a1075cdfd1474cbd275d7a84dc732d59.ppt
- Количество слайдов: 60

PROTEOMICS OF OBESITY Part II Prof. Dr. E. C. M. Mariman

Contents Slides Proteomics of obesity and adipose tissue: overview 1 -11 Protein profiling methodologies 12 – 23 Protein secretion by adipocytes 24 – 47 Molecular mechanisms for obesity-related physiology 48 – 59 Proteomics in obesity: summary 60 Some literature references 61 Abbreviations used 62
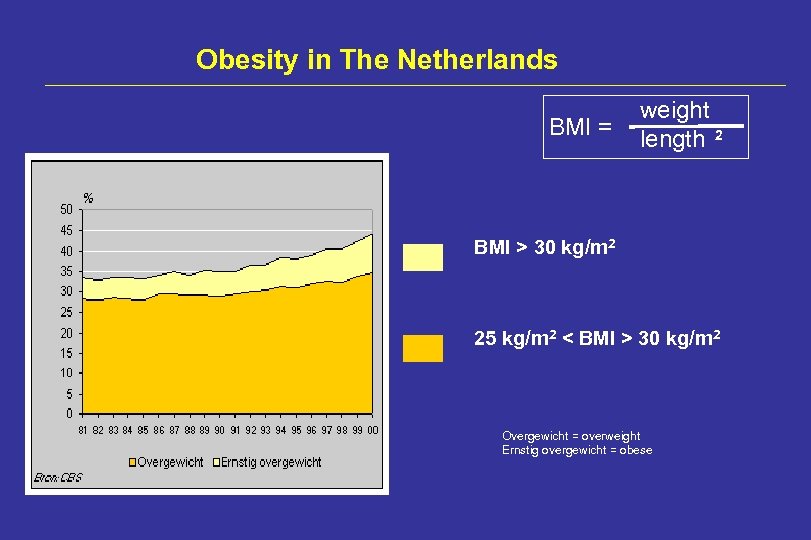
Obesity in The Netherlands BMI = weight length 2 BMI > 30 kg/m 2 25 kg/m 2 < BMI > 30 kg/m 2 Overgewicht = overweight Ernstig overgewicht = obese

CHILD OBESITY

Obesity linked with diseases Diabetes Reduced fertility Obesity cardiovascular diseases Cancer
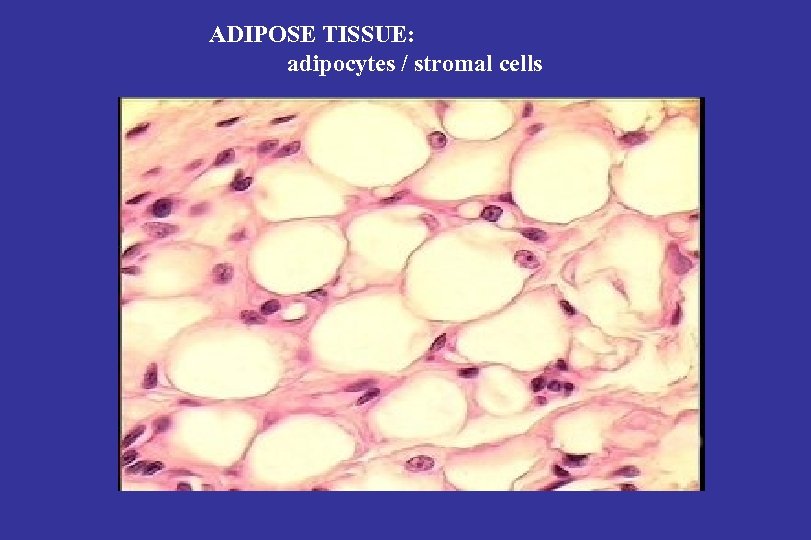
ADIPOSE TISSUE: adipocytes / stromal cells
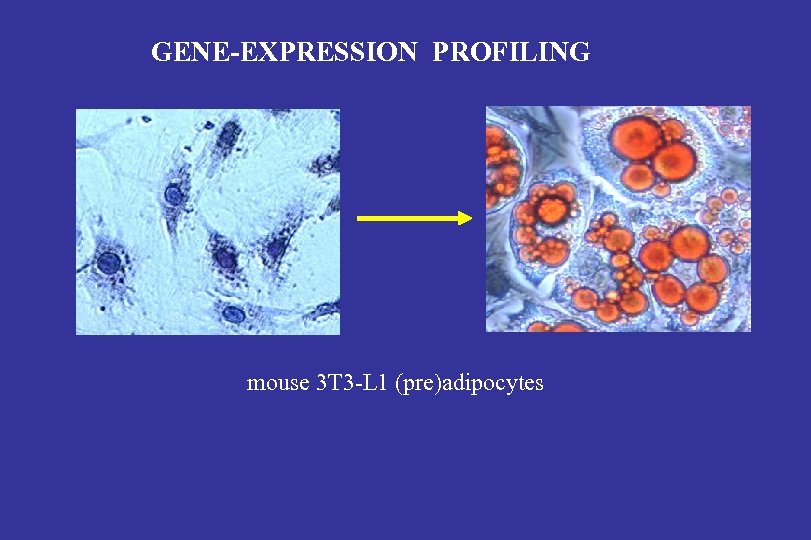
GENE-EXPRESSION PROFILING mouse 3 T 3 -L 1 (pre)adipocytes
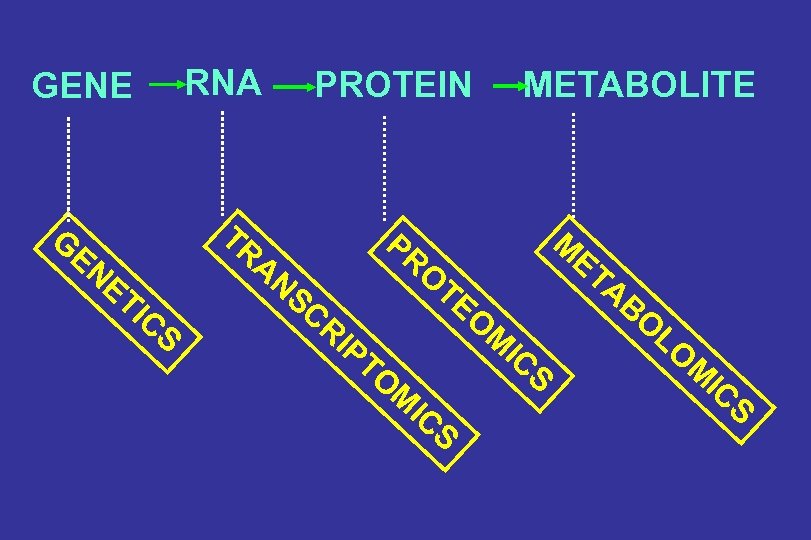
RNA GENE PROTEIN TR G EN ET I CS METABOLITE M AN SC PR O RI PT O TE O ET AB O M M IC S LO M IC S
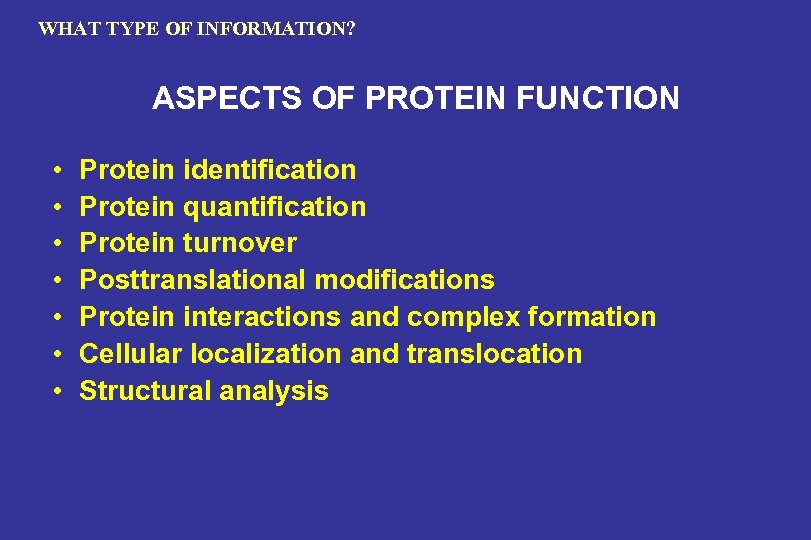
WHAT TYPE OF INFORMATION? ASPECTS OF PROTEIN FUNCTION • • Protein identification Protein quantification Protein turnover Posttranslational modifications Protein interactions and complex formation Cellular localization and translocation Structural analysis
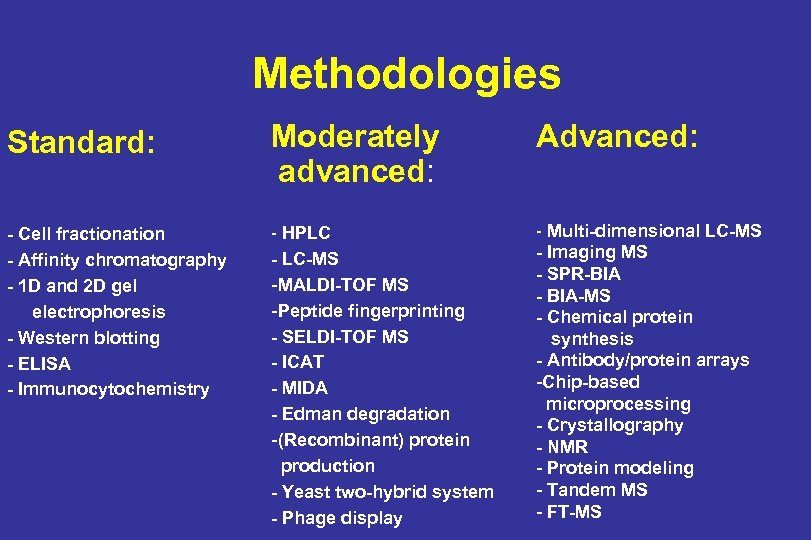
