPR studies
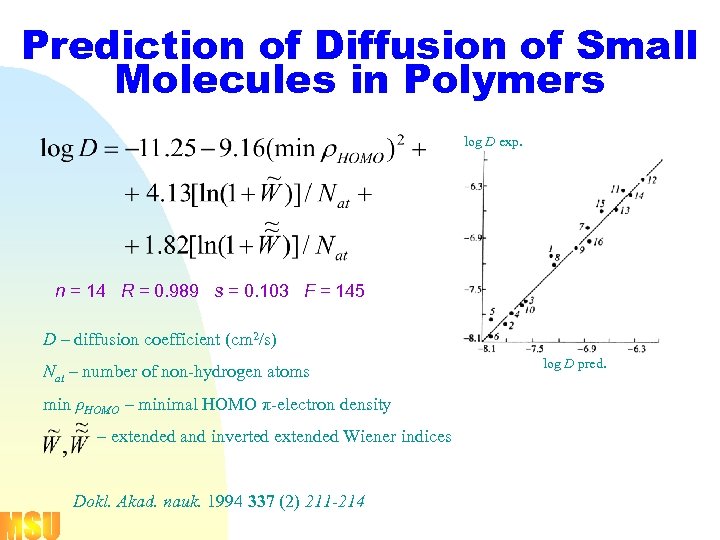 Prediction of Diffusion of Small Molecules in Polymers log D exp. n = 14 R = 0. 989 s = 0. 103 F = 145 D – diffusion coefficient (cm 2/s) Nat – number of non-hydrogen atoms min ρHOMO – minimal HOMO π-electron density – extended and inverted extended Wiener indices Dokl. Akad. nauk. 1994 337 (2) 211 -214 log D pred.
Prediction of Diffusion of Small Molecules in Polymers log D exp. n = 14 R = 0. 989 s = 0. 103 F = 145 D – diffusion coefficient (cm 2/s) Nat – number of non-hydrogen atoms min ρHOMO – minimal HOMO π-electron density – extended and inverted extended Wiener indices Dokl. Akad. nauk. 1994 337 (2) 211 -214 log D pred.
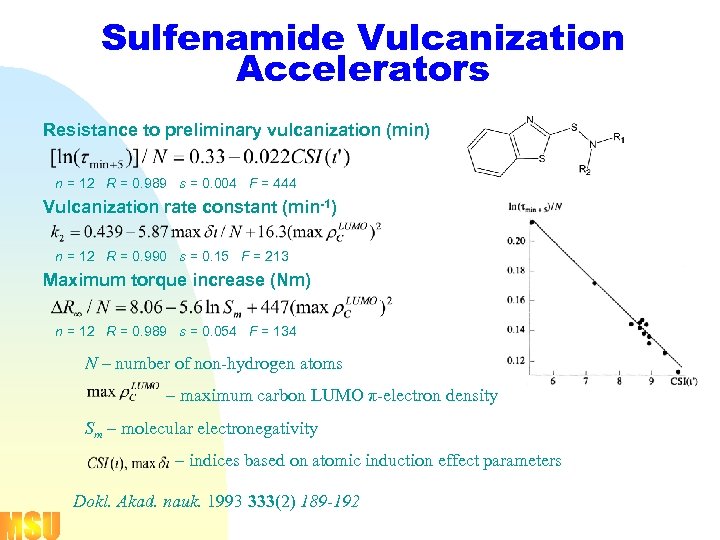 Sulfenamide Vulcanization Accelerators Resistance to preliminary vulcanization (min) n = 12 R = 0. 989 s = 0. 004 F = 444 Vulcanization rate constant (min-1) n = 12 R = 0. 990 s = 0. 15 F = 213 Maximum torque increase (Nm) n = 12 R = 0. 989 s = 0. 054 F = 134 N – number of non-hydrogen atoms – maximum carbon LUMO π-electron density Sm – molecular electronegativity – indices based on atomic induction effect parameters Dokl. Akad. nauk. 1993 333(2) 189 -192
Sulfenamide Vulcanization Accelerators Resistance to preliminary vulcanization (min) n = 12 R = 0. 989 s = 0. 004 F = 444 Vulcanization rate constant (min-1) n = 12 R = 0. 990 s = 0. 15 F = 213 Maximum torque increase (Nm) n = 12 R = 0. 989 s = 0. 054 F = 134 N – number of non-hydrogen atoms – maximum carbon LUMO π-electron density Sm – molecular electronegativity – indices based on atomic induction effect parameters Dokl. Akad. nauk. 1993 333(2) 189 -192
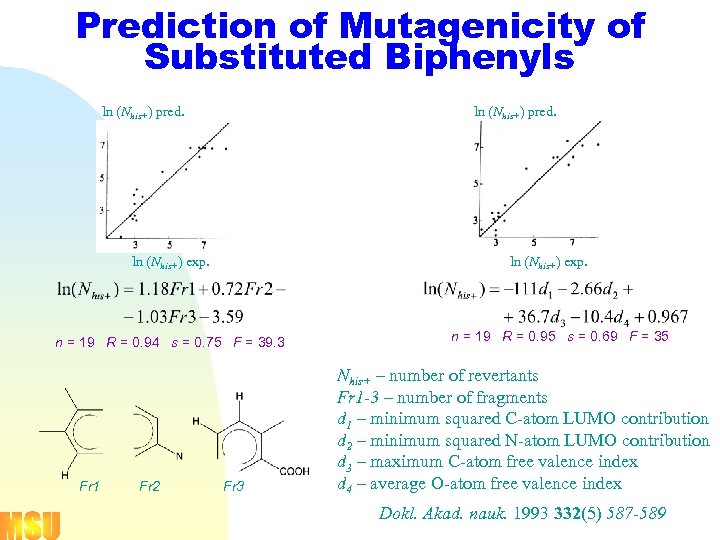 Prediction of Mutagenicity of Substituted Biphenyls ln (Nhis+) pred. ln (Nhis+) exp. n = 19 R = 0. 94 s = 0. 75 F = 39. 3 Fr 1 Fr 2 Fr 3 n = 19 R = 0. 95 s = 0. 69 F = 35 Nhis+ – number of revertants Fr 1 -3 – number of fragments d 1 – minimum squared C-atom LUMO contribution d 2 – minimum squared N-atom LUMO contribution d 3 – maximum C-atom free valence index d 4 – average O-atom free valence index Dokl. Akad. nauk. 1993 332(5) 587 -589
Prediction of Mutagenicity of Substituted Biphenyls ln (Nhis+) pred. ln (Nhis+) exp. n = 19 R = 0. 94 s = 0. 75 F = 39. 3 Fr 1 Fr 2 Fr 3 n = 19 R = 0. 95 s = 0. 69 F = 35 Nhis+ – number of revertants Fr 1 -3 – number of fragments d 1 – minimum squared C-atom LUMO contribution d 2 – minimum squared N-atom LUMO contribution d 3 – maximum C-atom free valence index d 4 – average O-atom free valence index Dokl. Akad. nauk. 1993 332(5) 587 -589
 Fragmental Descriptors The numbers of fragments of various kind and various size (chains, cycles, branched fragments) in a molecule with several levels of classification of atoms. For each molecule hundreds of fragmental descriptors can be computed. If a structure-property data set is sufficiently large to allow building statistically significant models, then any topological index can be replaced with a set of substructural (or fragmental) descriptors.
