d9b1bd5d8e6b3ae63d09a7a3a6254aac.ppt
- Количество слайдов: 35
 Proteomics
Proteomics
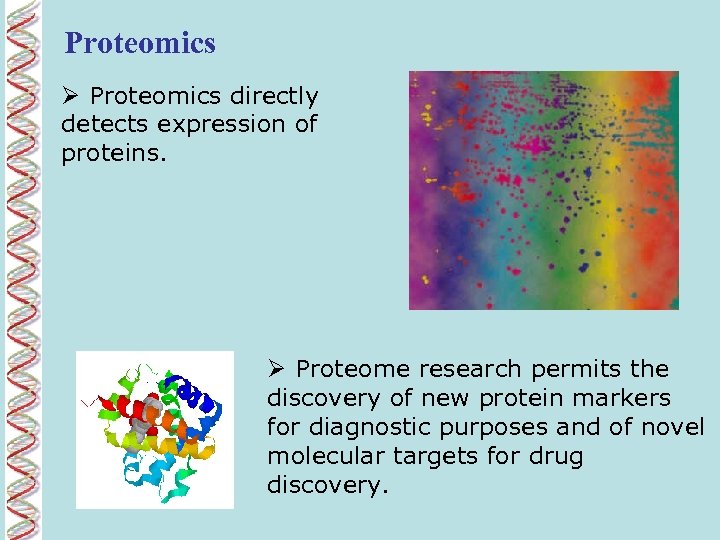 Proteomics Ø Proteomics directly detects expression of proteins. Ø Proteome research permits the discovery of new protein markers for diagnostic purposes and of novel molecular targets for drug discovery.
Proteomics Ø Proteomics directly detects expression of proteins. Ø Proteome research permits the discovery of new protein markers for diagnostic purposes and of novel molecular targets for drug discovery.
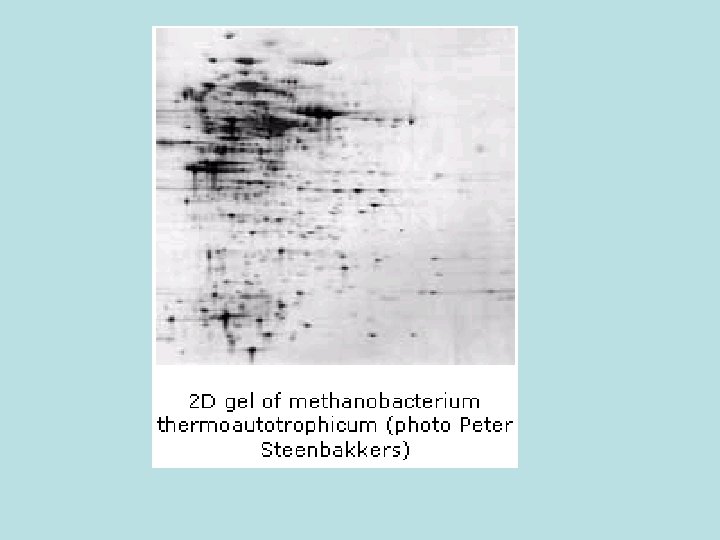
 . 1 SWISS-2 DPAGE database • SWISS-2 DPAGE is an annotated twodimensional polyacrylamide gel electrophoresis (2 -D PAGE) database established in 1993. • The SWISS-2 DPAGE database is maintained by the Swiss Institute of Bioinformatics, in collaboration with the Central Clinical Chemistry Laboratory of the Geneva University Hospital.
. 1 SWISS-2 DPAGE database • SWISS-2 DPAGE is an annotated twodimensional polyacrylamide gel electrophoresis (2 -D PAGE) database established in 1993. • The SWISS-2 DPAGE database is maintained by the Swiss Institute of Bioinformatics, in collaboration with the Central Clinical Chemistry Laboratory of the Geneva University Hospital.
