8c7220bc7bca681ed653e095f9176b36.ppt
- Количество слайдов: 43
 PROTEOMICS collateral update: Targeted Peptide Quantitation- a sales guide with applications perspectives Application support EU and USA, with input from Factories, Demo labs and Strategic teams Scott Peterman, Michaela Scigelova and Madalina Oppermann October 2012
PROTEOMICS collateral update: Targeted Peptide Quantitation- a sales guide with applications perspectives Application support EU and USA, with input from Factories, Demo labs and Strategic teams Scott Peterman, Michaela Scigelova and Madalina Oppermann October 2012
 Overview: targeted peptide quantitation • • • 2 Application perspectives Customer profiles Application options Examples Conclusions
Overview: targeted peptide quantitation • • • 2 Application perspectives Customer profiles Application options Examples Conclusions
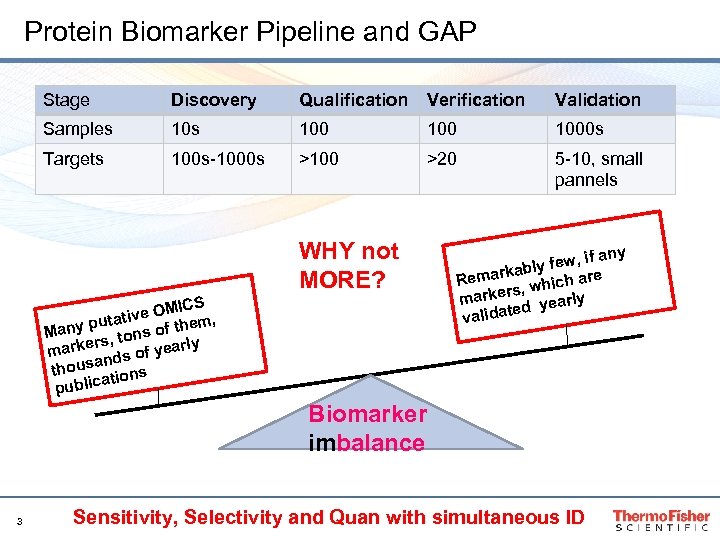 Protein Biomarker Pipeline and GAP Stage Discovery Qualification Verification Validation Samples 100 1000 s Targets 100 s-1000 s >100 >20 5 -10, small pannels WHY not MORE? MICS tive O em, puta f th Many tons o rly rs, marke ds of yea an thous ions at public if any y few, kabl Remar h are s, whic r marke arly ted ye valida Biomarker imbalance 3 Sensitivity, Selectivity and Quan with simultaneous ID
Protein Biomarker Pipeline and GAP Stage Discovery Qualification Verification Validation Samples 100 1000 s Targets 100 s-1000 s >100 >20 5 -10, small pannels WHY not MORE? MICS tive O em, puta f th Many tons o rly rs, marke ds of yea an thous ions at public if any y few, kabl Remar h are s, whic r marke arly ted ye valida Biomarker imbalance 3 Sensitivity, Selectivity and Quan with simultaneous ID
 Issues Related to Biomarker Research 4 Google images
Issues Related to Biomarker Research 4 Google images
 Experimental Roadblocks to succeed Targeted Protein Quantitation – Where are customers struggling? (see notes section for more info) 1. Target selection: Assumption: determination of protein of interest is easy • • Target peptide determination? Precursor or product ions? Which product ions? How many at each level for data confidence? 2. Data acquisition strategy • Defined from the above answers • Need to determine LC methods to fit target list? 3. Data interrogation • Methodology for qual/quan 4. Method refinement • Sample prep methods needed? • Experimental methods changed? • Incorporation of synthetic peptides/recombinant proteins? 5
Experimental Roadblocks to succeed Targeted Protein Quantitation – Where are customers struggling? (see notes section for more info) 1. Target selection: Assumption: determination of protein of interest is easy • • Target peptide determination? Precursor or product ions? Which product ions? How many at each level for data confidence? 2. Data acquisition strategy • Defined from the above answers • Need to determine LC methods to fit target list? 3. Data interrogation • Methodology for qual/quan 4. Method refinement • Sample prep methods needed? • Experimental methods changed? • Incorporation of synthetic peptides/recombinant proteins? 5
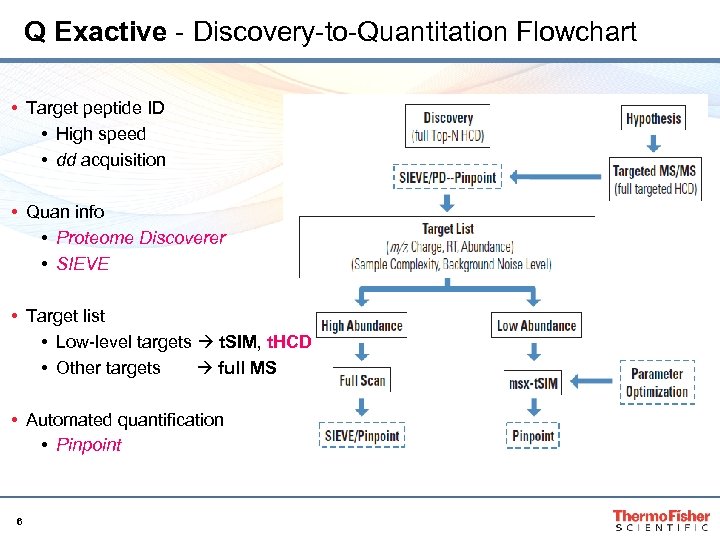 Q Exactive - Discovery-to-Quantitation Flowchart • Target peptide ID • High speed • dd acquisition • Quan info • Proteome Discoverer • SIEVE • Target list • Low-level targets t. SIM, t. HCD • Other targets full MS • Automated quantification • Pinpoint 6
Q Exactive - Discovery-to-Quantitation Flowchart • Target peptide ID • High speed • dd acquisition • Quan info • Proteome Discoverer • SIEVE • Target list • Low-level targets t. SIM, t. HCD • Other targets full MS • Automated quantification • Pinpoint 6
 Questions to gather information on which mass spec is the right one for your customer? • Thoughts to consider -these are questions to help guide, not hard set • How many protein targets are you routinely monitoring per assay? • A tipping point may be 20 proteins - >20 leads to QE, <20 either instrument • How long is your typical LC-MS experimental analysis time for targeted quantitation methods? <40 min triples are in play, >40 min and throughput is not an issue for QE • How many different assays/studies do you anticipate working on per month, quarter, year? <3 triples are in play, >4 favors QE • Do you plan on incorporating sample enrichment/purification off-line? Yes, then favors triples; No, then favors QE • How many samples do you plan on analyzing per study? Large numbers tend to favor triples as they would most likely have some level of sample prep. • How many people in the lab will be running the system? Many favors QE • What is the staff turnover? A response of “high turnover” favors QE -------------------------------------------------------- • Do you have HRAM technology in your lab today? What are the new benefits sought next? • Do you have Qq. Q technology in your lab today? What are the new benefits sought next? 7
Questions to gather information on which mass spec is the right one for your customer? • Thoughts to consider -these are questions to help guide, not hard set • How many protein targets are you routinely monitoring per assay? • A tipping point may be 20 proteins - >20 leads to QE, <20 either instrument • How long is your typical LC-MS experimental analysis time for targeted quantitation methods? <40 min triples are in play, >40 min and throughput is not an issue for QE • How many different assays/studies do you anticipate working on per month, quarter, year? <3 triples are in play, >4 favors QE • Do you plan on incorporating sample enrichment/purification off-line? Yes, then favors triples; No, then favors QE • How many samples do you plan on analyzing per study? Large numbers tend to favor triples as they would most likely have some level of sample prep. • How many people in the lab will be running the system? Many favors QE • What is the staff turnover? A response of “high turnover” favors QE -------------------------------------------------------- • Do you have HRAM technology in your lab today? What are the new benefits sought next? • Do you have Qq. Q technology in your lab today? What are the new benefits sought next? 7