Fragmental Descriptors The numbers of fragments of various kind and various size (chains, cycles, branched fragments) in a molecule with several levels of classification of atoms. For each molecule hundreds of fragmental descriptors can be computed. If a structure-property data set is sufficiently large to allow building statistically significant models, then any topological index can be replaced with a set of substructural (or fragmental) descriptors.
 NEURAL NETWORK SOFTWARE: NASAWIN
NEURAL NETWORK SOFTWARE: NASAWIN
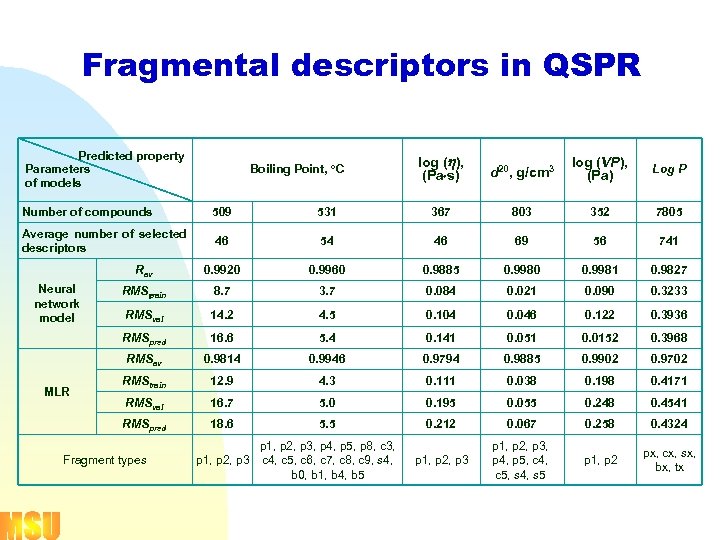 Fragmental descriptors in QSPR Predicted property Parameters of models Boiling Point, о. С log ( ), (Pa s) d 20, g/cm 3 log (VP), (Pa) Log P Number of compounds 509 531 367 803 352 7805 Average number of selected descriptors 46 54 46 69 56 741 Rav 0. 9920 0. 9960 0. 9885 0. 9980 0. 9981 0. 9827 RMStrain 8. 7 3. 7 0. 084 0. 021 0. 090 0. 3233 RMSval 14. 2 4. 5 0. 104 0. 046 0. 122 0. 3936 RMSpred 16. 6 5. 4 0. 141 0. 051 0. 0152 0. 3968 RMSav 0. 9814 0. 9946 0. 9794 0. 9885 0. 9902 0. 9702 RMStrain 12. 9 4. 3 0. 111 0. 038 0. 198 0. 4171 RMSval 16. 7 5. 0 0. 195 0. 055 0. 248 0. 4541 RMSpred 18. 6 5. 5 0. 212 0. 067 0. 258 0. 4324 p 1, p 2, p 3, p 4, p 5, c 4, c 5, s 4, s 5 p 1, p 2 px, cx, sx, bx, tx Neural network model MLR Fragment types p 1, p 2, p 3, p 4, p 5, p 8, c 3, p 1, p 2, p 3 c 4, c 5, c 6, c 7, c 8, c 9, s 4, b 0, b 1, b 4, b 5
Fragmental descriptors in QSPR Predicted property Parameters of models Boiling Point, о. С log ( ), (Pa s) d 20, g/cm 3 log (VP), (Pa) Log P Number of compounds 509 531 367 803 352 7805 Average number of selected descriptors 46 54 46 69 56 741 Rav 0. 9920 0. 9960 0. 9885 0. 9980 0. 9981 0. 9827 RMStrain 8. 7 3. 7 0. 084 0. 021 0. 090 0. 3233 RMSval 14. 2 4. 5 0. 104 0. 046 0. 122 0. 3936 RMSpred 16. 6 5. 4 0. 141 0. 051 0. 0152 0. 3968 RMSav 0. 9814 0. 9946 0. 9794 0. 9885 0. 9902 0. 9702 RMStrain 12. 9 4. 3 0. 111 0. 038 0. 198 0. 4171 RMSval 16. 7 5. 0 0. 195 0. 055 0. 248 0. 4541 RMSpred 18. 6 5. 5 0. 212 0. 067 0. 258 0. 4324 p 1, p 2, p 3, p 4, p 5, c 4, c 5, s 4, s 5 p 1, p 2 px, cx, sx, bx, tx Neural network model MLR Fragment types p 1, p 2, p 3, p 4, p 5, p 8, c 3, p 1, p 2, p 3 c 4, c 5, c 6, c 7, c 8, c 9, s 4, b 0, b 1, b 4, b 5
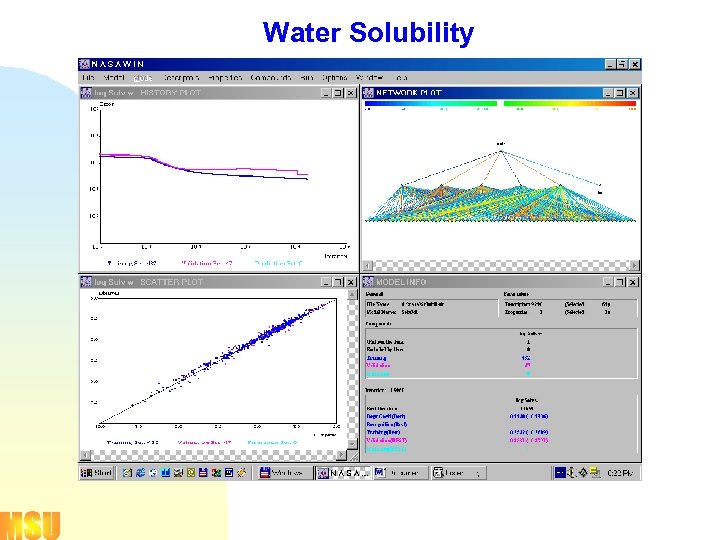 Water Solubility
Water Solubility
![Boiling point [1] (diverse set of 885 compounds) fragment types p 1, p 2, Boiling point [1] (diverse set of 885 compounds) fragment types p 1, p 2,](https://present5.com/presentation/a9cc945610e63825758231ba07354053/image-18.jpg) Boiling point [1] (diverse set of 885 compounds) fragment types p 1, p 2, p 3, p 4, p 5, p 6, c 3, c 4, c 5, c 6, s 4, s 5, s 6