 SWISS-2 DPAGE Search Page
SWISS-2 DPAGE Search Page
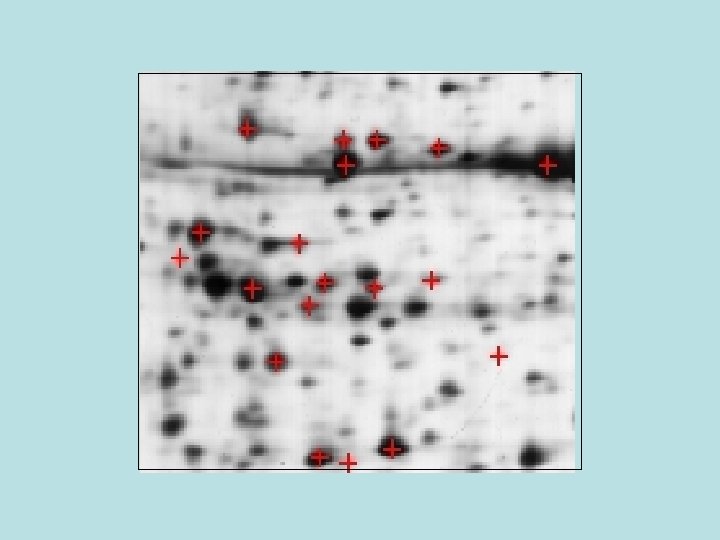
 View entry in original SWISS-2 DPAGE format Entry name: VSP 2_ARATH Primary accession number: 82122 Entered in SWISS-2 DPAGE in. Release 13, December 2000 Last modified in Release 14, October 2001 Name and origin of the protein: Description. Vegetative storage protein 2. Gene name(s)VSP 2 OR AT 5 G 24770 From Arabidopsis thaliana (Mouse-ear cress). [Tax. ID: 3702[ Taxonomy. Eukaryota; Viridiplantae; Streptophyta; Embryophyta; Tracheophyta; Spermatophyta; Magnoliophyta; eudicotyledons; core eudicots; Rosidae; eurosids II; Brassicales; Brassicaceae; Arabidopsis. References[1] MAPPING ON GEL. Sarazin B. , Tonella L. , Marques K. , Paesano S. , Chane-Favre L. , Sanchez J. -C. , Hochstrasser D. F. , Thiellement H. ; Submitted (OCT-2000) to the SWISS-2 DPAGE database. 2 D PAGE maps for identified proteins Compute theoretical p. I/Mw How to interpret a protein map Arabidopsis thaliana MAP LOCATIONS: SPOT 2 D-001 KKV: p. I=6. 47, Mw=29849 *** the clicked spot*** MAPPING: MASS SPECTROMETRY [1. [
View entry in original SWISS-2 DPAGE format Entry name: VSP 2_ARATH Primary accession number: 82122 Entered in SWISS-2 DPAGE in. Release 13, December 2000 Last modified in Release 14, October 2001 Name and origin of the protein: Description. Vegetative storage protein 2. Gene name(s)VSP 2 OR AT 5 G 24770 From Arabidopsis thaliana (Mouse-ear cress). [Tax. ID: 3702[ Taxonomy. Eukaryota; Viridiplantae; Streptophyta; Embryophyta; Tracheophyta; Spermatophyta; Magnoliophyta; eudicotyledons; core eudicots; Rosidae; eurosids II; Brassicales; Brassicaceae; Arabidopsis. References[1] MAPPING ON GEL. Sarazin B. , Tonella L. , Marques K. , Paesano S. , Chane-Favre L. , Sanchez J. -C. , Hochstrasser D. F. , Thiellement H. ; Submitted (OCT-2000) to the SWISS-2 DPAGE database. 2 D PAGE maps for identified proteins Compute theoretical p. I/Mw How to interpret a protein map Arabidopsis thaliana MAP LOCATIONS: SPOT 2 D-001 KKV: p. I=6. 47, Mw=29849 *** the clicked spot*** MAPPING: MASS SPECTROMETRY [1. [
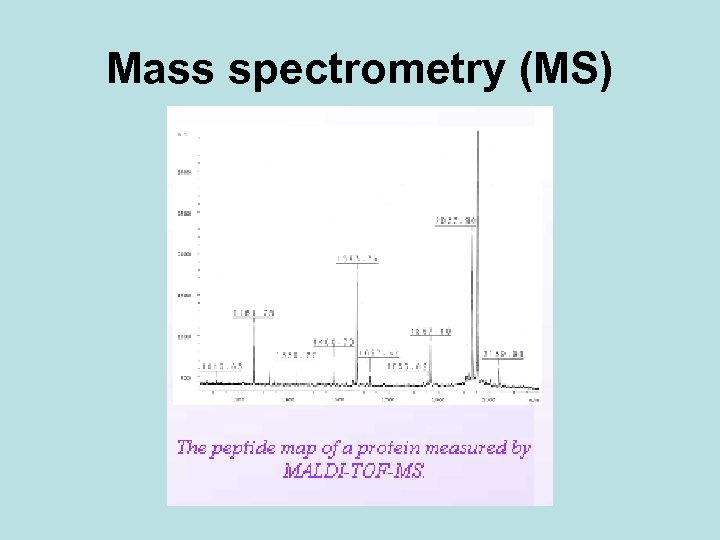 Mass spectrometry (MS)
Mass spectrometry (MS)
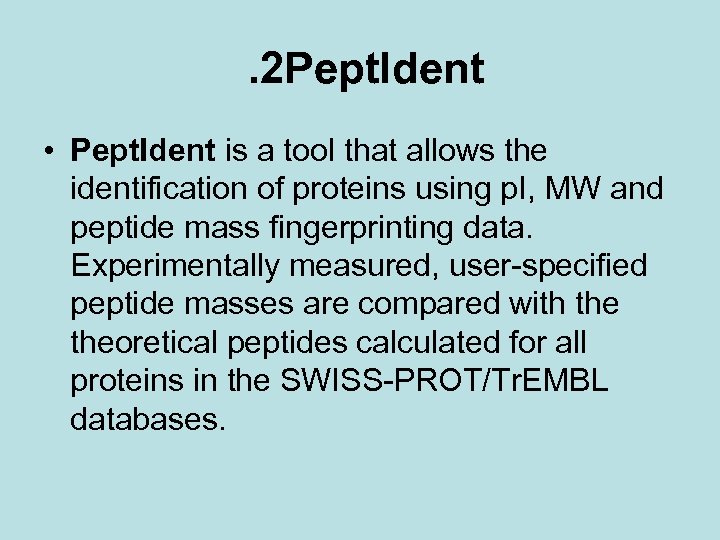 . 2 Pept. Ident • Pept. Ident is a tool that allows the identification of proteins using p. I, MW and peptide mass fingerprinting data. Experimentally measured, user-specified peptide masses are compared with theoretical peptides calculated for all proteins in the SWISS-PROT/Tr. EMBL databases.
. 2 Pept. Ident • Pept. Ident is a tool that allows the identification of proteins using p. I, MW and peptide mass fingerprinting data. Experimentally measured, user-specified peptide masses are compared with theoretical peptides calculated for all proteins in the SWISS-PROT/Tr. EMBL databases.
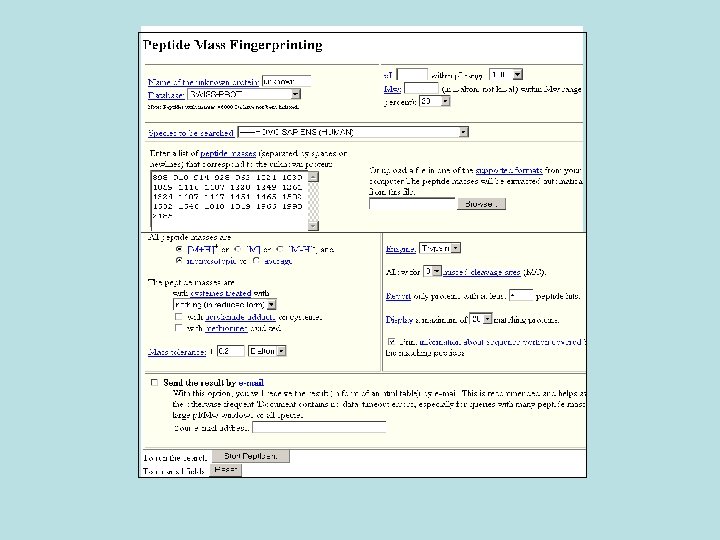
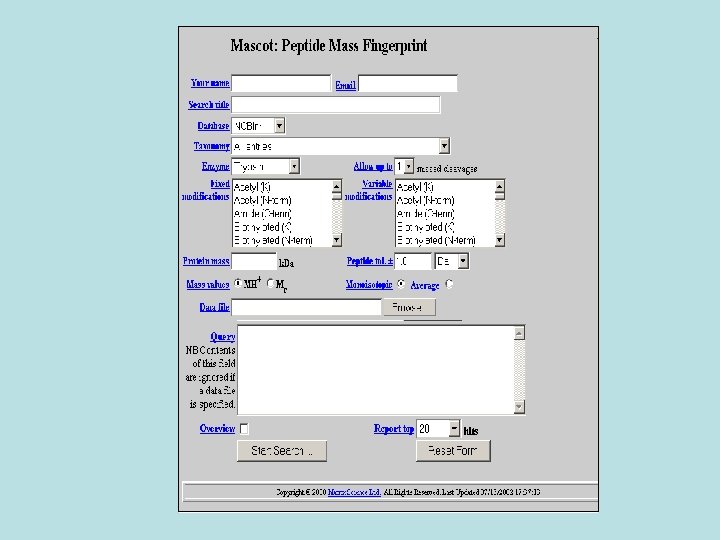
 . 3 Mascot • Mascot is a powerful search engine that uses mass spectrometry data to identify proteins from primary sequence databases.