Methodologies Standard: Moderately advanced: Advanced: - Cell fractionation - Affinity chromatography - 1 D and 2 D gel electrophoresis - Western blotting - ELISA - Immunocytochemistry - HPLC - LC-MS - MALDI-TOF MS - Peptide fingerprinting - SELDI-TOF MS - ICAT - MIDA - Edman degradation - (Recombinant) protein production - Yeast two-hybrid system - Phage display - Multi-dimensional LC-MS - Imaging MS - SPR-BIA - BIA-MS - Chemical protein synthesis - Antibody/protein arrays -Chip-based microprocessing - Crystallography - NMR - Protein modeling - Tandem MS - FT-MS

Proteomics, why? • “ Gene structure alone tells us very little about the physiological function of proteins since it ignores the co- and posttranslational modifications to which they are subjected. ” (Edmund Fisher, 1997, Nobel Laureate Physiology & Medicine 1992) • Proteins are the “working horses” in the cell • Proteins are still preferred targets for drug- and nutritional intervention • Body fluids contain proteins and usually not RNA

PROTEIN PROFILING

PROTEINS Cy 3 Cy 5 MIX INCUBATE
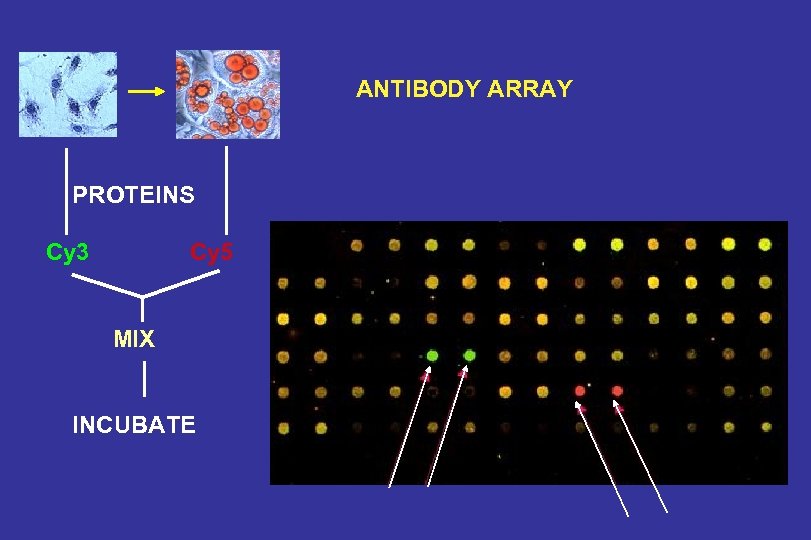
ANTIBODY ARRAY PROTEINS Cy 3 Cy 5 MIX INCUBATE
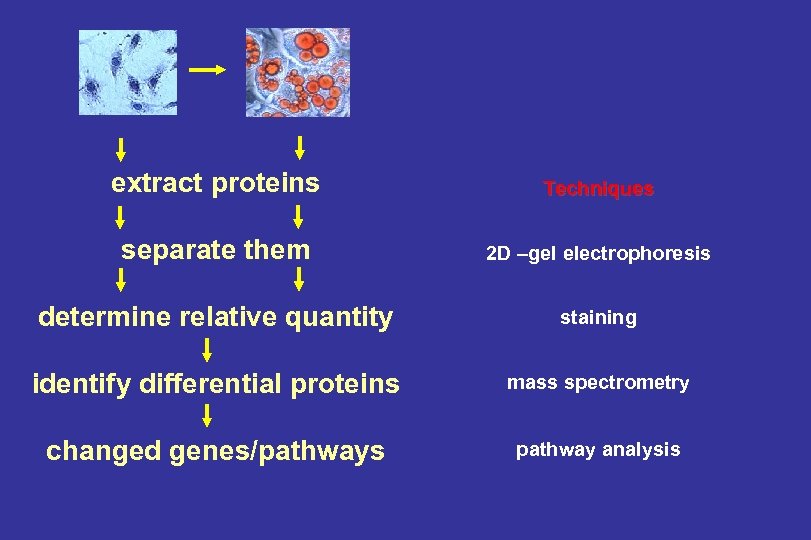
extract proteins Techniques separate them 2 D –gel electrophoresis determine relative quantity staining identify differential proteins mass spectrometry changed genes/pathways pathway analysis
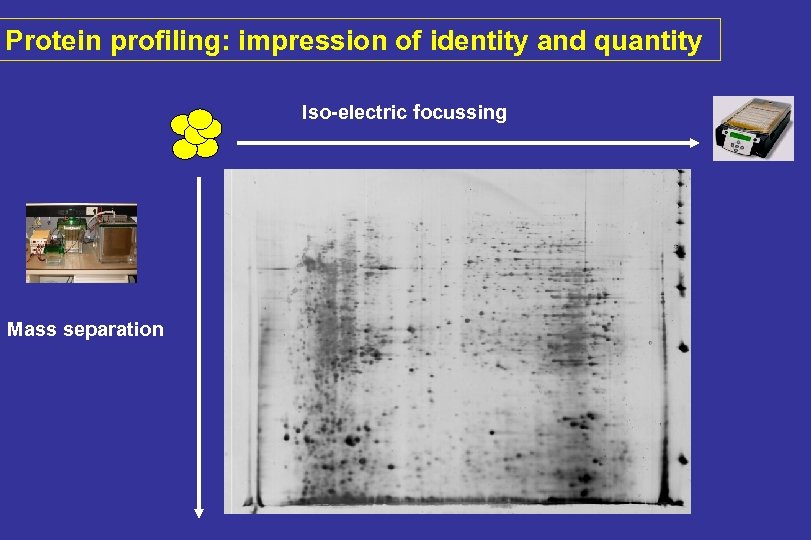
Protein profiling: impression of identity and quantity Iso-electric focussing Mass separation