Boiling point [1] (diverse set of 885 compounds) fragment types p 1, p 2, p 3, p 4, p 5, p 6, c 3, c 4, c 5, c 6, s 4, s 5, s 6
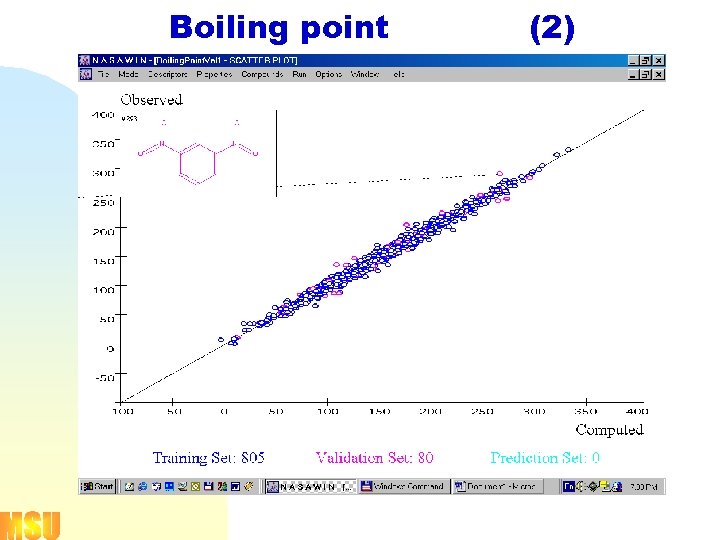 Boiling point (2)
Boiling point (2)
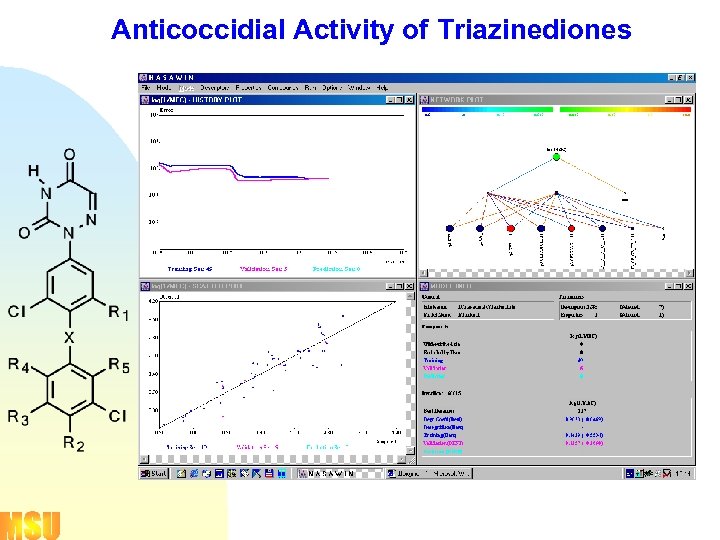 Anticoccidial Activity of Triazinediones
Anticoccidial Activity of Triazinediones
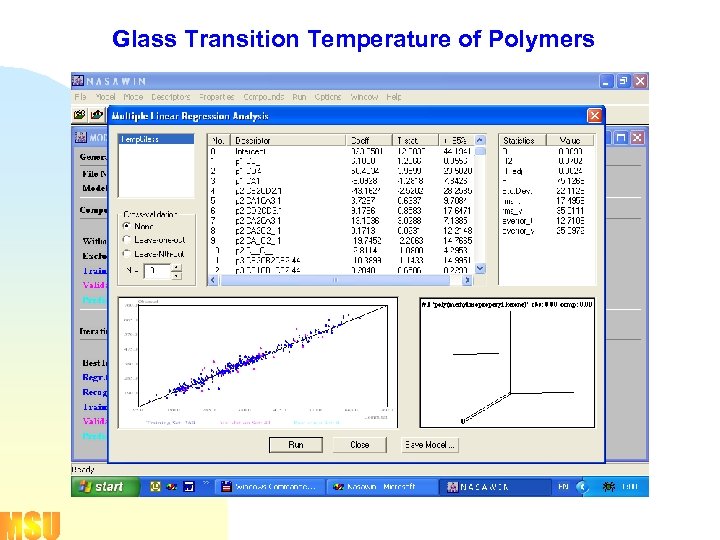 Glass Transition Temperature of Polymers
Glass Transition Temperature of Polymers
 Molar Heat Capacity of Polymers in the Liquid State
Molar Heat Capacity of Polymers in the Liquid State
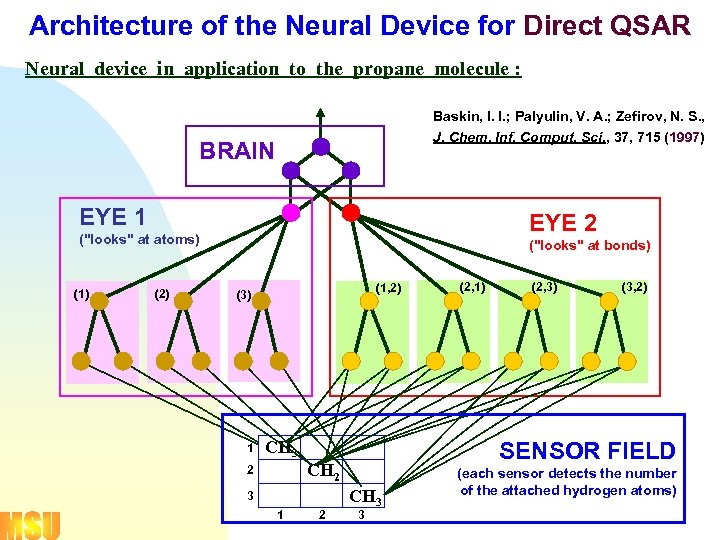 Architecture of the Neural Device for Direct QSAR Neural device in application to the propane molecule : Baskin, I. I. ; Palyulin, V. A. ; Zefirov, N. S. , J. Chem. Inf. Comput. Sci. , 37, 715 (1997) BRAIN EYE 1 EYE 2 ("looks" at atoms) (1) (2) ("looks" at bonds) (1, 2) (3) 1 CH 3 3 1 2 (2, 3) (3, 2) SENSOR FIELD CH 2 2 (2, 1) CH 3 3 (each sensor detects the number of the attached hydrogen atoms)
Architecture of the Neural Device for Direct QSAR Neural device in application to the propane molecule : Baskin, I. I. ; Palyulin, V. A. ; Zefirov, N. S. , J. Chem. Inf. Comput. Sci. , 37, 715 (1997) BRAIN EYE 1 EYE 2 ("looks" at atoms) (1) (2) ("looks" at bonds) (1, 2) (3) 1 CH 3 3 1 2 (2, 3) (3, 2) SENSOR FIELD CH 2 2 (2, 1) CH 3 3 (each sensor detects the number of the attached hydrogen atoms)
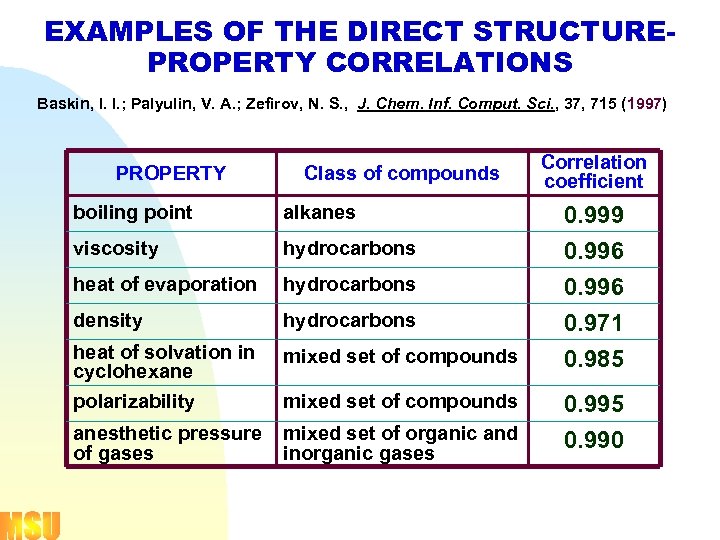 EXAMPLES OF THE DIRECT STRUCTUREPROPERTY CORRELATIONS Baskin, I. I. ; Palyulin, V. A. ; Zefirov, N. S. , J. Chem. Inf. Comput. Sci. , 37, 715 (1997) PROPERTY Class of compounds boiling point alkanes viscosity hydrocarbons heat of evaporation hydrocarbons density hydrocarbons heat of solvation in cyclohexane mixed set of compounds polarizability mixed set of compounds anesthetic pressure mixed set of organic and of gases inorganic gases Correlation coefficient 0. 999 0. 996 0. 971 0. 985 0. 990
EXAMPLES OF THE DIRECT STRUCTUREPROPERTY CORRELATIONS Baskin, I. I. ; Palyulin, V. A. ; Zefirov, N. S. , J. Chem. Inf. Comput. Sci. , 37, 715 (1997) PROPERTY Class of compounds boiling point alkanes viscosity hydrocarbons heat of evaporation hydrocarbons density hydrocarbons heat of solvation in cyclohexane mixed set of compounds polarizability mixed set of compounds anesthetic pressure mixed set of organic and of gases inorganic gases Correlation coefficient 0. 999 0. 996 0. 971 0. 985 0. 990