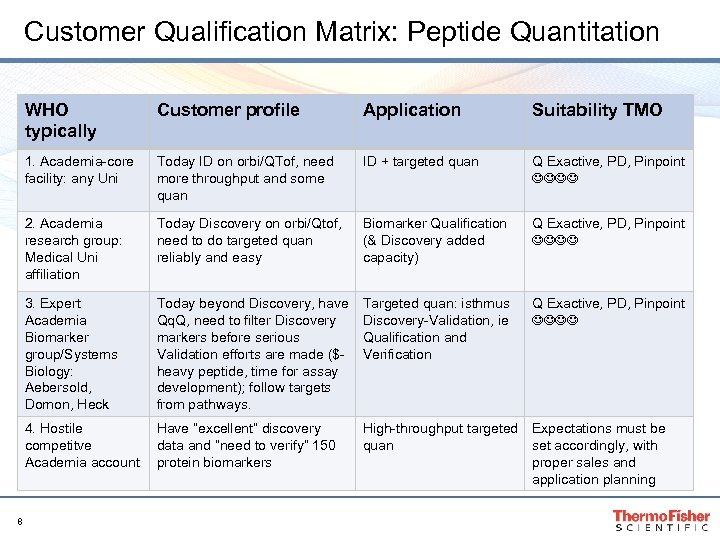 Customer Qualification Matrix: Peptide Quantitation WHO typically Application Suitability TMO 1. Academia-core facility: any Uni Today ID on orbi/QTof, need more throughput and some quan ID + targeted quan Q Exactive, PD, Pinpoint 2. Academia research group: Medical Uni affiliation Today Discovery on orbi/Qtof, need to do targeted quan reliably and easy Biomarker Qualification (& Discovery added capacity) Q Exactive, PD, Pinpoint 3. Expert Academia Biomarker group/Systems Biology: Aebersold, Domon, Heck Today beyond Discovery, have Qq. Q, need to filter Discovery markers before serious Validation efforts are made ($heavy peptide, time for assay development); follow targets from pathways. Targeted quan: isthmus Discovery-Validation, ie Qualification and Verification Q Exactive, PD, Pinpoint 4. Hostile competitve Academia account 8 Customer profile Have ”excellent” discovery data and ”need to verify” 150 protein biomarkers High-throughput targeted Expectations must be quan set accordingly, with proper sales and application planning
Customer Qualification Matrix: Peptide Quantitation WHO typically Application Suitability TMO 1. Academia-core facility: any Uni Today ID on orbi/QTof, need more throughput and some quan ID + targeted quan Q Exactive, PD, Pinpoint 2. Academia research group: Medical Uni affiliation Today Discovery on orbi/Qtof, need to do targeted quan reliably and easy Biomarker Qualification (& Discovery added capacity) Q Exactive, PD, Pinpoint 3. Expert Academia Biomarker group/Systems Biology: Aebersold, Domon, Heck Today beyond Discovery, have Qq. Q, need to filter Discovery markers before serious Validation efforts are made ($heavy peptide, time for assay development); follow targets from pathways. Targeted quan: isthmus Discovery-Validation, ie Qualification and Verification Q Exactive, PD, Pinpoint 4. Hostile competitve Academia account 8 Customer profile Have ”excellent” discovery data and ”need to verify” 150 protein biomarkers High-throughput targeted Expectations must be quan set accordingly, with proper sales and application planning
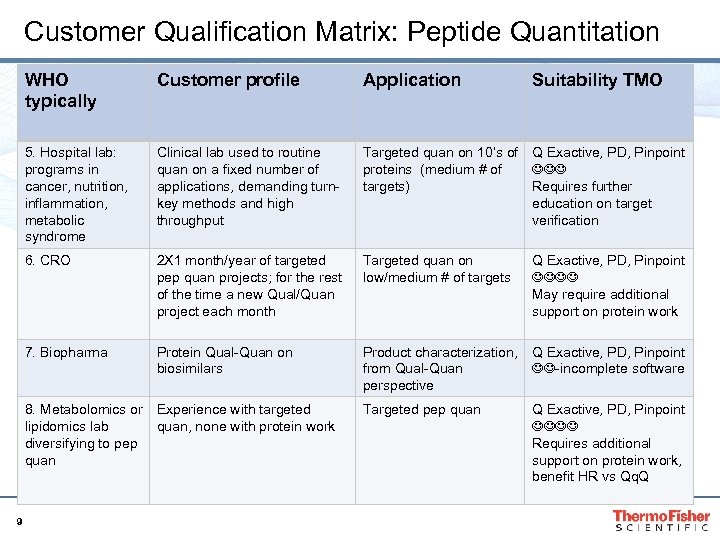 Customer Qualification Matrix: Peptide Quantitation WHO typically Customer profile Application 5. Hospital lab: programs in cancer, nutrition, inflammation, metabolic syndrome Clinical lab used to routine quan on a fixed number of applications, demanding turnkey methods and high throughput Targeted quan on 10’s of Q Exactive, PD, Pinpoint proteins (medium # of targets) Requires further education on target verification 6. CRO 2 X 1 month/year of targeted pep quan projects; for the rest of the time a new Qual/Quan project each month Targeted quan on low/medium # of targets 7. Biopharma Protein Qual-Quan on biosimilars Product characterization, Q Exactive, PD, Pinpoint -incomplete software from Qual-Quan perspective 8. Metabolomics or Experience with targeted lipidomics lab quan, none with protein work diversifying to pep quan 9 Targeted pep quan Suitability TMO Q Exactive, PD, Pinpoint May require additional support on protein work Q Exactive, PD, Pinpoint Requires additional support on protein work, benefit HR vs Qq. Q
Customer Qualification Matrix: Peptide Quantitation WHO typically Customer profile Application 5. Hospital lab: programs in cancer, nutrition, inflammation, metabolic syndrome Clinical lab used to routine quan on a fixed number of applications, demanding turnkey methods and high throughput Targeted quan on 10’s of Q Exactive, PD, Pinpoint proteins (medium # of targets) Requires further education on target verification 6. CRO 2 X 1 month/year of targeted pep quan projects; for the rest of the time a new Qual/Quan project each month Targeted quan on low/medium # of targets 7. Biopharma Protein Qual-Quan on biosimilars Product characterization, Q Exactive, PD, Pinpoint -incomplete software from Qual-Quan perspective 8. Metabolomics or Experience with targeted lipidomics lab quan, none with protein work diversifying to pep quan 9 Targeted pep quan Suitability TMO Q Exactive, PD, Pinpoint May require additional support on protein work Q Exactive, PD, Pinpoint Requires additional support on protein work, benefit HR vs Qq. Q
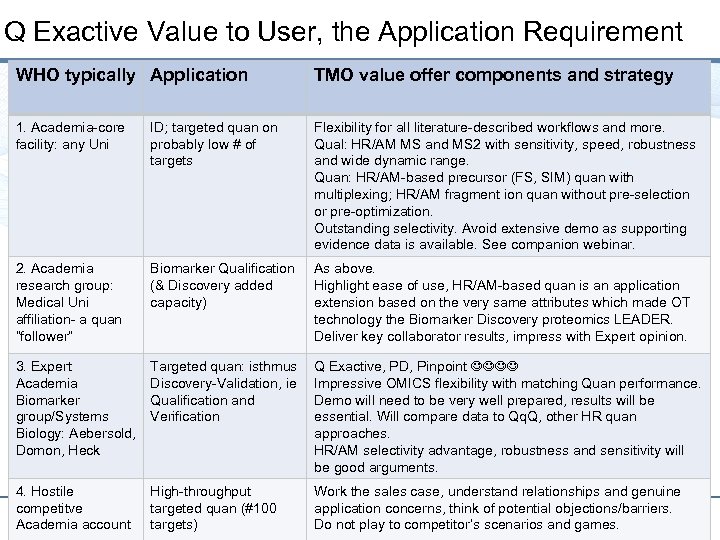 Q Exactive Value to User, the Application Requirement WHO typically Application TMO value offer components and strategy 1. Academia-core facility: any Uni ID; targeted quan on probably low # of targets Flexibility for all literature-described workflows and more. Qual: HR/AM MS and MS 2 with sensitivity, speed, robustness and wide dynamic range. Quan: HR/AM-based precursor (FS, SIM) quan with multiplexing; HR/AM fragment ion quan without pre-selection or pre-optimization. Outstanding selectivity. Avoid extensive demo as supporting evidence data is available. See companion webinar. 2. Academia research group: Medical Uni affiliation- a quan ”follower” Biomarker Qualification As above. (& Discovery added Highlight ease of use, HR/AM-based quan is an application capacity) extension based on the very same attributes which made OT technology the Biomarker Discovery proteomics LEADER. Deliver key collaborator results, impress with Expert opinion. 3. Expert Academia Biomarker group/Systems Biology: Aebersold, Domon, Heck Targeted quan: isthmus Discovery-Validation, ie Qualification and Verification Q Exactive, PD, Pinpoint Impressive OMICS flexibility with matching Quan performance. Demo will need to be very well prepared, results will be essential. Will compare data to Qq. Q, other HR quan approaches. HR/AM selectivity advantage, robustness and sensitivity will be good arguments. 4. Hostile competitve 10 Academia account High-throughput targeted quan (#100 targets) Work the sales case, understand relationships and genuine application concerns, think of potential objections/barriers. Do not play to competitor’s scenarios and games.