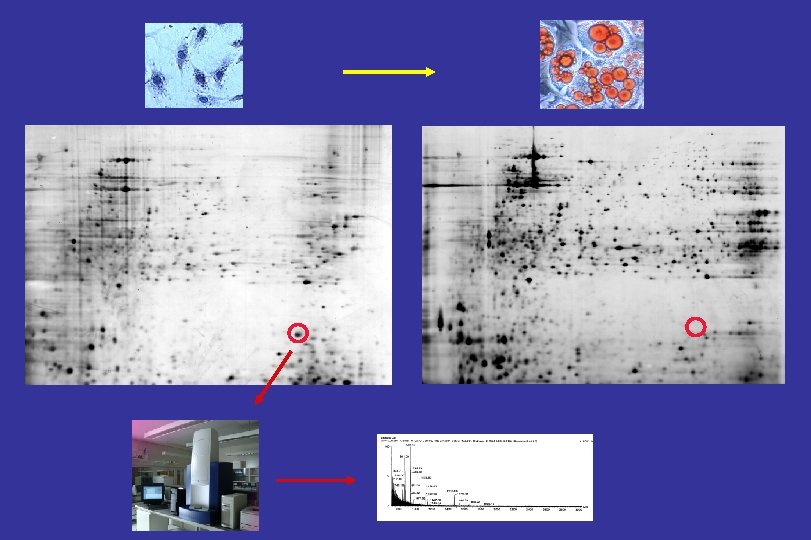
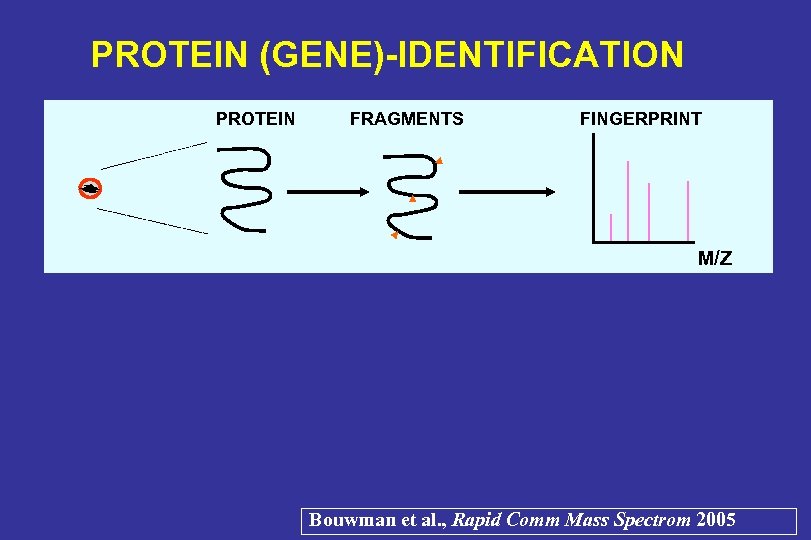
PROTEIN (GENE)-IDENTIFICATION PROTEIN FRAGMENTS FINGERPRINT M/Z Bouwman et al. , Rapid Comm Mass Spectrom 2005
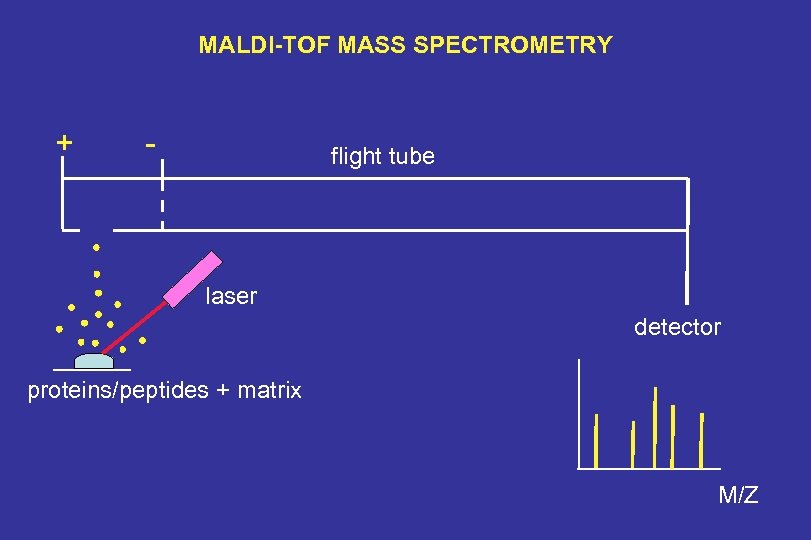
MALDI-TOF MASS SPECTROMETRY + - flight tube laser detector proteins/peptides + matrix M/Z
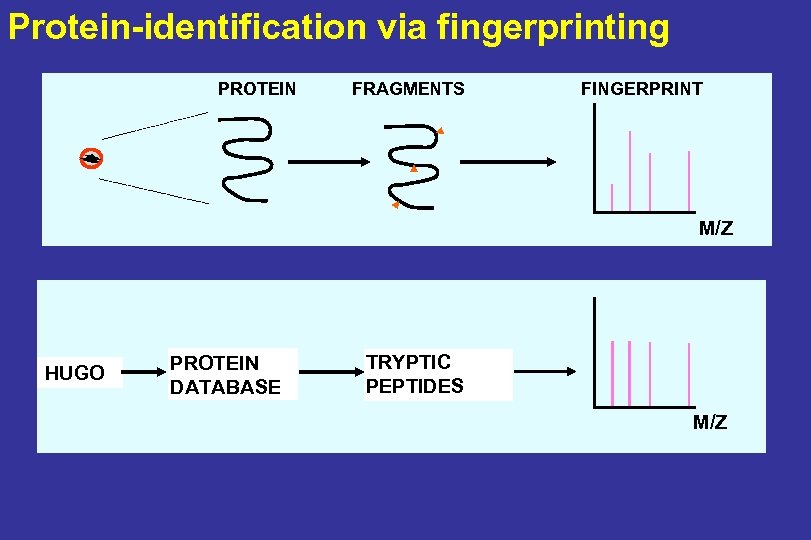
Protein-identification via fingerprinting PROTEIN FRAGMENTS FINGERPRINT M/Z HUGO PROTEIN DATABASE TRYPTIC PEPTIDES M/Z
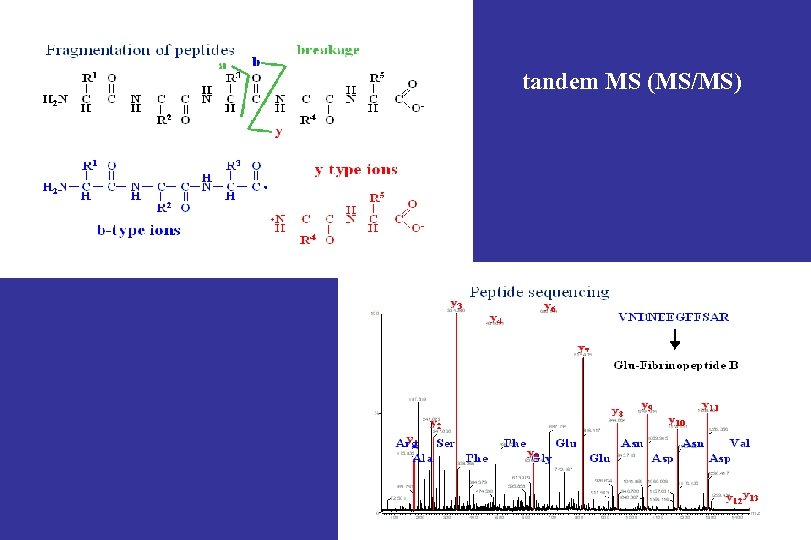
tandem MS (MS/MS)
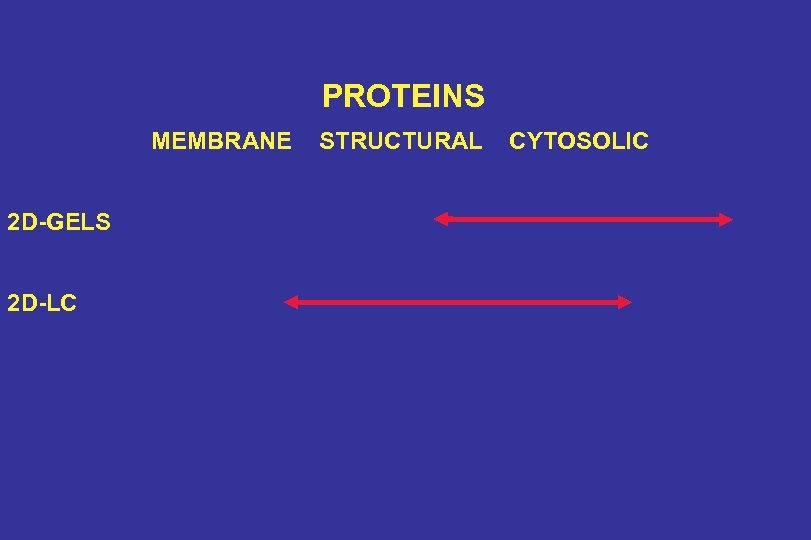
PROTEINS MEMBRANE 2 D-GELS 2 D-LC STRUCTURAL CYTOSOLIC

ADIPOCYTE FUNCTIONING IN OBESITY Protein secretion by adipocytes
Obesity at cellular level THE SECRETOME
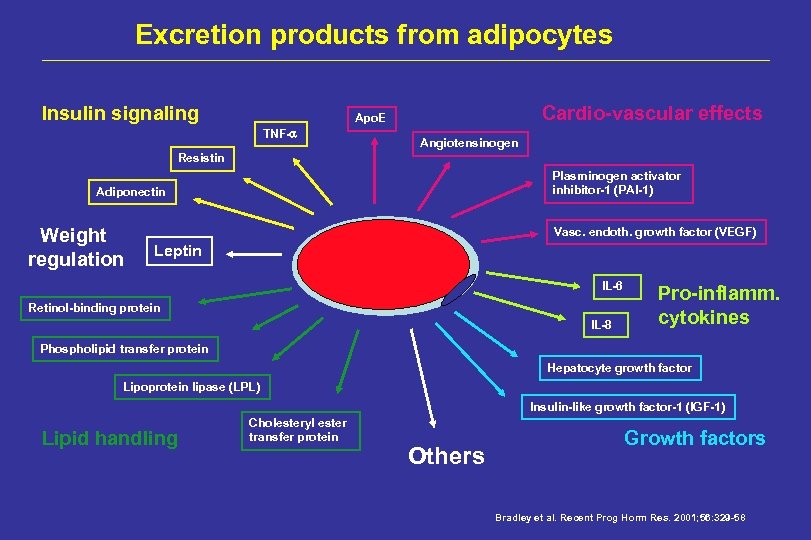
Excretion products from adipocytes Insulin signaling Cardio-vascular effects Apo. E TNF- Angiotensinogen Resistin Plasminogen activator inhibitor-1 (PAI-1) Adiponectin Weight regulation Vasc. endoth. growth factor (VEGF) Leptin IL-6 Retinol-binding protein IL-8 Pro-inflamm. cytokines Phospholipid transfer protein Hepatocyte growth factor Lipoprotein lipase (LPL) Insulin-like growth factor-1 (IGF-1) Lipid handling Cholesteryl ester transfer protein Others Growth factors Bradley et al. Recent Prog Horm Res. 2001; 56: 329 -58
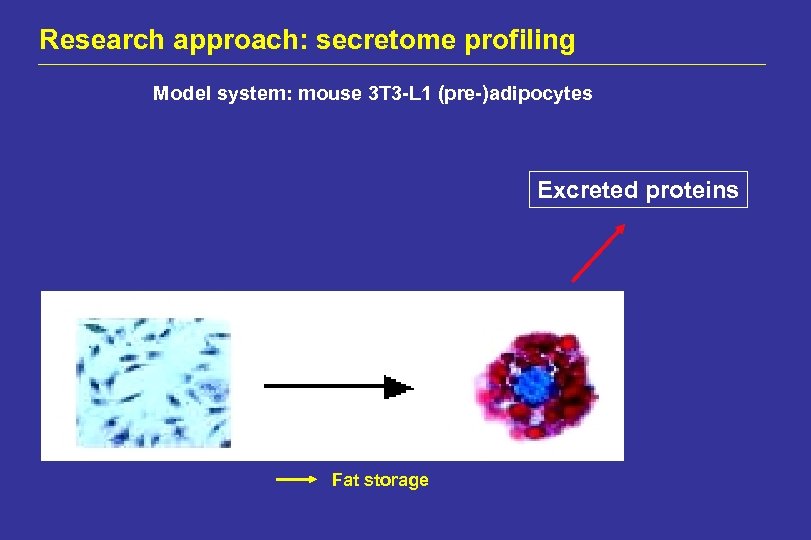