. 3 Mascot • Mascot is a powerful search engine that uses mass spectrometry data to identify proteins from primary sequence databases.
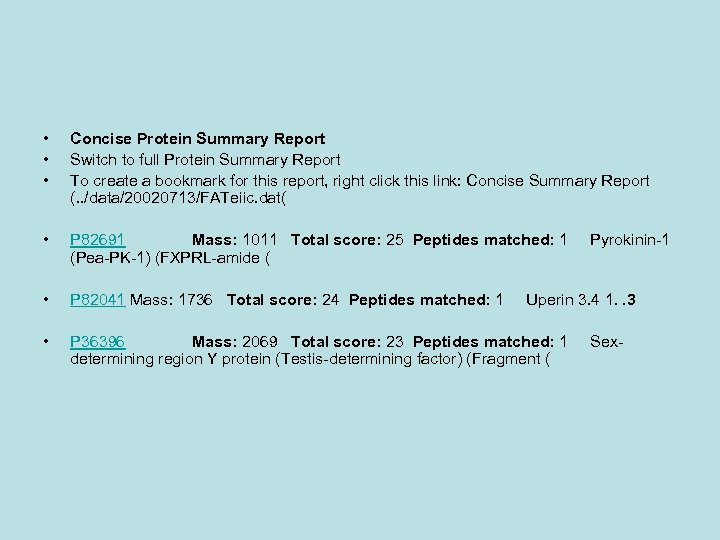 • • • Concise Protein Summary Report Switch to full Protein Summary Report To create a bookmark for this report, right click this link: Concise Summary Report (. . /data/20020713/FATeiic. dat( • P 82691 Mass: 1011 Total score: 25 Peptides matched: 1 Pyrokinin-1 (Pea-PK-1) (FXPRL-amide ( • P 82041 Mass: 1736 Total score: 24 Peptides matched: 1 Uperin 3. 4 1. . 3 • P 36396 Mass: 2069 Total score: 23 Peptides matched: 1 Sexdetermining region Y protein (Testis-determining factor) (Fragment (
• • • Concise Protein Summary Report Switch to full Protein Summary Report To create a bookmark for this report, right click this link: Concise Summary Report (. . /data/20020713/FATeiic. dat( • P 82691 Mass: 1011 Total score: 25 Peptides matched: 1 Pyrokinin-1 (Pea-PK-1) (FXPRL-amide ( • P 82041 Mass: 1736 Total score: 24 Peptides matched: 1 Uperin 3. 4 1. . 3 • P 36396 Mass: 2069 Total score: 23 Peptides matched: 1 Sexdetermining region Y protein (Testis-determining factor) (Fragment (
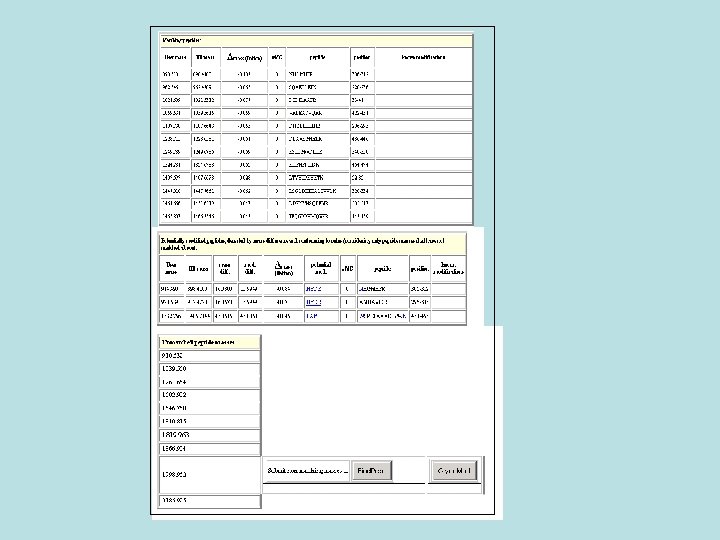