Q Exactive Value to User, the Application Requirement WHO typically Application TMO value offer components and strategy 1. Academia-core facility: any Uni ID; targeted quan on probably low # of targets Flexibility for all literature-described workflows and more. Qual: HR/AM MS and MS 2 with sensitivity, speed, robustness and wide dynamic range. Quan: HR/AM-based precursor (FS, SIM) quan with multiplexing; HR/AM fragment ion quan without pre-selection or pre-optimization. Outstanding selectivity. Avoid extensive demo as supporting evidence data is available. See companion webinar. 2. Academia research group: Medical Uni affiliation- a quan ”follower” Biomarker Qualification As above. (& Discovery added Highlight ease of use, HR/AM-based quan is an application capacity) extension based on the very same attributes which made OT technology the Biomarker Discovery proteomics LEADER. Deliver key collaborator results, impress with Expert opinion. 3. Expert Academia Biomarker group/Systems Biology: Aebersold, Domon, Heck Targeted quan: isthmus Discovery-Validation, ie Qualification and Verification Q Exactive, PD, Pinpoint Impressive OMICS flexibility with matching Quan performance. Demo will need to be very well prepared, results will be essential. Will compare data to Qq. Q, other HR quan approaches. HR/AM selectivity advantage, robustness and sensitivity will be good arguments. 4. Hostile competitve 10 Academia account High-throughput targeted quan (#100 targets) Work the sales case, understand relationships and genuine application concerns, think of potential objections/barriers. Do not play to competitor’s scenarios and games.
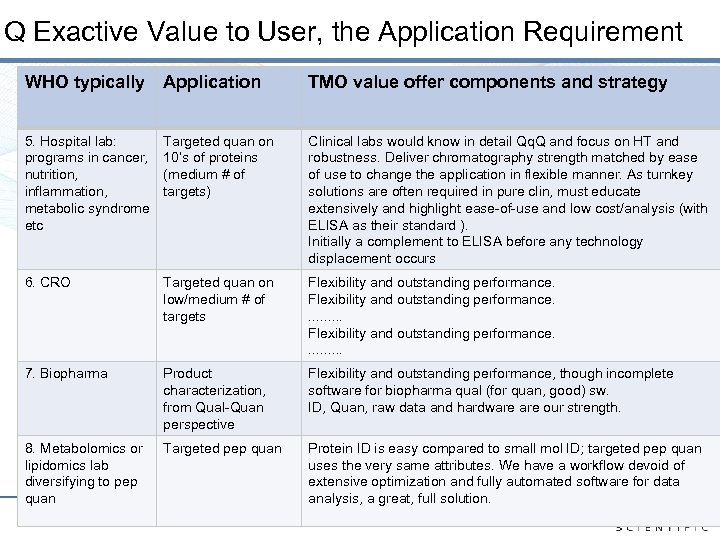 Q Exactive Value to User, the Application Requirement WHO typically Application TMO value offer components and strategy 5. Hospital lab: programs in cancer, nutrition, inflammation, metabolic syndrome etc Targeted quan on 10’s of proteins (medium # of targets) Clinical labs would know in detail Qq. Q and focus on HT and robustness. Deliver chromatography strength matched by ease of use to change the application in flexible manner. As turnkey solutions are often required in pure clin, must educate extensively and highlight ease-of-use and low cost/analysis (with ELISA as their standard ). Initially a complement to ELISA before any technology displacement occurs 6. CRO Targeted quan on low/medium # of targets Flexibility and outstanding performance. . . . . 7. Biopharma Product characterization, from Qual-Quan perspective Flexibility and outstanding performance, though incomplete software for biopharma qual (for quan, good) sw. ID, Quan, raw data and hardware our strength. 8. Metabolomics or lipidomics lab diversifying to pep quan Targeted pep quan Protein ID is easy compared to small mol ID; targeted pep quan uses the very same attributes. We have a workflow devoid of extensive optimization and fully automated software for data analysis, a great, full solution. 11
Q Exactive Value to User, the Application Requirement WHO typically Application TMO value offer components and strategy 5. Hospital lab: programs in cancer, nutrition, inflammation, metabolic syndrome etc Targeted quan on 10’s of proteins (medium # of targets) Clinical labs would know in detail Qq. Q and focus on HT and robustness. Deliver chromatography strength matched by ease of use to change the application in flexible manner. As turnkey solutions are often required in pure clin, must educate extensively and highlight ease-of-use and low cost/analysis (with ELISA as their standard ). Initially a complement to ELISA before any technology displacement occurs 6. CRO Targeted quan on low/medium # of targets Flexibility and outstanding performance. . . . . 7. Biopharma Product characterization, from Qual-Quan perspective Flexibility and outstanding performance, though incomplete software for biopharma qual (for quan, good) sw. ID, Quan, raw data and hardware our strength. 8. Metabolomics or lipidomics lab diversifying to pep quan Targeted pep quan Protein ID is easy compared to small mol ID; targeted pep quan uses the very same attributes. We have a workflow devoid of extensive optimization and fully automated software for data analysis, a great, full solution. 11