Research approach: secretome profiling Model system: mouse 3 T 3 -L 1 (pre-)adipocytes Excreted proteins Fat storage
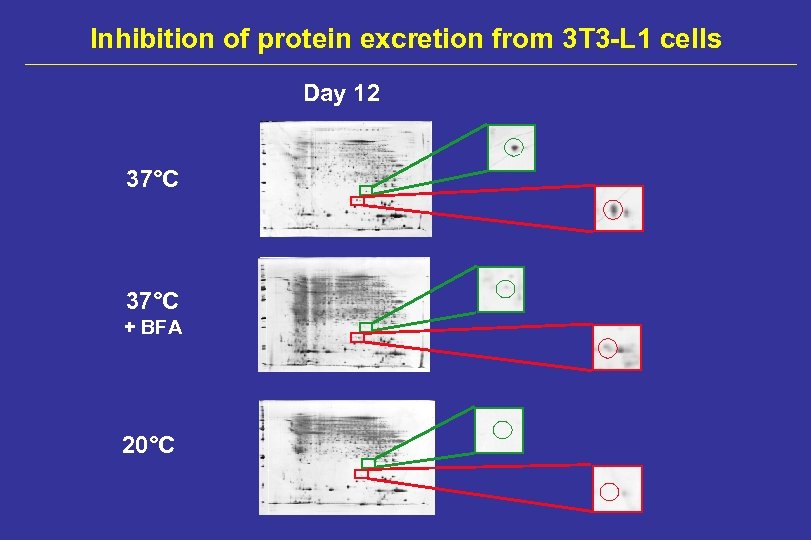
Inhibition of protein excretion from 3 T 3 -L 1 cells Day 12 37°C + BFA 20°C
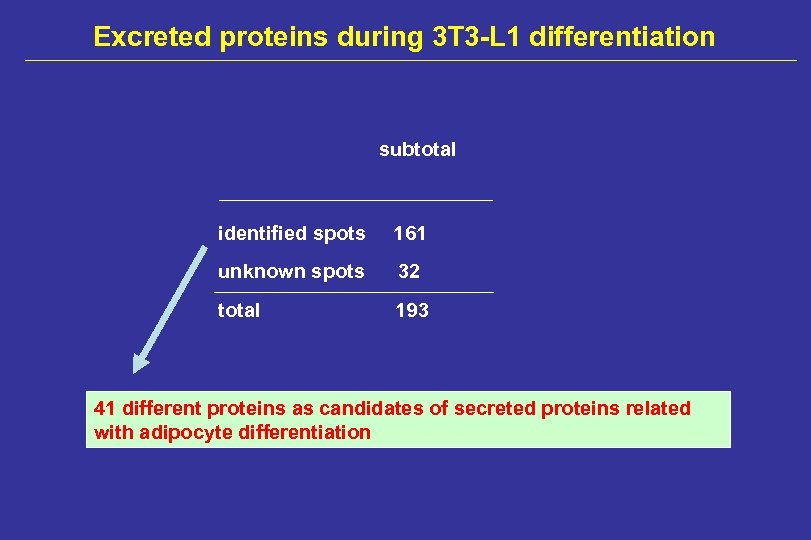
Excreted proteins during 3 T 3 -L 1 differentiation subtotal identified spots 161 unknown spots 32 total 193 41 different proteins as candidates of secreted proteins related with adipocyte differentiation
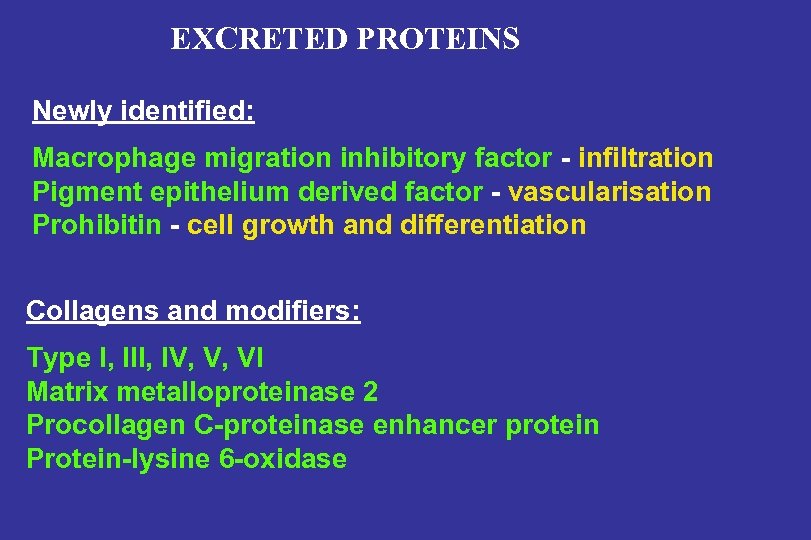
EXCRETED PROTEINS Newly identified: Macrophage migration inhibitory factor - infiltration Pigment epithelium derived factor - vascularisation Prohibitin - cell growth and differentiation Collagens and modifiers: Type I, III, IV, V, VI Matrix metalloproteinase 2 Procollagen C-proteinase enhancer protein Protein-lysine 6 -oxidase
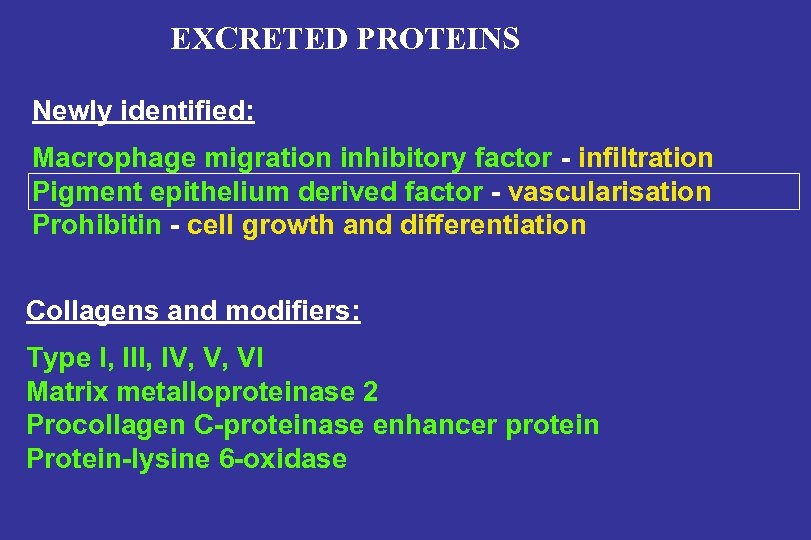
EXCRETED PROTEINS Newly identified: Macrophage migration inhibitory factor - infiltration Pigment epithelium derived factor - vascularisation Prohibitin - cell growth and differentiation Collagens and modifiers: Type I, III, IV, V, VI Matrix metalloproteinase 2 Procollagen C-proteinase enhancer protein Protein-lysine 6 -oxidase
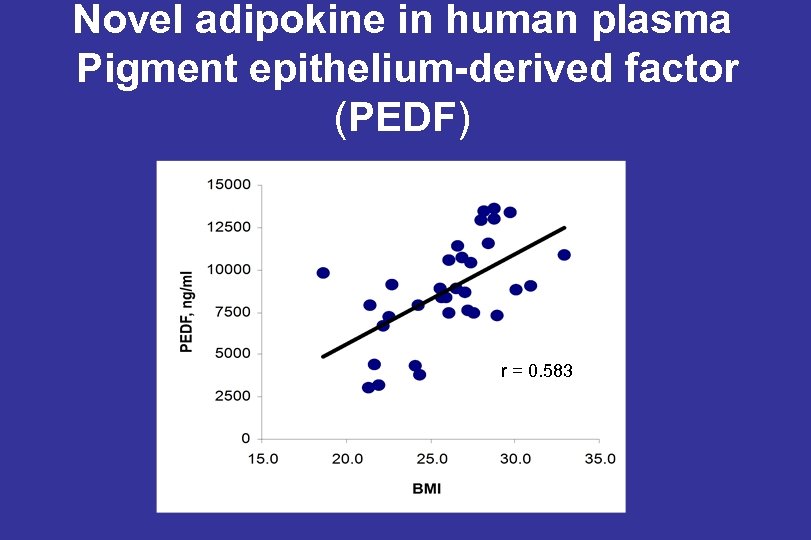
Novel adipokine in human plasma Pigment epithelium-derived factor (PEDF) r = 0. 583