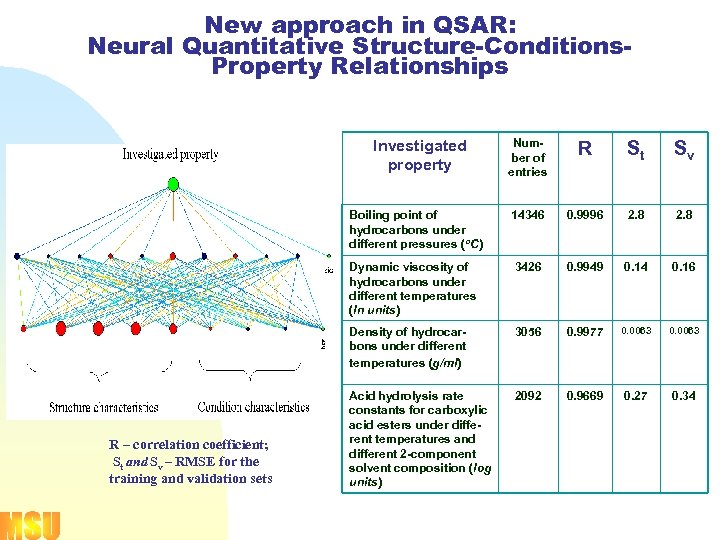 New approach in QSAR: Neural Quantitative Structure-Conditions. Property Relationships Number of entries R St Sv Boiling point of hydrocarbons under different pressures (о. С) 14346 0. 9996 2. 8 Dynamic viscosity of hydrocarbons under different temperatures (ln units) 3426 0. 9949 0. 14 0. 16 Density of hydrocarbons under different temperatures (g/ml) 3056 0. 9977 0. 0063 Acid hydrolysis rate constants for carboxylic acid esters under different temperatures and different 2 -component solvent composition (log units) 2092 0. 9669 0. 27 0. 34 Investigated property R – correlation coefficient; St and Sv – RMSE for the training and validation sets
New approach in QSAR: Neural Quantitative Structure-Conditions. Property Relationships Number of entries R St Sv Boiling point of hydrocarbons under different pressures (о. С) 14346 0. 9996 2. 8 Dynamic viscosity of hydrocarbons under different temperatures (ln units) 3426 0. 9949 0. 14 0. 16 Density of hydrocarbons under different temperatures (g/ml) 3056 0. 9977 0. 0063 Acid hydrolysis rate constants for carboxylic acid esters under different temperatures and different 2 -component solvent composition (log units) 2092 0. 9669 0. 27 0. 34 Investigated property R – correlation coefficient; St and Sv – RMSE for the training and validation sets
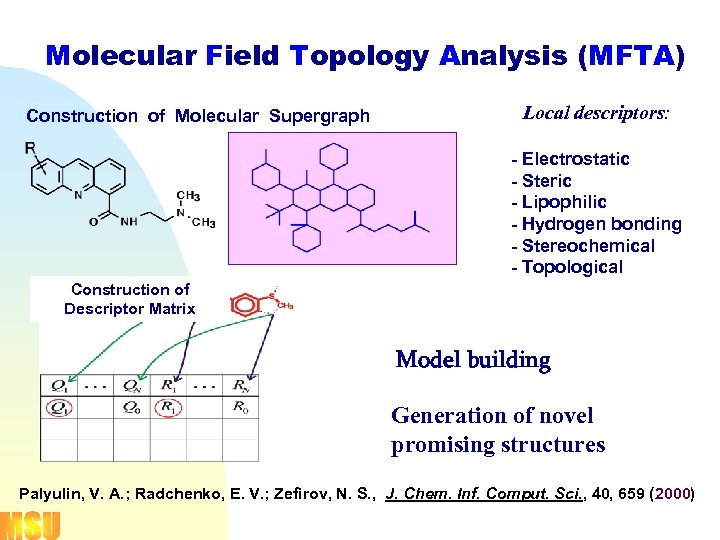 Molecular Field Topology Analysis (MFTA) Construction of Molecular Supergraph Local descriptors: - Electrostatic - Steric - Lipophilic - Hydrogen bonding - Stereochemical - Topological Construction of Descriptor Matrix Model building Generation of novel promising structures Palyulin, V. A. ; Radchenko, E. V. ; Zefirov, N. S. , J. Chem. Inf. Comput. Sci. , 40, 659 (2000)
Molecular Field Topology Analysis (MFTA) Construction of Molecular Supergraph Local descriptors: - Electrostatic - Steric - Lipophilic - Hydrogen bonding - Stereochemical - Topological Construction of Descriptor Matrix Model building Generation of novel promising structures Palyulin, V. A. ; Radchenko, E. V. ; Zefirov, N. S. , J. Chem. Inf. Comput. Sci. , 40, 659 (2000)
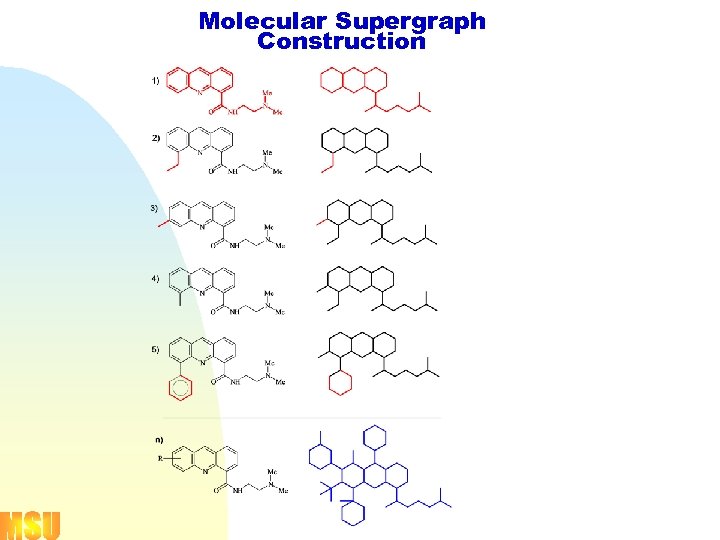 Molecular Supergraph Construction
Molecular Supergraph Construction
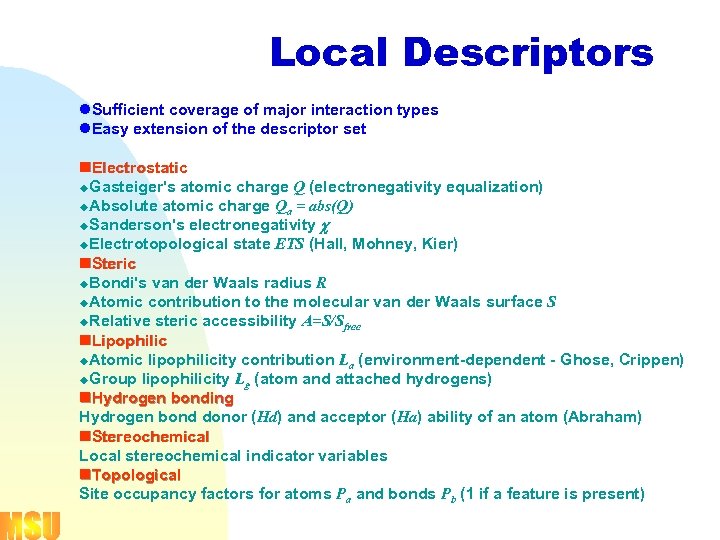 Local Descriptors l. Sufficient coverage of major interaction types l. Easy extension of the descriptor set n. Electrostatic u. Gasteiger's atomic charge Q (electronegativity equalization) u. Absolute atomic charge Qa = abs(Q) u. Sanderson's electronegativity u. Electrotopological state ETS (Hall, Mohney, Kier) n. Steric u. Bondi's van der Waals radius R u. Atomic contribution to the molecular van der Waals surface S u. Relative steric accessibility A=S/Sfree n. Lipophilic u. Atomic lipophilicity contribution La (environment-dependent - Ghose, Crippen) u. Group lipophilicity Lg (atom and attached hydrogens) n. Hydrogen bonding Hydrogen bond donor (Hd) and acceptor (Ha) ability of an atom (Abraham) n. Stereochemical Local stereochemical indicator variables n. Topological Site occupancy factors for atoms Pa and bonds Pb (1 if a feature is present)