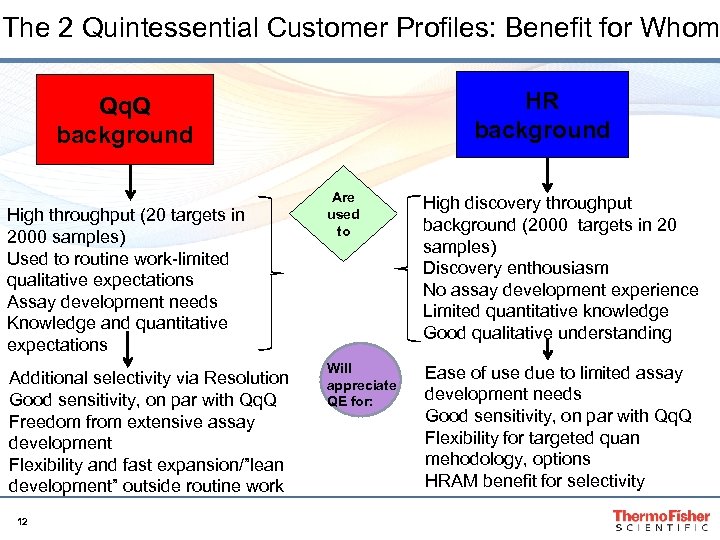 The 2 Quintessential Customer Profiles: Benefit for Whom HR background Qq. Q background High throughput (20 targets in 2000 samples) Used to routine work-limited qualitative expectations Assay development needs Knowledge and quantitative expectations Additional selectivity via Resolution Good sensitivity, on par with Qq. Q Freedom from extensive assay development Flexibility and fast expansion/”lean development” outside routine work 12 Are used to High discovery throughput background (2000 targets in 20 samples) Discovery enthousiasm No assay development experience Limited quantitative knowledge Good qualitative understanding Will appreciate QE for: Ease of use due to limited assay development needs Good sensitivity, on par with Qq. Q Flexibility for targeted quan mehodology, options HRAM benefit for selectivity
The 2 Quintessential Customer Profiles: Benefit for Whom HR background Qq. Q background High throughput (20 targets in 2000 samples) Used to routine work-limited qualitative expectations Assay development needs Knowledge and quantitative expectations Additional selectivity via Resolution Good sensitivity, on par with Qq. Q Freedom from extensive assay development Flexibility and fast expansion/”lean development” outside routine work 12 Are used to High discovery throughput background (2000 targets in 20 samples) Discovery enthousiasm No assay development experience Limited quantitative knowledge Good qualitative understanding Will appreciate QE for: Ease of use due to limited assay development needs Good sensitivity, on par with Qq. Q Flexibility for targeted quan mehodology, options HRAM benefit for selectivity
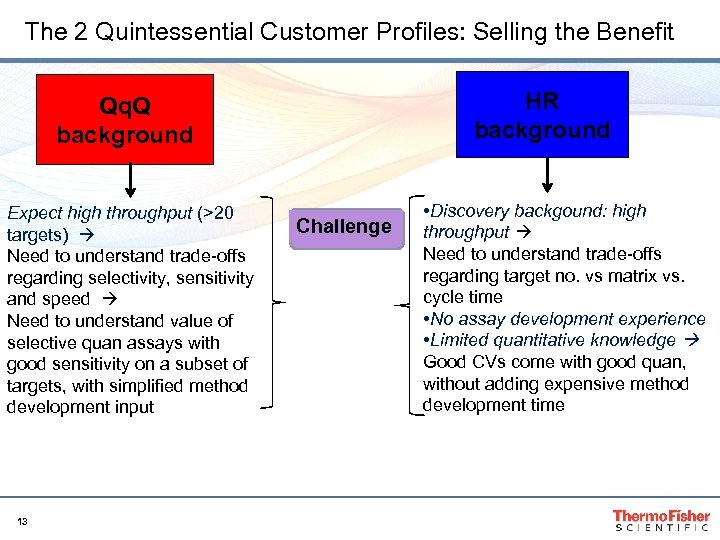 The 2 Quintessential Customer Profiles: Selling the Benefit HR background Qq. Q background Expect high throughput (>20 targets) Need to understand trade-offs regarding selectivity, sensitivity and speed Need to understand value of selective quan assays with good sensitivity on a subset of targets, with simplified method development input 13 Challenge • Discovery backgound: high throughput Need to understand trade-offs regarding target no. vs matrix vs. cycle time • No assay development experience • Limited quantitative knowledge Good CVs come with good quan, without adding expensive method development time
The 2 Quintessential Customer Profiles: Selling the Benefit HR background Qq. Q background Expect high throughput (>20 targets) Need to understand trade-offs regarding selectivity, sensitivity and speed Need to understand value of selective quan assays with good sensitivity on a subset of targets, with simplified method development input 13 Challenge • Discovery backgound: high throughput Need to understand trade-offs regarding target no. vs matrix vs. cycle time • No assay development experience • Limited quantitative knowledge Good CVs come with good quan, without adding expensive method development time
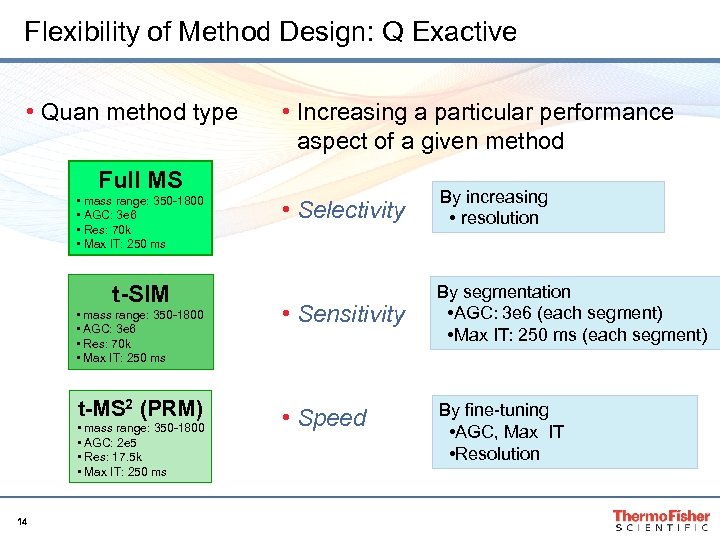 Flexibility of Method Design: Q Exactive • Quan method type • Increasing a particular performance aspect of a given method Full MS • mass range: 350 -1800 • AGC: 3 e 6 • Res: 70 k • Max IT: 250 ms t-SIM • mass range: 350 -1800 • AGC: 3 e 6 • Res: 70 k • Max IT: 250 ms t-MS 2 (PRM) • mass range: 350 -1800 • AGC: 2 e 5 • Res: 17. 5 k • Max IT: 250 ms 14 • Selectivity By increasing • resolution • Sensitivity By segmentation • AGC: 3 e 6 (each segment) • Max IT: 250 ms (each segment) • Speed By fine-tuning • AGC, Max IT • Resolution
Flexibility of Method Design: Q Exactive • Quan method type • Increasing a particular performance aspect of a given method Full MS • mass range: 350 -1800 • AGC: 3 e 6 • Res: 70 k • Max IT: 250 ms t-SIM • mass range: 350 -1800 • AGC: 3 e 6 • Res: 70 k • Max IT: 250 ms t-MS 2 (PRM) • mass range: 350 -1800 • AGC: 2 e 5 • Res: 17. 5 k • Max IT: 250 ms 14 • Selectivity By increasing • resolution • Sensitivity By segmentation • AGC: 3 e 6 (each segment) • Max IT: 250 ms (each segment) • Speed By fine-tuning • AGC, Max IT • Resolution
 Practical Aspects of HR/AM-based Targeted Peptide Quantitation 15 The world leader in serving science
Practical Aspects of HR/AM-based Targeted Peptide Quantitation 15 The world leader in serving science
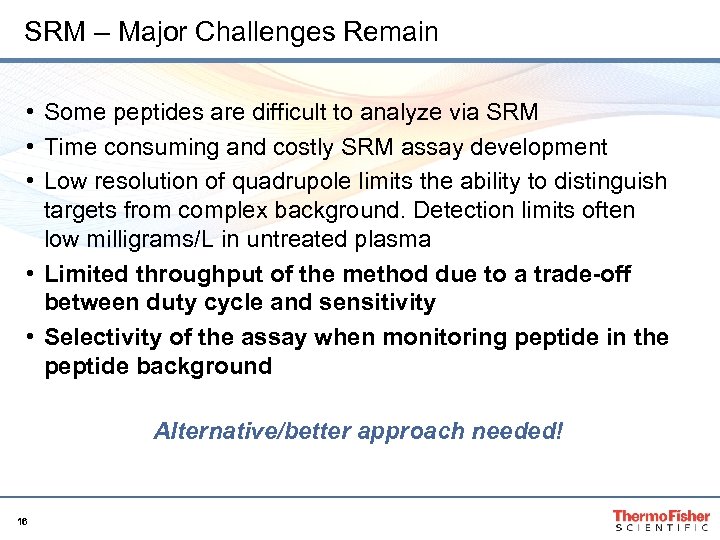 SRM – Major Challenges Remain • Some peptides are difficult to analyze via SRM • Time consuming and costly SRM assay development • Low resolution of quadrupole limits the ability to distinguish targets from complex background. Detection limits often low milligrams/L in untreated plasma • Limited throughput of the method due to a trade-off between duty cycle and sensitivity • Selectivity of the assay when monitoring peptide in the peptide background Alternative/better approach needed! 16