EXCRETED PROTEINS Newly identified: Macrophage migration inhibitory factor - infiltration Pigment epithelium derived factor - vascularisation Prohibitin - cell growth and differentiation Collagens and modifiers: Type I, III, IV, V, VI Matrix metalloproteinase 2 Procollagen C-proteinase enhancer protein Protein-lysine 6 -oxidase
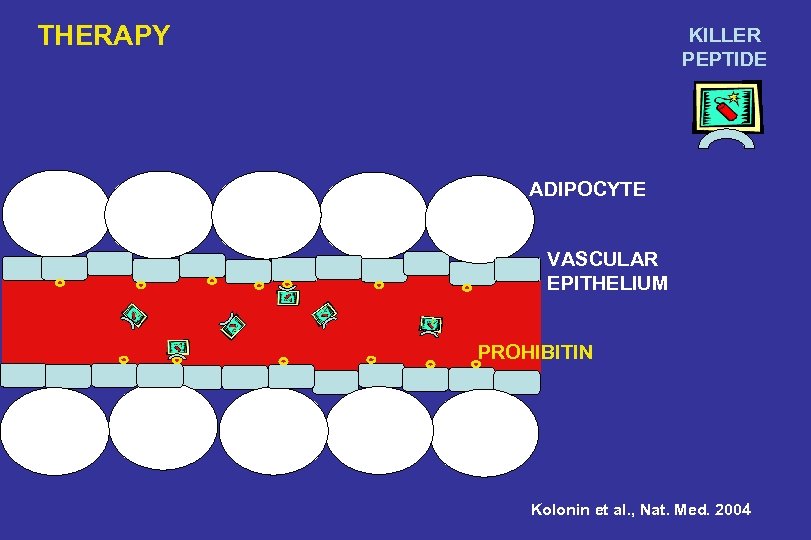
THERAPY KILLER PEPTIDE ADIPOCYTE VASCULAR EPITHELIUM PROHIBITIN Kolonin et al. , Nat. Med. 2004
EXCRETED PROTEINS Newly identified: Macrophage migration inhibitory factor - infiltration Pigment epithelium derived factor - vascularisation Prohibitin - cell growth and differentiation Collagens and modifiers: Type I, III, IV, V, VI Matrix metalloproteinase 2 Procollagen C-proteinase enhancer protein Protein-lysine 6 -oxidase
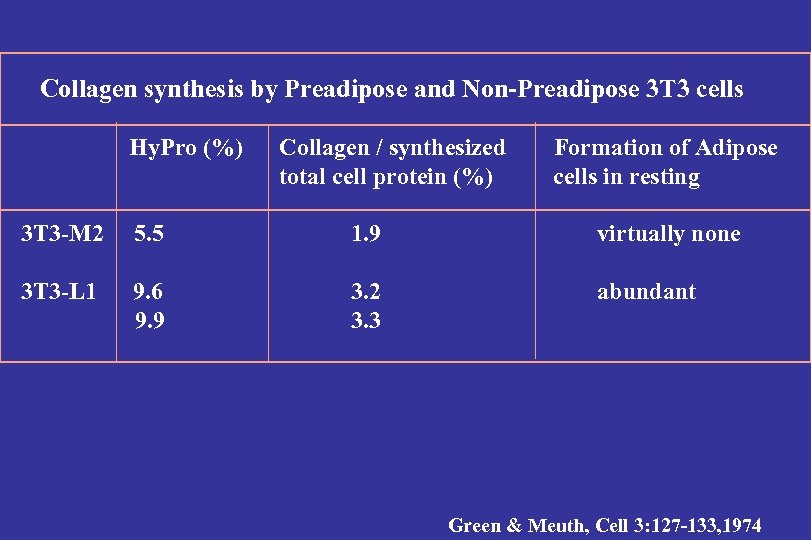
Collagen synthesis by Preadipose and Non-Preadipose 3 T 3 cells Hy. Pro (%) Collagen / synthesized total cell protein (%) Formation of Adipose cells in resting 3 T 3 -M 2 5. 5 1. 9 virtually none 3 T 3 -L 1 9. 6 9. 9 3. 2 3. 3 abundant Green & Meuth, Cell 3: 127 -133, 1974
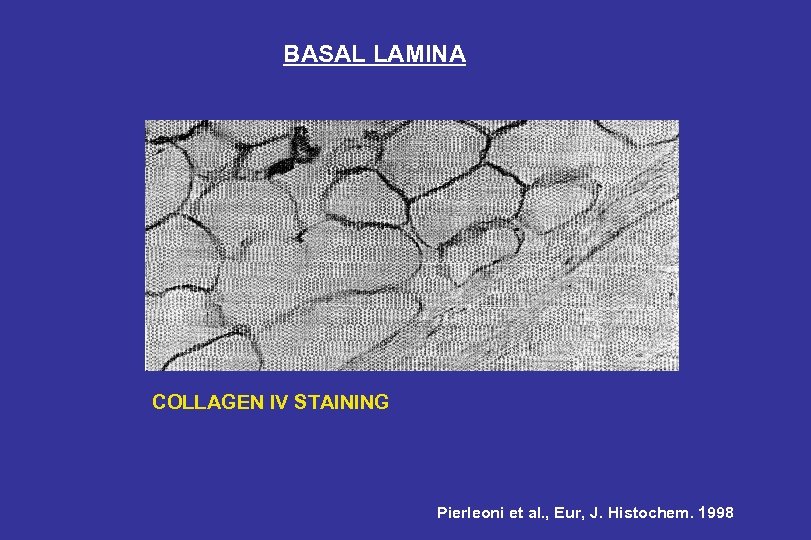
BASAL LAMINA COLLAGEN IV STAINING Pierleoni et al. , Eur, J. Histochem. 1998
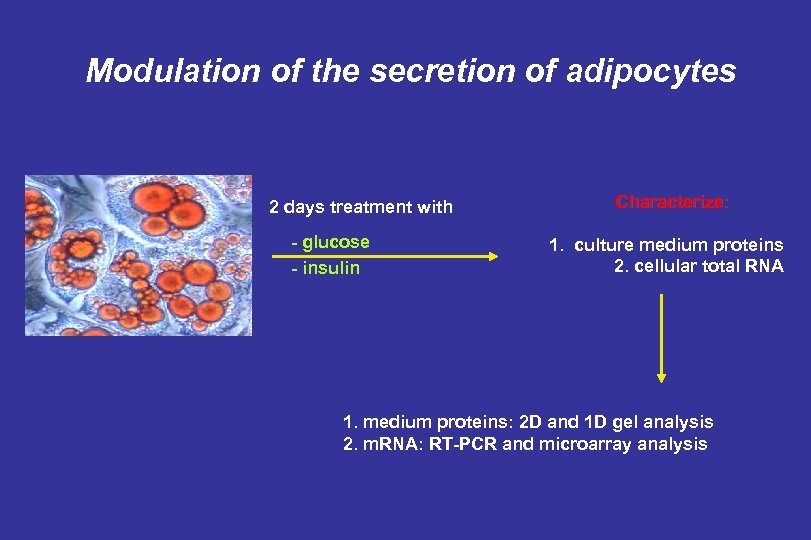
Modulation of the secretion of adipocytes 2 days treatment with - glucose - insulin Characterize: 1. culture medium proteins 2. cellular total RNA 1. medium proteins: 2 D and 1 D gel analysis 2. m. RNA: RT-PCR and microarray analysis
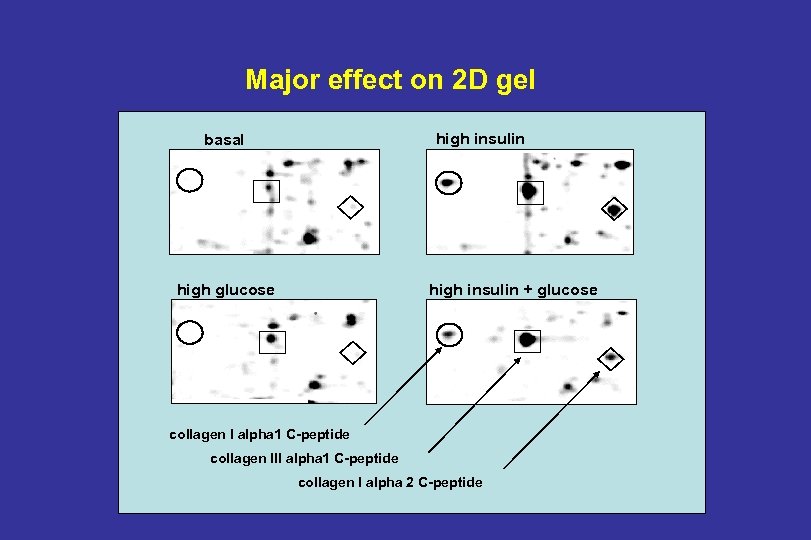