Local Descriptors l. Sufficient coverage of major interaction types l. Easy extension of the descriptor set n. Electrostatic u. Gasteiger's atomic charge Q (electronegativity equalization) u. Absolute atomic charge Qa = abs(Q) u. Sanderson's electronegativity u. Electrotopological state ETS (Hall, Mohney, Kier) n. Steric u. Bondi's van der Waals radius R u. Atomic contribution to the molecular van der Waals surface S u. Relative steric accessibility A=S/Sfree n. Lipophilic u. Atomic lipophilicity contribution La (environment-dependent - Ghose, Crippen) u. Group lipophilicity Lg (atom and attached hydrogens) n. Hydrogen bonding Hydrogen bond donor (Hd) and acceptor (Ha) ability of an atom (Abraham) n. Stereochemical Local stereochemical indicator variables n. Topological Site occupancy factors for atoms Pa and bonds Pb (1 if a feature is present)
![Affinity of substituted 2, 5 -diazabicyclo[2. 2. 1]heptanes to nicotinic acetylcholine receptor Training set: Affinity of substituted 2, 5 -diazabicyclo[2. 2. 1]heptanes to nicotinic acetylcholine receptor Training set:](https://present5.com/presentation/a9cc945610e63825758231ba07354053/image-29.jpg) Affinity of substituted 2, 5 -diazabicyclo[2. 2. 1]heptanes to nicotinic acetylcholine receptor Training set: 31 compounds R 1 = H, Me, CH 2 CN R 2 = R = H, Me, F, Cl, Br, OH, NH 2, OMe, CN, CH 2 NH 2, CONH 2, NO 2, Ph. COO
Affinity of substituted 2, 5 -diazabicyclo[2. 2. 1]heptanes to nicotinic acetylcholine receptor Training set: 31 compounds R 1 = H, Me, CH 2 CN R 2 = R = H, Me, F, Cl, Br, OH, NH 2, OMe, CN, CH 2 NH 2, CONH 2, NO 2, Ph. COO
![Affinity of substituted 2, 5 -diazabicyclo[2. 2. 1]heptanes to nicotinic acetylcholine receptor Ki – Affinity of substituted 2, 5 -diazabicyclo[2. 2. 1]heptanes to nicotinic acetylcholine receptor Ki –](https://present5.com/presentation/a9cc945610e63825758231ba07354053/image-30.jpg) Affinity of substituted 2, 5 -diazabicyclo[2. 2. 1]heptanes to nicotinic acetylcholine receptor Ki – inhibition of competitive binding MED – minimum effective dose (hot plate test) Predicted lg(1/Ki) lg(1/MED) Q, R, Ha, Hd, Lg F=7 R=0. 960 Q 2=0. 850 Q, R F=4 R=0. 977 Q 2=0. 918 Experimental
Affinity of substituted 2, 5 -diazabicyclo[2. 2. 1]heptanes to nicotinic acetylcholine receptor Ki – inhibition of competitive binding MED – minimum effective dose (hot plate test) Predicted lg(1/Ki) lg(1/MED) Q, R, Ha, Hd, Lg F=7 R=0. 960 Q 2=0. 850 Q, R F=4 R=0. 977 Q 2=0. 918 Experimental