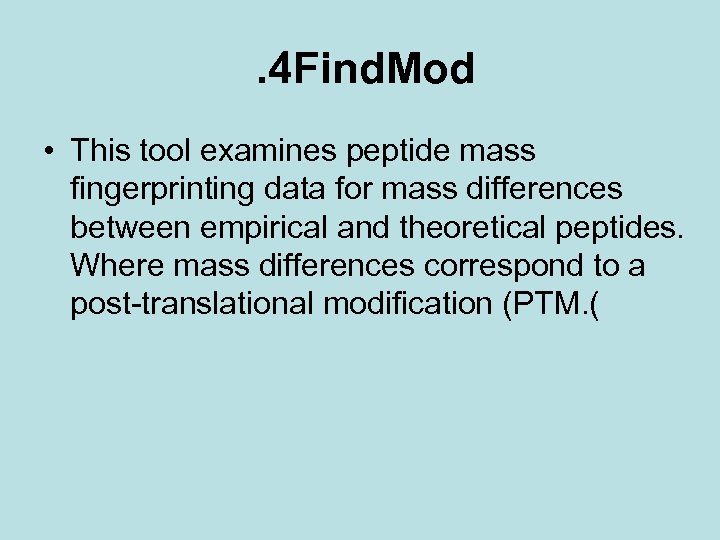 . 4 Find. Mod • This tool examines peptide mass fingerprinting data for mass differences between empirical and theoretical peptides. Where mass differences correspond to a post-translational modification (PTM. (
. 4 Find. Mod • This tool examines peptide mass fingerprinting data for mass differences between empirical and theoretical peptides. Where mass differences correspond to a post-translational modification (PTM. (
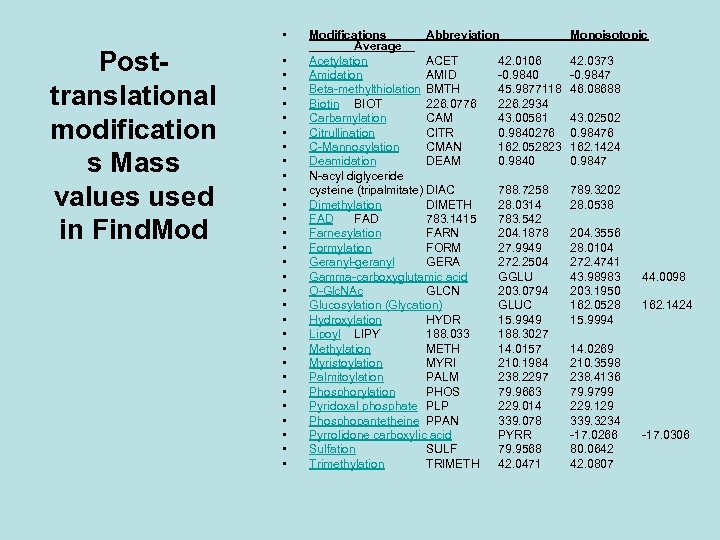 • Posttranslational modification s Mass values used in Find. Mod • • • • • • • • Modifications Abbreviation Average__ Acetylation ACET 42. 0106 Amidation AMID -0. 9840 Beta-methylthiolation BMTH 45. 9877118 Biotin BIOT 226. 0776 226. 2934 Carbamylation CAM 43. 00581 Citrullination CITR 0. 9840276 C-Mannosylation CMAN 162. 052823 Deamidation DEAM 0. 9840 N-acyl diglyceride cysteine (tripalmitate) DIAC 788. 7258 Dimethylation DIMETH 28. 0314 FAD 783. 1415 783. 542 Farnesylation FARN 204. 1878 Formylation FORM 27. 9949 Geranyl-geranyl GERA 272. 2504 Gamma-carboxyglutamic acid GGLU O-Glc. NAc GLCN 203. 0794 Glucosylation (Glycation) GLUC Hydroxylation HYDR 15. 9949 Lipoyl LIPY 188. 033 188. 3027 Methylation METH 14. 0157 Myristoylation MYRI 210. 1984 Palmitoylation PALM 238. 