Major effect on 2 D gel high insulin basal high glucose high insulin + glucose collagen I alpha 1 C-peptide collagen III alpha 1 C-peptide collagen I alpha 2 C-peptide
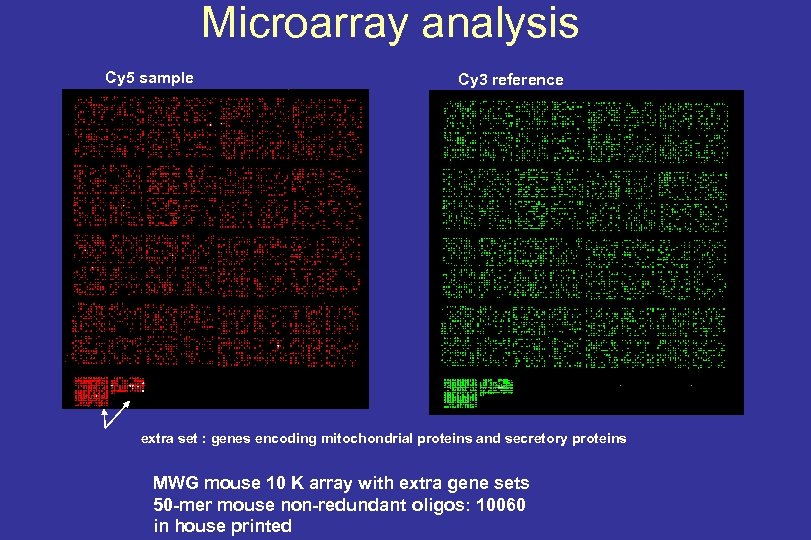
Microarray analysis Cy 5 sample Cy 3 reference extra set : genes encoding mitochondrial proteins and secretory proteins MWG mouse 10 K array with extra gene sets 50 -mer mouse non-redundant oligos: 10060 in house printed
Comparison on transcriptome and proteome of secretory genes
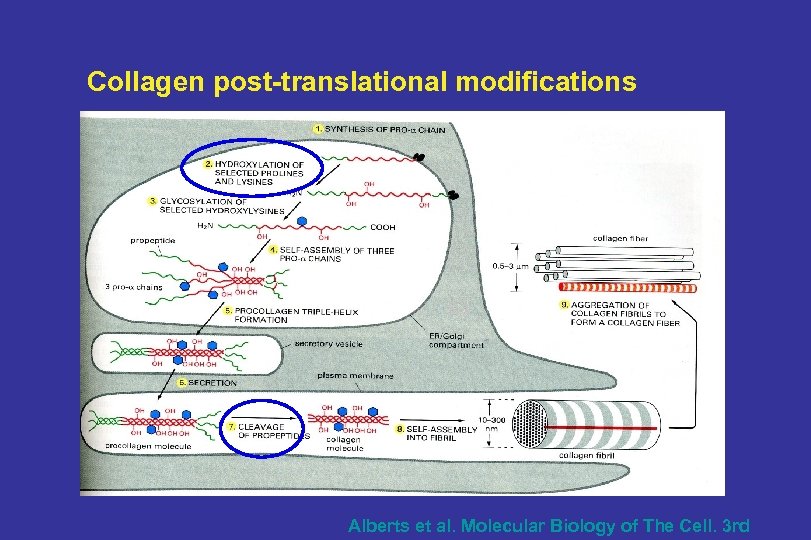
Collagen post-translational modifications Alberts et al. Molecular Biology of The Cell. 3 rd
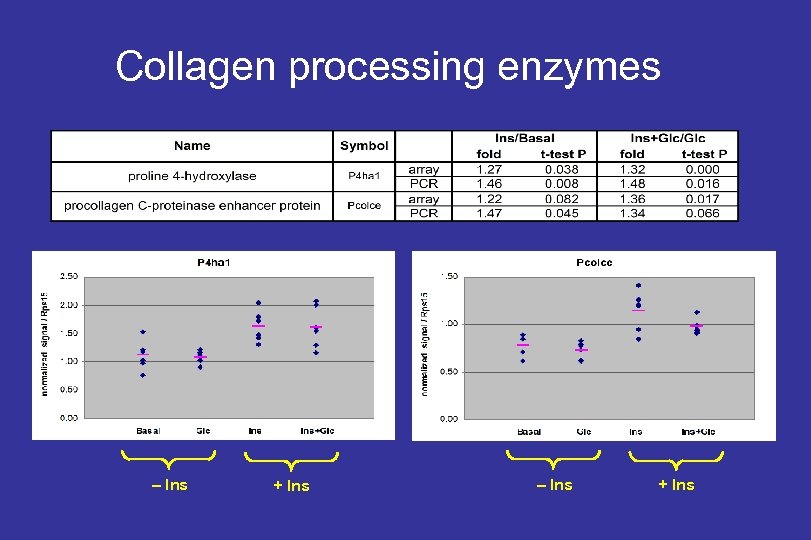
Collagen processing enzymes – Ins + Ins
• from a proteomics viewpoint, insulin has a significant impact on protein secretion of 3 T 3 -L 1 adipocytes • transcription is not the major regulation point for these secreted proteins • the discrepancy of the insulin effect between transcript and secretion level of collagens, may be partly explained by the transcriptional regulation of processing enzymes. Wang et al. , Diabetologia 2006
SECRETOME: SECRETED PROTEINS INTERVENTION TARGETS ECM COMPONENTS INSULIN-REGULATED
MOLECULAR MECHANISMS FOR OBESITY-RELATED PHYSIOLOGY EC Framework Programme 5 project NUGENOB : Low Fat-Burners vs High Fat-Burners
Whole body nutrient oxidation: carbohydrates fatty acids Different fatty acid flux LFO > HFO visible in adipose proteome?