![Affinity of substituted 2, 5 -diazabicyclo[2. 2. 1]heptanes to nicotinic acetylcholine receptor Q H Affinity of substituted 2, 5 -diazabicyclo[2. 2. 1]heptanes to nicotinic acetylcholine receptor Q H](https://present5.com/presentation/a9cc945610e63825758231ba07354053/image-31.jpg) Affinity of substituted 2, 5 -diazabicyclo[2. 2. 1]heptanes to nicotinic acetylcholine receptor Q H a Ki – inhibition of competitive binding R L g
Affinity of substituted 2, 5 -diazabicyclo[2. 2. 1]heptanes to nicotinic acetylcholine receptor Q H a Ki – inhibition of competitive binding R L g
![Affinity of substituted 2, 5 -diazabicyclo[2. 2. 1]heptanes to nicotinic acetylcholine receptor Construction of Affinity of substituted 2, 5 -diazabicyclo[2. 2. 1]heptanes to nicotinic acetylcholine receptor Construction of](https://present5.com/presentation/a9cc945610e63825758231ba07354053/image-32.jpg) Affinity of substituted 2, 5 -diazabicyclo[2. 2. 1]heptanes to nicotinic acetylcholine receptor Construction of novel potentially active structures Total generated structures: 171 5 best structures wrt lg(1/Ki) R 1 = Me, Et, CN, Pr, i-Pr, t-Bu, Ph, 4. 01 3. 69 где R = CH 3, Cl, Br, NO 2 R 2 = Me, Et, Pr, CN, i-Pr, t-Bu 3. 44 3. 66 3. 69 Activity range in training set -3. 41. . . 2. 05
Affinity of substituted 2, 5 -diazabicyclo[2. 2. 1]heptanes to nicotinic acetylcholine receptor Construction of novel potentially active structures Total generated structures: 171 5 best structures wrt lg(1/Ki) R 1 = Me, Et, CN, Pr, i-Pr, t-Bu, Ph, 4. 01 3. 69 где R = CH 3, Cl, Br, NO 2 R 2 = Me, Et, Pr, CN, i-Pr, t-Bu 3. 44 3. 66 3. 69 Activity range in training set -3. 41. . . 2. 05
![Bradycardic activity of 3, 7, 9, 9 -tetraalkyl 3, 7 -diazabicyclo[3. 3. 1]nonanes Training Bradycardic activity of 3, 7, 9, 9 -tetraalkyl 3, 7 -diazabicyclo[3. 3. 1]nonanes Training](https://present5.com/presentation/a9cc945610e63825758231ba07354053/image-33.jpg) Bradycardic activity of 3, 7, 9, 9 -tetraalkyl 3, 7 -diazabicyclo[3. 3. 1]nonanes Training set: 26 compounds R 1, R 2 = Me, Pr, i-Pr, Bu, i-Bu, C 5 H 11, C 6 H 13, C 10 H 21, CH 2 -c-Pr, CH 2 -c-C 6 H 11, CH=CH 2, CH 2 CH=CH 2 R 3, R 4 = Me, Et, Pr, Bu, -(CH 2)3 -, -(CH 2)4 -, -(CH 2)5 -
Bradycardic activity of 3, 7, 9, 9 -tetraalkyl 3, 7 -diazabicyclo[3. 3. 1]nonanes Training set: 26 compounds R 1, R 2 = Me, Pr, i-Pr, Bu, i-Bu, C 5 H 11, C 6 H 13, C 10 H 21, CH 2 -c-Pr, CH 2 -c-C 6 H 11, CH=CH 2, CH 2 CH=CH 2 R 3, R 4 = Me, Et, Pr, Bu, -(CH 2)3 -, -(CH 2)4 -, -(CH 2)5 -