2297 Phosphorylation PHOS 79. 9663 Pyridoxal phosphate PLP 229. 014 Phosphopantetheine PPAN 339. 078 Pyrrolidone carboxylic acid PYRR Sulfation SULF 79. 9568 Trimethylation TRIMETH 42. 0471 Monoisotopic 42. 0373 -0. 9847 46. 08688 43. 02502 0. 98476 162. 1424 0. 9847 789. 3202 28. 0538 204. 3556 28. 0104 272. 4741 43. 98983 203. 1950 162. 0528 15. 9994 14. 0269 210. 3598 238. 4136 79. 9799 229. 129 339. 3234 -17. 0266 80. 0642 42. 0807 44. 0098 162. 1424 -17. 0306
• Posttranslational modification s Mass values used in Find. Mod • • • • • • • • Modifications Abbreviation Average__ Acetylation ACET 42. 0106 Amidation AMID -0. 9840 Beta-methylthiolation BMTH 45. 9877118 Biotin BIOT 226. 0776 226. 2934 Carbamylation CAM 43. 00581 Citrullination CITR 0. 9840276 C-Mannosylation CMAN 162. 052823 Deamidation DEAM 0. 9840 N-acyl diglyceride cysteine (tripalmitate) DIAC 788. 7258 Dimethylation DIMETH 28. 0314 FAD 783. 1415 783. 542 Farnesylation FARN 204. 1878 Formylation FORM 27. 9949 Geranyl-geranyl GERA 272. 2504 Gamma-carboxyglutamic acid GGLU O-Glc. NAc GLCN 203. 0794 Glucosylation (Glycation) GLUC Hydroxylation HYDR 15. 9949 Lipoyl LIPY 188. 033 188. 3027 Methylation METH 14. 0157 Myristoylation MYRI 210. 