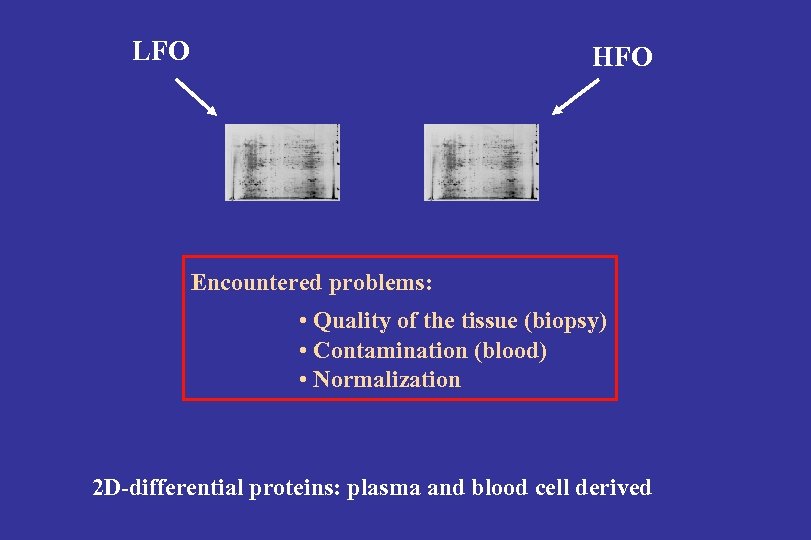
LFO HFO Encountered problems: • Quality of the tissue (biopsy) • Contamination (blood) • Normalization 2 D-differential proteins: plasma and blood cell derived
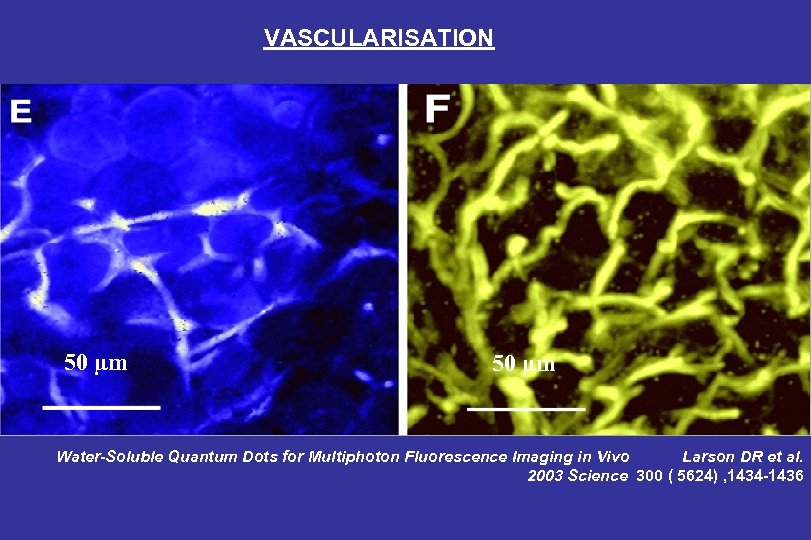
VASCULARISATION 50 µm Water-Soluble Quantum Dots for Multiphoton Fluorescence Imaging in Vivo Larson DR et al. 2003 Science 300 ( 5624) , 1434 -1436
courtesy of Peter Arner/Gabby Hul
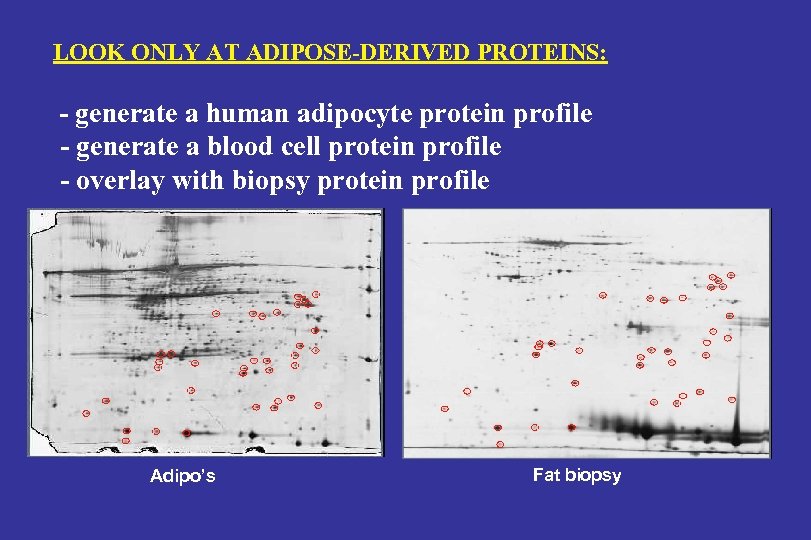
LOOK ONLY AT ADIPOSE-DERIVED PROTEINS: - generate a human adipocyte protein profile - generate a blood cell protein profile - overlay with biopsy protein profile Adipo’s Fat biopsy
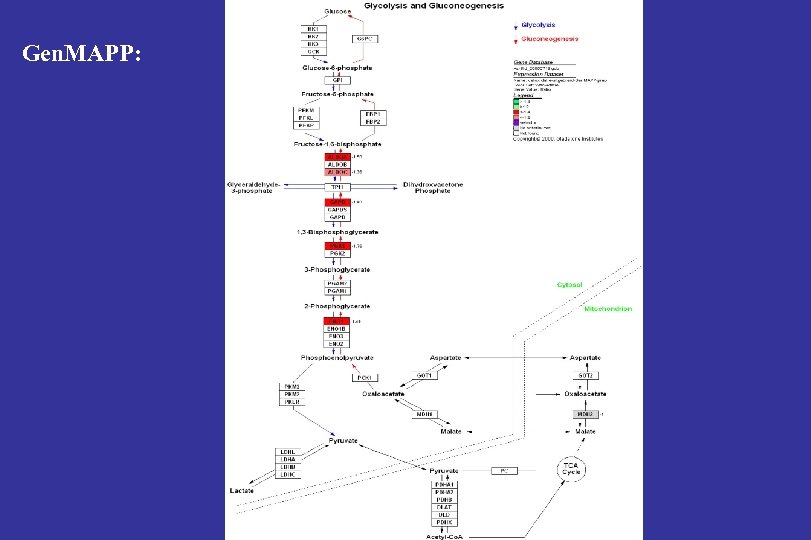
Gen. MAPP:
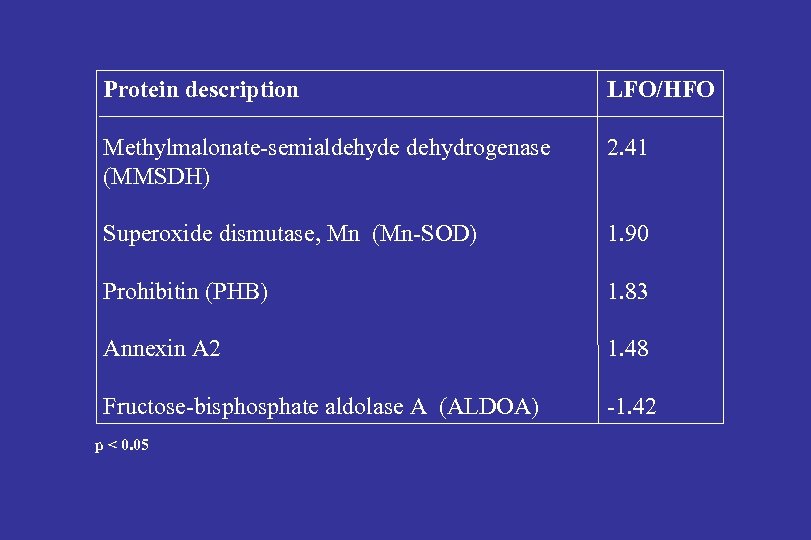
Protein description LFO/HFO Methylmalonate-semialdehyde dehydrogenase (MMSDH) 2. 41 Superoxide dismutase, Mn (Mn-SOD) 1. 90 Prohibitin (PHB) 1. 83 Annexin A 2 1. 48 Fructose-bisphosphate aldolase A (ALDOA) -1. 42 p < 0. 05
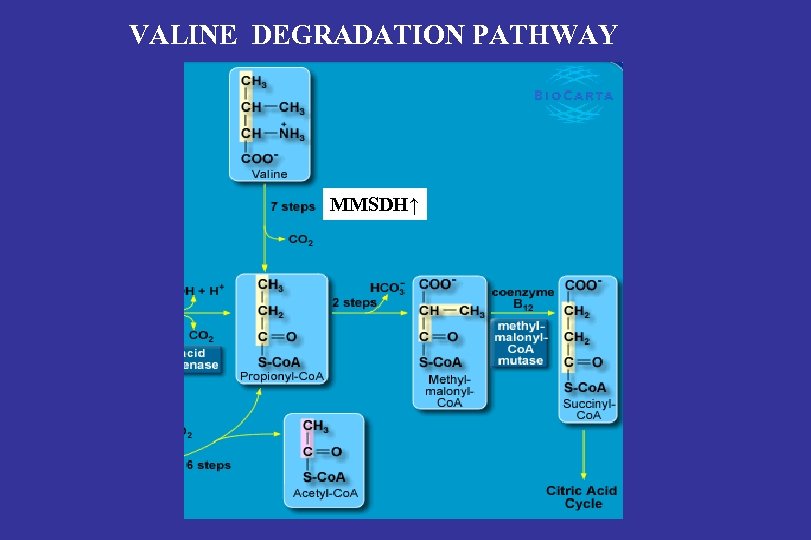
VALINE DEGRADATION PATHWAY MMSDH↑
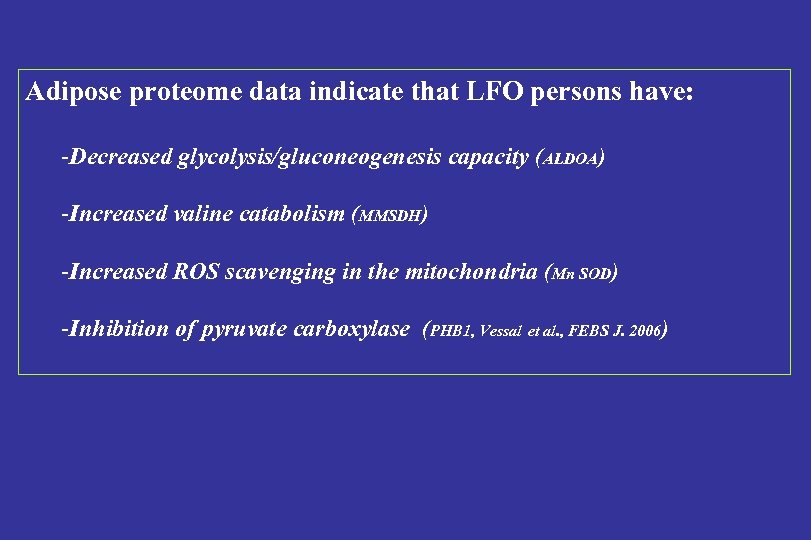
Adipose proteome data indicate that LFO persons have: -Decreased glycolysis/gluconeogenesis capacity (ALDOA) -Increased valine catabolism (MMSDH) -Increased ROS scavenging in the mitochondria (Mn SOD) -Inhibition of pyruvate carboxylase (PHB 1, Vessal et al. , FEBS J. 2006)
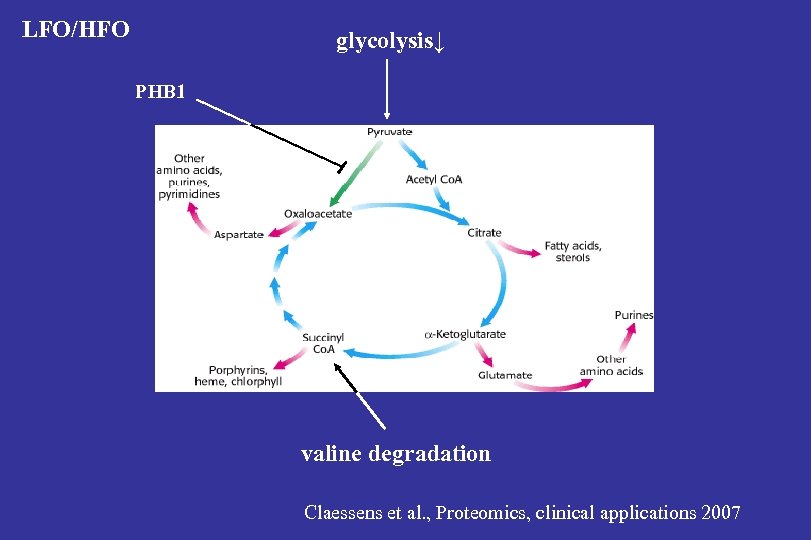
LFO/HFO glycolysis↓ PHB 1 valine degradation Claessens et al. , Proteomics, clinical applications 2007