![Bradicardic activity of 3, 7, 9, 9 -tetraalkyl 3, 7 -diazabicyclo[3. 3. 1]nonanes SR Bradicardic activity of 3, 7, 9, 9 -tetraalkyl 3, 7 -diazabicyclo[3. 3. 1]nonanes SR](https://present5.com/presentation/a9cc945610e63825758231ba07354053/image-34.jpg) Bradicardic activity of 3, 7, 9, 9 -tetraalkyl 3, 7 -diazabicyclo[3. 3. 1]nonanes SR 75 – ability to decrease pacemaker pulse frequency (target effect) F 75 – ability to decrease myocardium contraction force (side effect) Sel. F – selectivity wrt F FRP 75 – ability to increase refractory period (side effect) Sel. FRP – selectivity wrt FRP lg(1/SR 75) lg(1/FRP 75) Sel. FRP Q, R, Ha, Hd F=5 R=0. 976 Q 2=0. 830 Q, R, Ha, Hd F=3 R=0. 932 Q 2=0. 800 Q, R, Ha, Hd F=7 R=0. 952 Q 2=0. 510 Q, R, Ha, Hd F=6 R=0. 972 Q 2=0. 819 Q, R, Ha, Hd F=1 R=0. 310 Q 2=0. 022
Bradicardic activity of 3, 7, 9, 9 -tetraalkyl 3, 7 -diazabicyclo[3. 3. 1]nonanes SR 75 – ability to decrease pacemaker pulse frequency (target effect) F 75 – ability to decrease myocardium contraction force (side effect) Sel. F – selectivity wrt F FRP 75 – ability to increase refractory period (side effect) Sel. FRP – selectivity wrt FRP lg(1/SR 75) lg(1/FRP 75) Sel. FRP Q, R, Ha, Hd F=5 R=0. 976 Q 2=0. 830 Q, R, Ha, Hd F=3 R=0. 932 Q 2=0. 800 Q, R, Ha, Hd F=7 R=0. 952 Q 2=0. 510 Q, R, Ha, Hd F=6 R=0. 972 Q 2=0. 819 Q, R, Ha, Hd F=1 R=0. 310 Q 2=0. 022
![Bradicardic activity of 3, 7, 9, 9 -tetraalkyl 3, 7 -diazabicyclo[3. 3. 1]nonanes SR Bradicardic activity of 3, 7, 9, 9 -tetraalkyl 3, 7 -diazabicyclo[3. 3. 1]nonanes SR](https://present5.com/presentation/a9cc945610e63825758231ba07354053/image-35.jpg) Bradicardic activity of 3, 7, 9, 9 -tetraalkyl 3, 7 -diazabicyclo[3. 3. 1]nonanes SR 75 – ability to decrease pacemaker pulse frequency (target effect) Predicted Q R Experimental
Bradicardic activity of 3, 7, 9, 9 -tetraalkyl 3, 7 -diazabicyclo[3. 3. 1]nonanes SR 75 – ability to decrease pacemaker pulse frequency (target effect) Predicted Q R Experimental
![Bradicardic activity of 3, 7, 9, 9 -tetraalkyl 3, 7 -diazabicyclo[3. 3. 1]nonanes Sel. Bradicardic activity of 3, 7, 9, 9 -tetraalkyl 3, 7 -diazabicyclo[3. 3. 1]nonanes Sel.](https://present5.com/presentation/a9cc945610e63825758231ba07354053/image-36.jpg) Bradicardic activity of 3, 7, 9, 9 -tetraalkyl 3, 7 -diazabicyclo[3. 3. 1]nonanes Sel. F – selectivity of antiarrhythmic activity wrt myocardium contraction force Predicted Q R Experimental Ha
Bradicardic activity of 3, 7, 9, 9 -tetraalkyl 3, 7 -diazabicyclo[3. 3. 1]nonanes Sel. F – selectivity of antiarrhythmic activity wrt myocardium contraction force Predicted Q R Experimental Ha