SRM – Major Challenges Remain • Some peptides are difficult to analyze via SRM • Time consuming and costly SRM assay development • Low resolution of quadrupole limits the ability to distinguish targets from complex background. Detection limits often low milligrams/L in untreated plasma • Limited throughput of the method due to a trade-off between duty cycle and sensitivity • Selectivity of the assay when monitoring peptide in the peptide background Alternative/better approach needed! 16
 Given the immense benefit that high resolution and accurate mass (HR/AM) instruments have brought to the discovery proteomics field, could highly accurate mass measurement capabilities be leveraged to provide benefits in the targeted proteomics domain as well? If HR/AM approach is to substitute SRM one, its selectivity must be comparable while at the same time ensuring also similar sensitivity and dynamic range of quantitation. 17
Given the immense benefit that high resolution and accurate mass (HR/AM) instruments have brought to the discovery proteomics field, could highly accurate mass measurement capabilities be leveraged to provide benefits in the targeted proteomics domain as well? If HR/AM approach is to substitute SRM one, its selectivity must be comparable while at the same time ensuring also similar sensitivity and dynamic range of quantitation. 17
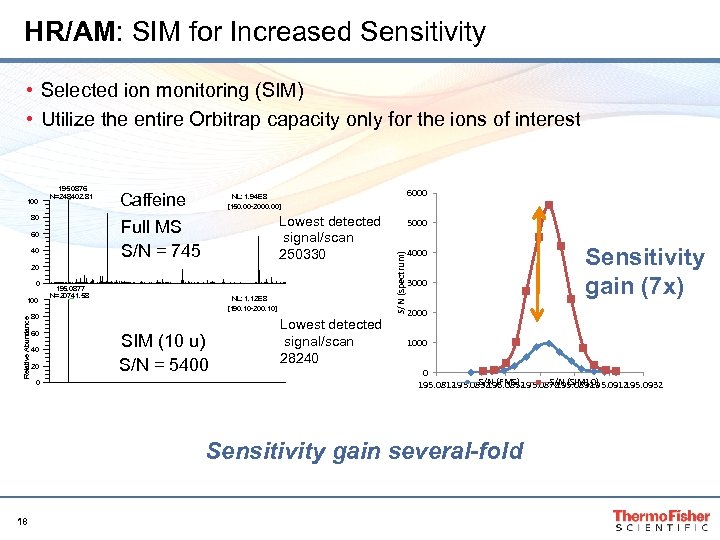 HR/AM: SIM for Increased Sensitivity • Selected ion monitoring (SIM) • Utilize the entire Orbitrap capacity only for the ions of interest 195. 0876 N=248402. 81 80 Caffeine Lowest detected signal/scan 250330 Full MS S/N = 745 60 40 20 0 Relative Abundance 100 195. 0877 N=20741. 58 NL: 1. 12 E 8 [190. 10 -200. 10] 80 60 40 20 0 6000 NL: 1. 94 E 8 [150. 00 -2000. 00] SIM (10 u) S/N = 5400 Lowest detected signal/scan 28240 5000 S/N (spectrum) 100 4000 3000 2000 1000 0 S/N 195. 0812 195. 0832 (FMS) 195. 0872 (SIM 10) 195. 0852 195. 0892 195. 0912 195. 0932 Sensitivity gain several-fold 18 Sensitivity gain (7 x)
HR/AM: SIM for Increased Sensitivity • Selected ion monitoring (SIM) • Utilize the entire Orbitrap capacity only for the ions of interest 195. 0876 N=248402. 81 80 Caffeine Lowest detected signal/scan 250330 Full MS S/N = 745 60 40 20 0 Relative Abundance 100 195. 0877 N=20741. 58 NL: 1. 12 E 8 [190. 10 -200. 10] 80 60 40 20 0 6000 NL: 1. 94 E 8 [150. 00 -2000. 00] SIM (10 u) S/N = 5400 Lowest detected signal/scan 28240 5000 S/N (spectrum) 100 4000 3000 2000 1000 0 S/N 195. 0812 195. 0832 (FMS) 195. 0872 (SIM 10) 195. 0852 195. 0892 195. 0912 195. 0932 Sensitivity gain several-fold 18 Sensitivity gain (7 x)
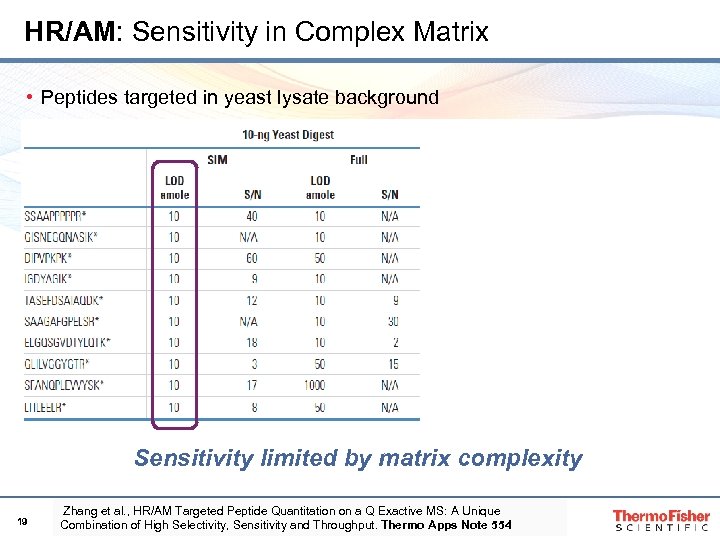 HR/AM: Sensitivity in Complex Matrix • Peptides targeted in yeast lysate background Sensitivity limited by matrix complexity 19 Zhang et al. , HR/AM Targeted Peptide Quantitation on a Q Exactive MS: A Unique Combination of High Selectivity, Sensitivity and Throughput. Thermo Apps Note 554
HR/AM: Sensitivity in Complex Matrix • Peptides targeted in yeast lysate background Sensitivity limited by matrix complexity 19 Zhang et al. , HR/AM Targeted Peptide Quantitation on a Q Exactive MS: A Unique Combination of High Selectivity, Sensitivity and Throughput. Thermo Apps Note 554
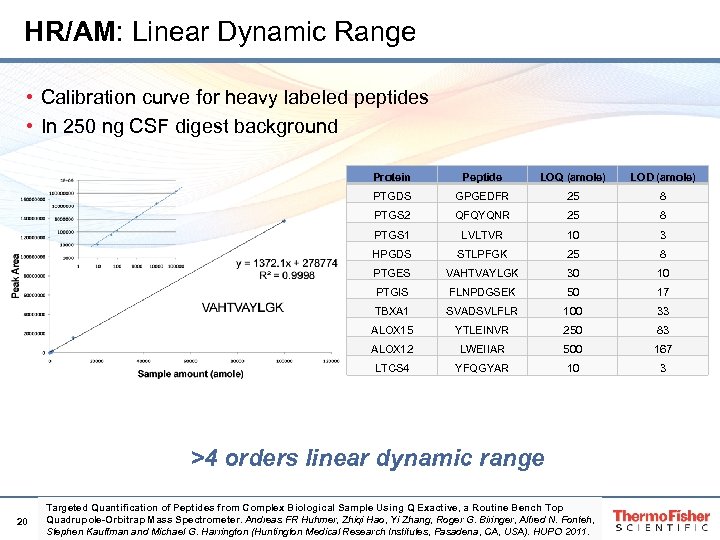 HR/AM: Linear Dynamic Range • Calibration curve for heavy labeled peptides • In 250 ng CSF digest background Protein Peptide LOQ (amole) LOD (amole) PTGDS GPGEDFR 25 8 PTGS 2 QFQYQNR 25 8 PTGS 1 LVLTVR 10 3 HPGDS STLPFGK 25 8 PTGES VAHTVAYLGK 30 10 PTGIS FLNPDGSEK 50 17 TBXA 1 SVADSVLFLR 100 33 ALOX 15 YTLEINVR 250 83 ALOX 12 LWEIIAR 500 167 LTCS 4 YFQGYAR 10 3 >4 orders linear dynamic range 20 Targeted Quantification of Peptides from Complex Biological Sample Using Q Exactive, a Routine Bench Top Quadrupole-Orbitrap Mass Spectrometer. Andreas FR Huhmer, Zhiqi Hao, Yi Zhang, Roger G. Biringer, Alfred N. Fonteh, Stephen Kauffman and Michael G. Harrington (Huntington Medical Research Institutes, Pasadena, CA, USA). HUPO 2011.
HR/AM: Linear Dynamic Range • Calibration curve for heavy labeled peptides • In 250 ng CSF digest background Protein Peptide LOQ (amole) LOD (amole) PTGDS GPGEDFR 25 8 PTGS 2 QFQYQNR 25 8 PTGS 1 LVLTVR 10 3 HPGDS STLPFGK 25 8 PTGES VAHTVAYLGK 30 10 PTGIS FLNPDGSEK 50 17 TBXA 1 SVADSVLFLR 100 33 ALOX 15 YTLEINVR 250 83 ALOX 12 LWEIIAR 500 167 LTCS 4 YFQGYAR 10 3 >4 orders linear dynamic range 20 Targeted Quantification of Peptides from Complex Biological Sample Using Q Exactive, a Routine Bench Top Quadrupole-Orbitrap Mass Spectrometer. Andreas FR Huhmer, Zhiqi Hao, Yi Zhang, Roger G. Biringer, Alfred N. Fonteh, Stephen Kauffman and Michael G. Harrington (Huntington Medical Research Institutes, Pasadena, CA, USA). HUPO 2011.