PROTEOMICS IN OBESITY • Proteomics can be used to identify proteins related to human obesity using in vitro or animal models • The influence of nutrition and nutritional hormones on cell function can be studied • Novel targets for intervention can be found • Mechanistic models are provided for physiological differences in humans
Recommended literature: Nutritional Proteomics: methods and concepts for research in nutritional science. Schweigert FJ. Ann Nutr Metab 51: 99107, 2007. Nutrigenomics and nutrigenetics: the 'omics' revolution in nutritional science. Mariman E. Biotechnol Appl Biochem 44: 119 -28, 2006. Proteomics in nutrition research: principles, technologies and applications. Fuchs D, Winkelmann I, Johnson IT, Mariman E, Wenzel U, Daniel H. Br J Nutr 94: 302 -14, 2005. Nutriproteomics: identifying the molecular targets of nutritive And non-nutritive components of the diet. Barnes S, Kim H. J Biochem Mol Biol 37: 59 -74, 2004.
Abbreviations used BFA Brefeldin-A BIA-MS Biomolecular interaction analysis – mass spectrometry Cy-3, Cy-5 Fluorescent cyanine dyes ECM Extra-cellular matrix ELISA Enzyme-Linked immunosorbent assay FC Fold change FT-MS Fourier transform mass spectrometry HPLC High performance liquid chromatography HUGO Human Genome Organisation ICAT Isotope-coded affinity tags IL- 6 (etc) Interleukin -6 (etc) M/Z Mass /charge ratio MALDI-TOF Matrix-assisted laser desorptio/ionisation – time of flight MIDA Mass isotopomer distribution analysis MWG Biotech AG PHB Prohibitin ROS Reactive oxygen species SELDI-TOF Surface enhanced laser desorption ionization – time of flight SPR-BIA Surface plasmon resonance – biomolecular interaction analysis
a1075cdfd1474cbd275d7a84dc732d59.ppt