1984 Palmitoylation PALM 238. 2297 Phosphorylation PHOS 79. 9663 Pyridoxal phosphate PLP 229. 014 Phosphopantetheine PPAN 339. 078 Pyrrolidone carboxylic acid PYRR Sulfation SULF 79. 9568 Trimethylation TRIMETH 42. 0471 Monoisotopic 42. 0373 -0. 9847 46. 08688 43. 02502 0. 98476 162. 1424 0. 9847 789. 3202 28. 0538 204. 3556 28. 0104 272. 4741 43. 98983 203. 1950 162. 0528 15. 9994 14. 0269 210. 3598 238. 4136 79. 9799 229. 129 339. 3234 -17. 0266 80. 0642 42. 0807 44. 0098 162. 1424 -17. 0306

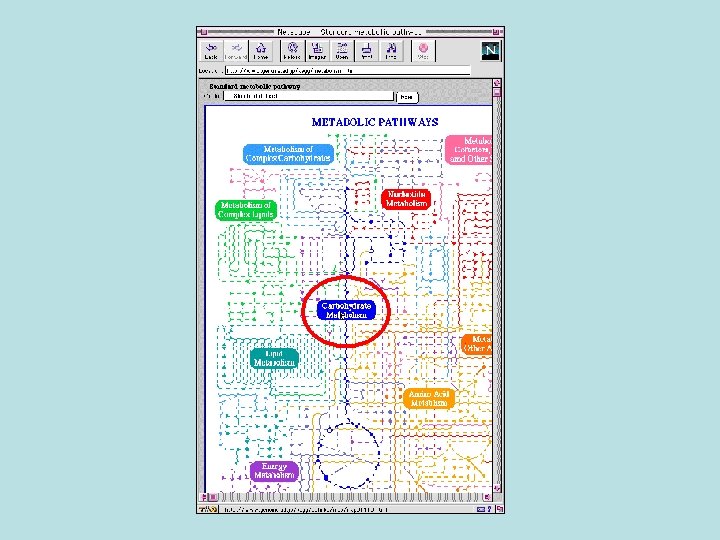
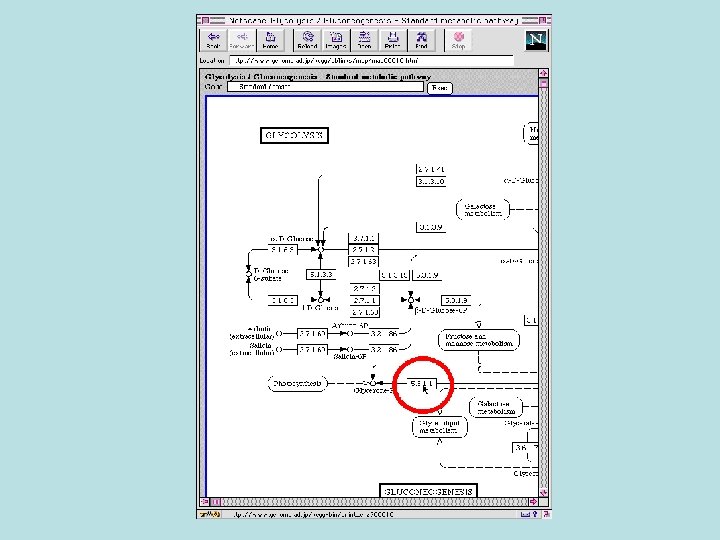
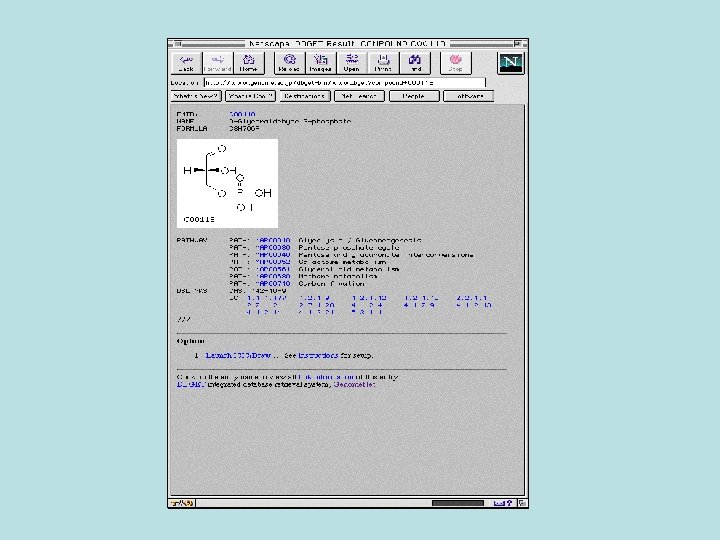
 Biochemical Pathway Databases • Linking the biochemical pathways together and integration with the genomic data are the great tasks of biochemical pathway databases.
Biochemical Pathway Databases • Linking the biochemical pathways together and integration with the genomic data are the great tasks of biochemical pathway databases.
 Metabolomics: From Genes to Pathways:
Metabolomics: From Genes to Pathways:
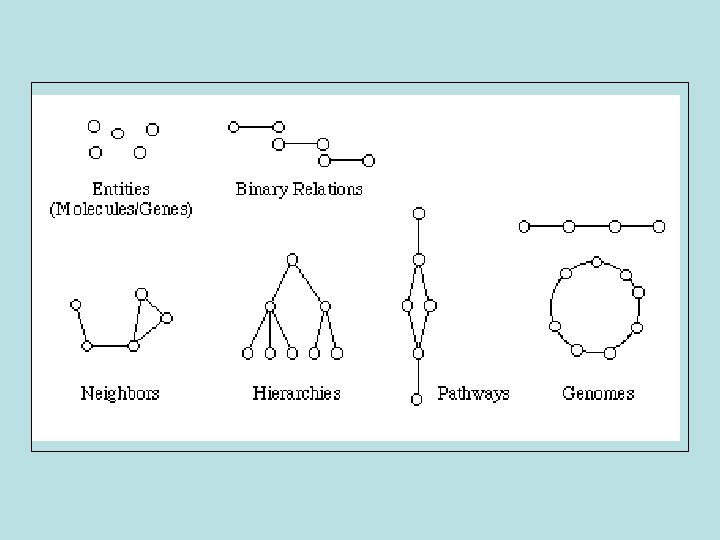
 Where do we go? “Deconstruction of biological processes into their molecular components”.
Where do we go? “Deconstruction of biological processes into their molecular components”.