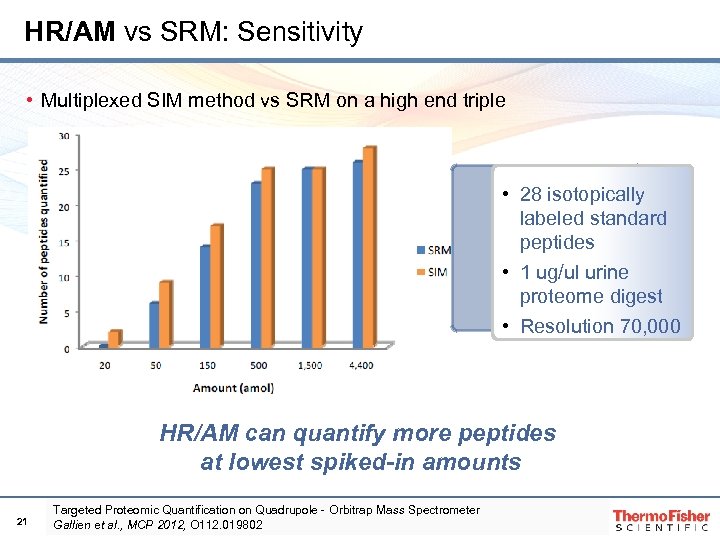 HR/AM vs SRM: Sensitivity • Multiplexed SIM method vs SRM on a high end triple • 28 isotopically labeled standard peptides • 1 ug/ul urine proteome digest • Resolution 70, 000 HR/AM can quantify more peptides at lowest spiked-in amounts 21 Targeted Proteomic Quantification on Quadrupole‐Orbitrap Mass Spectrometer Gallien et al. , MCP 2012, O 112. 019802
HR/AM vs SRM: Sensitivity • Multiplexed SIM method vs SRM on a high end triple • 28 isotopically labeled standard peptides • 1 ug/ul urine proteome digest • Resolution 70, 000 HR/AM can quantify more peptides at lowest spiked-in amounts 21 Targeted Proteomic Quantification on Quadrupole‐Orbitrap Mass Spectrometer Gallien et al. , MCP 2012, O 112. 019802
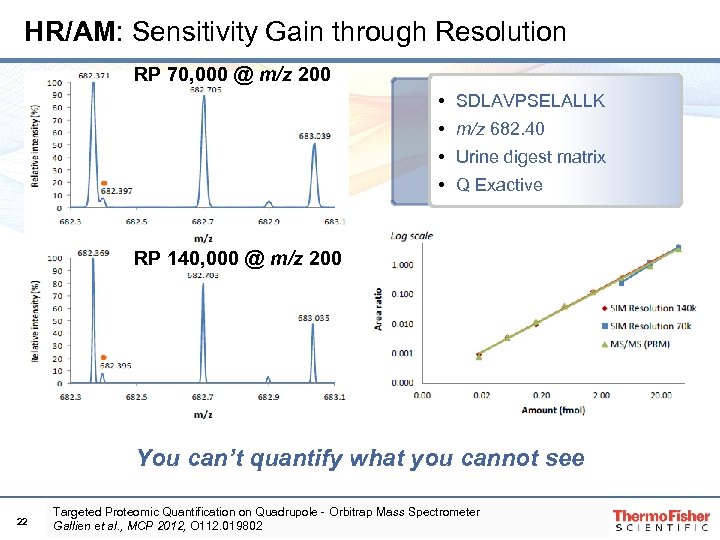 HR/AM: Sensitivity Gain through Resolution RP 70, 000 @ m/z 200 • SDLAVPSELALLK • m/z 682. 40 • Urine digest matrix • Q Exactive RP 140, 000 @ m/z 200 You can’t quantify what you cannot see 22 Targeted Proteomic Quantification on Quadrupole‐Orbitrap Mass Spectrometer Gallien et al. , MCP 2012, O 112. 019802
HR/AM: Sensitivity Gain through Resolution RP 70, 000 @ m/z 200 • SDLAVPSELALLK • m/z 682. 40 • Urine digest matrix • Q Exactive RP 140, 000 @ m/z 200 You can’t quantify what you cannot see 22 Targeted Proteomic Quantification on Quadrupole‐Orbitrap Mass Spectrometer Gallien et al. , MCP 2012, O 112. 019802
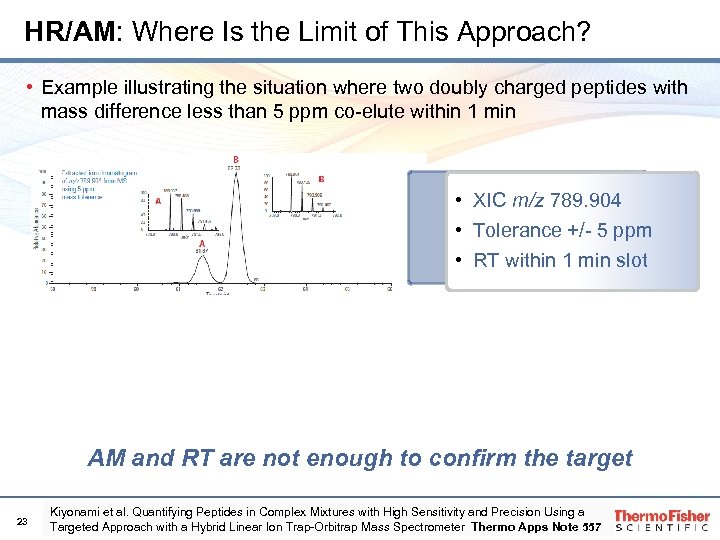 HR/AM: Where Is the Limit of This Approach? • Example illustrating the situation where two doubly charged peptides with mass difference less than 5 ppm co-elute within 1 min • XIC m/z 789. 904 • Tolerance +/- 5 ppm • RT within 1 min slot AM and RT are not enough to confirm the target 23 Kiyonami et al. Quantifying Peptides in Complex Mixtures with High Sensitivity and Precision Using a Targeted Approach with a Hybrid Linear Ion Trap-Orbitrap Mass Spectrometer Thermo Apps Note 557
HR/AM: Where Is the Limit of This Approach? • Example illustrating the situation where two doubly charged peptides with mass difference less than 5 ppm co-elute within 1 min • XIC m/z 789. 904 • Tolerance +/- 5 ppm • RT within 1 min slot AM and RT are not enough to confirm the target 23 Kiyonami et al. Quantifying Peptides in Complex Mixtures with High Sensitivity and Precision Using a Targeted Approach with a Hybrid Linear Ion Trap-Orbitrap Mass Spectrometer Thermo Apps Note 557
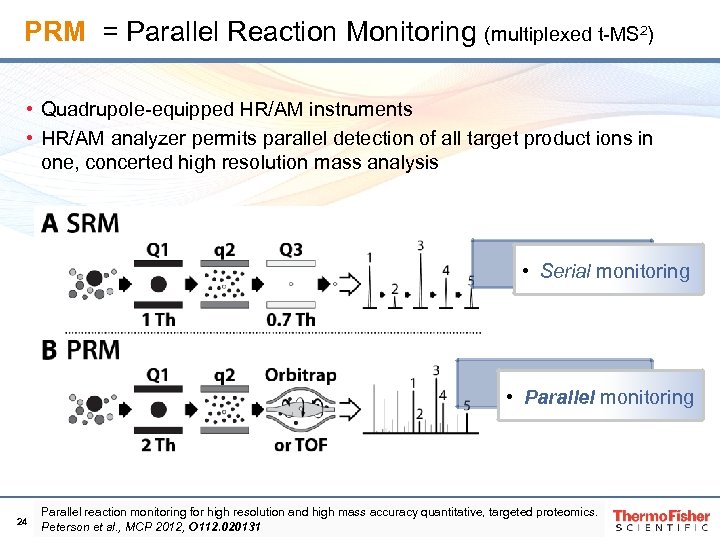 PRM = Parallel Reaction Monitoring (multiplexed t-MS 2) • Quadrupole-equipped HR/AM instruments • HR/AM analyzer permits parallel detection of all target product ions in one, concerted high resolution mass analysis • Serial monitoring • Parallel monitoring 24 Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Peterson et al. , MCP 2012, O 112. 020131