![Bradicardic activity of 3, 7, 9, 9 -tetraalkyl 3, 7 -diazabicyclo[3. 3. 1]nonanes Construction Bradicardic activity of 3, 7, 9, 9 -tetraalkyl 3, 7 -diazabicyclo[3. 3. 1]nonanes Construction](https://present5.com/presentation/a9cc945610e63825758231ba07354053/image-37.jpg) Bradicardic activity of 3, 7, 9, 9 -tetraalkyl 3, 7 -diazabicyclo[3. 3. 1]nonanes Construction of novel potentially active structures Total generated structures: 105 5 best structures wrt Sel. F R 1, R 3 = Me, Et, Pr, i-Pr, t-Bu, 70. 75 R 2 = Me, Et, Pr, i-Pr, t-Bu 70. 74 63. 83 63. 82 Activity range in training set 0. 4. . . 177 63. 12
Bradicardic activity of 3, 7, 9, 9 -tetraalkyl 3, 7 -diazabicyclo[3. 3. 1]nonanes Construction of novel potentially active structures Total generated structures: 105 5 best structures wrt Sel. F R 1, R 3 = Me, Et, Pr, i-Pr, t-Bu, 70. 75 R 2 = Me, Et, Pr, i-Pr, t-Bu 70. 74 63. 83 63. 82 Activity range in training set 0. 4. . . 177 63. 12
 Conclusions QSAR/QSPR (Quantitative structure-activity/property relationships) approaches can be considered as universal techniques for the modeling and prediction of nearly any properties of chemical compounds and many properties of materials. Some properties of materials can be predicted as dependent on the structure of small molecules used as additives (e. g. antioxidants, etc. ). A number of properties of polymers had been modelled as dependent of the chemical structure of monomeric unit (e. g. glass transition temperature, molar heat capacity for liquid and solid state, dielectric constant, refraction index).
Conclusions QSAR/QSPR (Quantitative structure-activity/property relationships) approaches can be considered as universal techniques for the modeling and prediction of nearly any properties of chemical compounds and many properties of materials. Some properties of materials can be predicted as dependent on the structure of small molecules used as additives (e. g. antioxidants, etc. ). A number of properties of polymers had been modelled as dependent of the chemical structure of monomeric unit (e. g. glass transition temperature, molar heat capacity for liquid and solid state, dielectric constant, refraction index).
 AMPA–receptor modulators (“ampakines”)
AMPA–receptor modulators (“ampakines”)
 The group of molecular design Academician N. S. Zefirov – Head of Organic Chemistry Division Dr. V. A. Palyulin – Head of Group Dr. I. I. Baskin Dr. A. A. Oliferenko Dr. E. V. Radchenko Dr. M. I. Skvortsova Dr. I. G. Tikhonova Dr. M. S. Belenikin Dr. A. A. Ivanov Dr. A. Yu. Zotov S. A. Pisarev A. A. Ivanova A. A. Melnikov
The group of molecular design Academician N. S. Zefirov – Head of Organic Chemistry Division Dr. V. A. Palyulin – Head of Group Dr. I. I. Baskin Dr. A. A. Oliferenko Dr. E. V. Radchenko Dr. M. I. Skvortsova Dr. I. G. Tikhonova Dr. M. S. Belenikin Dr. A. A. Ivanov Dr. A. Yu. Zotov S. A. Pisarev A. A. Ivanova A. A. Melnikov