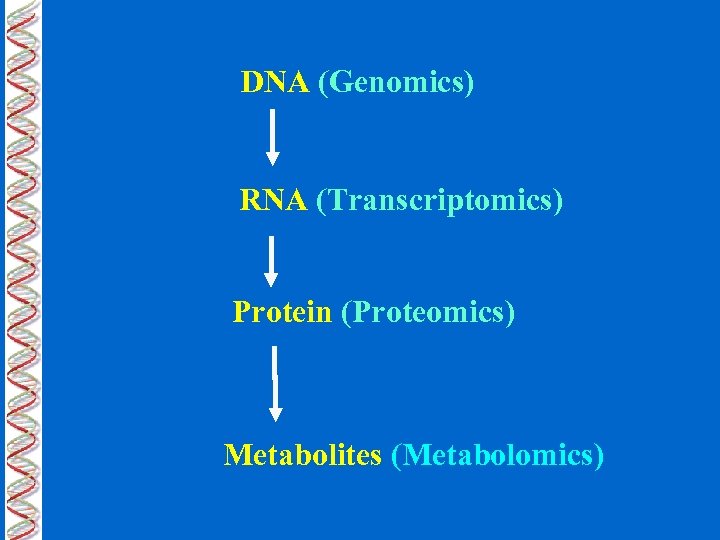 DNA (Genomics) RNA (Transcriptomics) Protein (Proteomics) Metabolites (Metabolomics)
DNA (Genomics) RNA (Transcriptomics) Protein (Proteomics) Metabolites (Metabolomics)
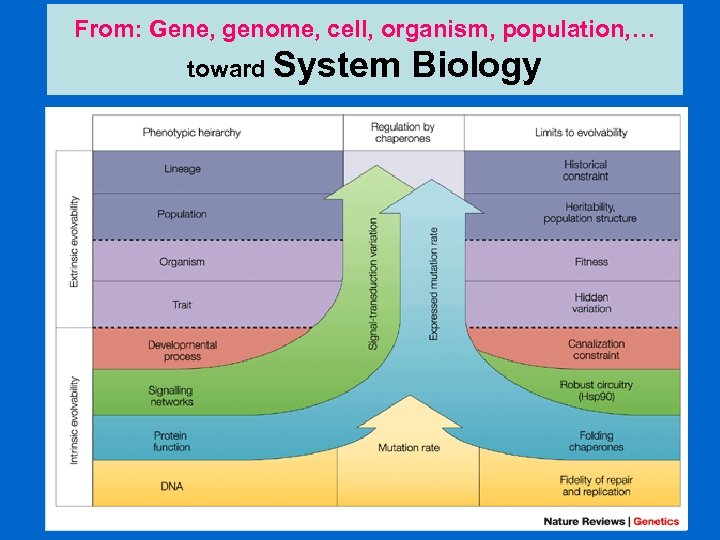 From: Gene, genome, cell, organism, population, … toward System Biology
From: Gene, genome, cell, organism, population, … toward System Biology
 What are we going to do?
What are we going to do?
 Fact: Individual research units would not work any more! Recommendation: Team up! Go beyond your own, your institute, and your country boundaries.
Fact: Individual research units would not work any more! Recommendation: Team up! Go beyond your own, your institute, and your country boundaries.
 Fact: Genomic data are suppose to reduce time and efforts for preparation of reagents, resources and information. Recommendation: Think big! • Search and use data intelligently. • Turn attention to complex biology from various angles, i. e. have all needed specialty in your team.
Fact: Genomic data are suppose to reduce time and efforts for preparation of reagents, resources and information. Recommendation: Think big! • Search and use data intelligently. • Turn attention to complex biology from various angles, i. e. have all needed specialty in your team.
 Fact: A mass of data is available freely! Recommendation: Learn how to use! Make use of them to develop technologies.
Fact: A mass of data is available freely! Recommendation: Learn how to use! Make use of them to develop technologies.
 Fact: Biology world is rapidly changing! Recommendation: Keep up with changes! • Re-establish systems with more flexibility and more freedom. • Loose regulations for funding, employment, etc. • Re-design your research project.
Fact: Biology world is rapidly changing! Recommendation: Keep up with changes! • Re-establish systems with more flexibility and more freedom. • Loose regulations for funding, employment, etc. • Re-design your research project.
 Thanks for Your Attention
Thanks for Your Attention
 Cautions: • One protein with different roles: • Alpha-enolase in liver • T-crystallin in eye lens • One structure in proteins with diverse functions: • TIM barrels in isomerases, oxidoreductase and hydrolases. • 30% error in automated annotations.
Cautions: • One protein with different roles: • Alpha-enolase in liver • T-crystallin in eye lens • One structure in proteins with diverse functions: • TIM barrels in isomerases, oxidoreductase and hydrolases. • 30% error in automated annotations.