PRM = Parallel Reaction Monitoring (multiplexed t-MS 2) • Quadrupole-equipped HR/AM instruments • HR/AM analyzer permits parallel detection of all target product ions in one, concerted high resolution mass analysis • Serial monitoring • Parallel monitoring 24 Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Peterson et al. , MCP 2012, O 112. 020131
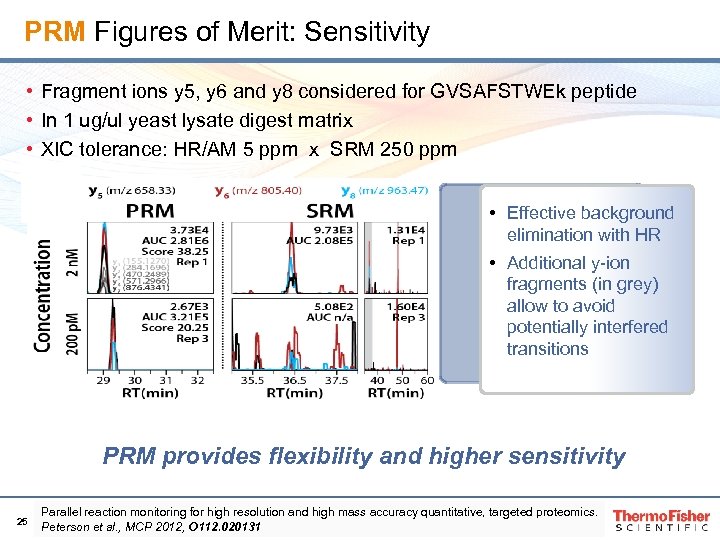 PRM Figures of Merit: Sensitivity • Fragment ions y 5, y 6 and y 8 considered for GVSAFSTWEk peptide • In 1 ug/ul yeast lysate digest matrix • XIC tolerance: HR/AM 5 ppm x SRM 250 ppm • Effective background elimination with HR • Additional y-ion fragments (in grey) allow to avoid potentially interfered transitions PRM provides flexibility and higher sensitivity 25 Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Peterson et al. , MCP 2012, O 112. 020131
PRM Figures of Merit: Sensitivity • Fragment ions y 5, y 6 and y 8 considered for GVSAFSTWEk peptide • In 1 ug/ul yeast lysate digest matrix • XIC tolerance: HR/AM 5 ppm x SRM 250 ppm • Effective background elimination with HR • Additional y-ion fragments (in grey) allow to avoid potentially interfered transitions PRM provides flexibility and higher sensitivity 25 Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Peterson et al. , MCP 2012, O 112. 020131
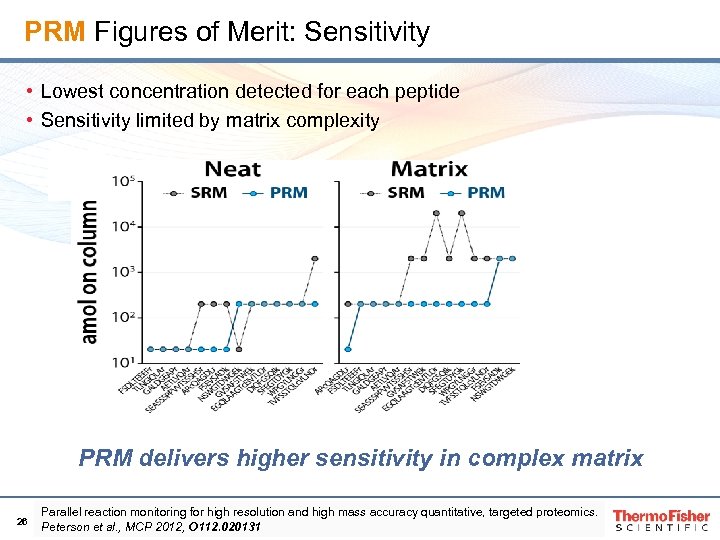 PRM Figures of Merit: Sensitivity • Lowest concentration detected for each peptide • Sensitivity limited by matrix complexity PRM delivers higher sensitivity in complex matrix 26 Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Peterson et al. , MCP 2012, O 112. 020131
PRM Figures of Merit: Sensitivity • Lowest concentration detected for each peptide • Sensitivity limited by matrix complexity PRM delivers higher sensitivity in complex matrix 26 Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Peterson et al. , MCP 2012, O 112. 020131
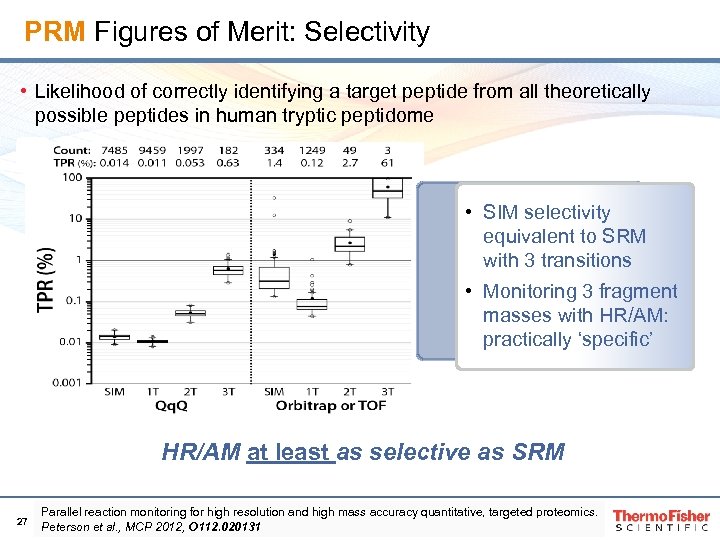 PRM Figures of Merit: Selectivity • Likelihood of correctly identifying a target peptide from all theoretically possible peptides in human tryptic peptidome • SIM selectivity equivalent to SRM with 3 transitions • Monitoring 3 fragment masses with HR/AM: practically ‘specific’ HR/AM at least as selective as SRM 27 Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Peterson et al. , MCP 2012, O 112. 020131
PRM Figures of Merit: Selectivity • Likelihood of correctly identifying a target peptide from all theoretically possible peptides in human tryptic peptidome • SIM selectivity equivalent to SRM with 3 transitions • Monitoring 3 fragment masses with HR/AM: practically ‘specific’ HR/AM at least as selective as SRM 27 Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Peterson et al. , MCP 2012, O 112. 020131
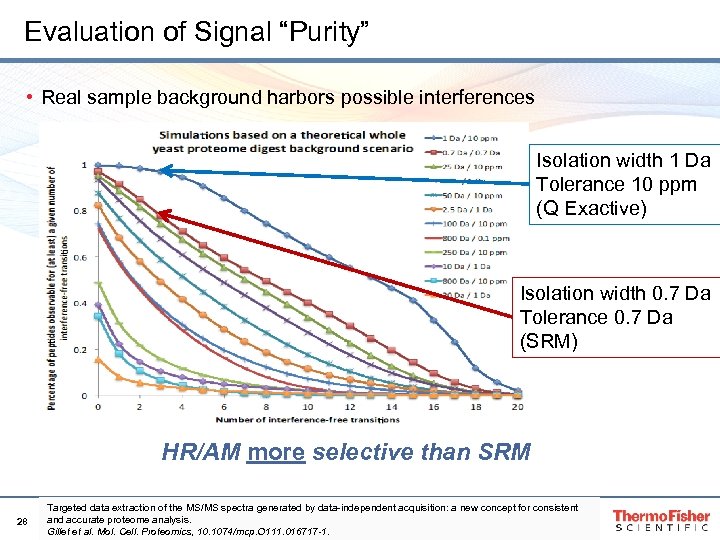 Evaluation of Signal “Purity” • Real sample background harbors possible interferences Isolation width 1 Da Tolerance 10 ppm (Q Exactive) Isolation width 0. 7 Da Tolerance 0. 7 Da (SRM) HR/AM more selective than SRM 28 Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Gillet et al. Mol. Cell. Proteomics, 10. 1074/mcp. O 111. 016717 -1.
Evaluation of Signal “Purity” • Real sample background harbors possible interferences Isolation width 1 Da Tolerance 10 ppm (Q Exactive) Isolation width 0. 7 Da Tolerance 0. 7 Da (SRM) HR/AM more selective than SRM 28 Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Gillet et al. Mol. Cell. Proteomics, 10. 1074/mcp. O 111. 016717 -1.
 PRM Summary • Sensitivity better than SRM • Selectivity higher than SRM • Dynamic range wider than SRM • Linearity equal to SRM • Minimal upfront method development • Facile and highly flexible automated data analysis 29
PRM Summary • Sensitivity better than SRM • Selectivity higher than SRM • Dynamic range wider than SRM • Linearity equal to SRM • Minimal upfront method development • Facile and highly flexible automated data analysis 29
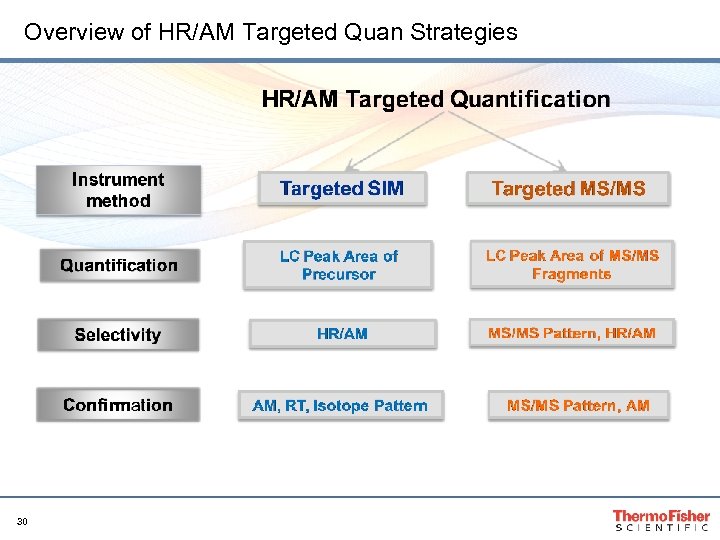 Overview of HR/AM Targeted Quan Strategies 30
Overview of HR/AM Targeted Quan Strategies 30
 Conclusions • HR/AM as a powerful solution for biomarker research: • Mainstream biomarker discovery tool in the form of Data Dependent Acquisition, DDA • Efficient way to reduce number of initial candidates engaging Accurate Inclusion Mass Screening strategy, AIMS, or datadirected strategies • Highly sensitive and selective targeted quantitation in complex matrices with confident confirmation of the target peptide by Parallel Reaction Monitoring, PRM Simultaneous ID, confirmation and quantitation with LODs at low amol/column level 31
Conclusions • HR/AM as a powerful solution for biomarker research: • Mainstream biomarker discovery tool in the form of Data Dependent Acquisition, DDA • Efficient way to reduce number of initial candidates engaging Accurate Inclusion Mass Screening strategy, AIMS, or datadirected strategies • Highly sensitive and selective targeted quantitation in complex matrices with confident confirmation of the target peptide by Parallel Reaction Monitoring, PRM Simultaneous ID, confirmation and quantitation with LODs at low amol/column level 31
 Why DIA Method? • Data-independent acquisition methods provide: • High throughput comprehensive quantification acquisition methods with qualitative confirmation. This is a method with very high multiplexing capabilities. • Data-independent acquisition strategy seeks to increase reproducibility and comprehensiveness of data collection. • Very simple and universal acquisition set-up. No requirement for detailed sample knowledge prior to the DIA-based analysis. • Data collection produce a complete record of quantitative data and a targeted data analysis strategy can be employed to mine additional analytes, retrospectively. • Limitations • Quantitative performances could be severely limited on very complex mixtures due to interfering background ions. • Not suitable for fast UHPLC separation. • DIA methods can not use conventional database search engines for data processing as the link between the precursor mass and the fragment ions is not recorded. A reference spectral library is needed. • Not as sensitive as targeted acquisition (SRM or HR/AM) approach. 32
Why DIA Method? • Data-independent acquisition methods provide: • High throughput comprehensive quantification acquisition methods with qualitative confirmation. This is a method with very high multiplexing capabilities. • Data-independent acquisition strategy seeks to increase reproducibility and comprehensiveness of data collection. • Very simple and universal acquisition set-up. No requirement for detailed sample knowledge prior to the DIA-based analysis. • Data collection produce a complete record of quantitative data and a targeted data analysis strategy can be employed to mine additional analytes, retrospectively. • Limitations • Quantitative performances could be severely limited on very complex mixtures due to interfering background ions. • Not suitable for fast UHPLC separation. • DIA methods can not use conventional database search engines for data processing as the link between the precursor mass and the fragment ions is not recorded. A reference spectral library is needed. • Not as sensitive as targeted acquisition (SRM or HR/AM) approach. 32
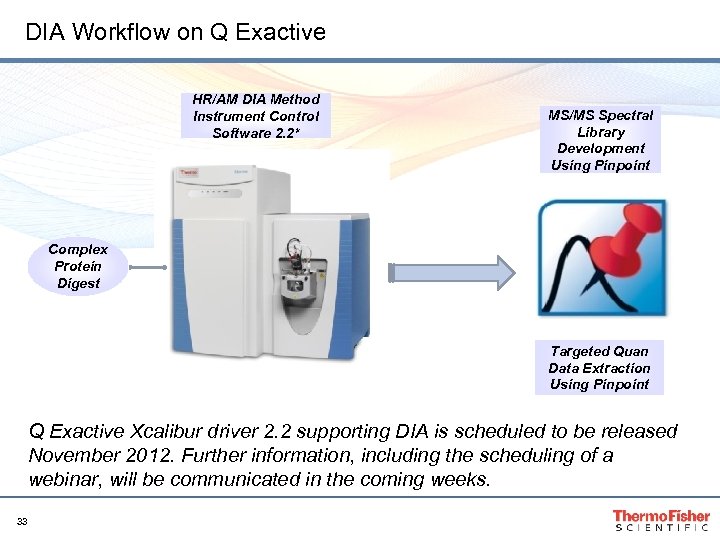 DIA Workflow on Q Exactive HR/AM DIA Method Instrument Control Software 2. 2* MS/MS Spectral Library Development Using Pinpoint Complex Protein Digest Targeted Quan Data Extraction Using Pinpoint Q Exactive Xcalibur driver 2. 2 supporting DIA is scheduled to be released November 2012. Further information, including the scheduling of a webinar, will be communicated in the coming weeks. 33
DIA Workflow on Q Exactive HR/AM DIA Method Instrument Control Software 2. 2* MS/MS Spectral Library Development Using Pinpoint Complex Protein Digest Targeted Quan Data Extraction Using Pinpoint Q Exactive Xcalibur driver 2. 2 supporting DIA is scheduled to be released November 2012. Further information, including the scheduling of a webinar, will be communicated in the coming weeks. 33
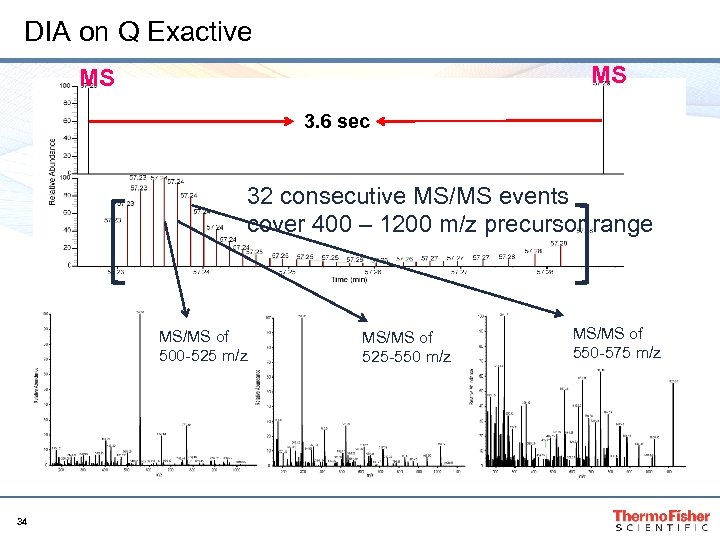 DIA on Q Exactive MS MS 3. 6 sec 32 consecutive MS/MS events cover 400 – 1200 m/z precursor range MS/MS of 500 -525 m/z 34 MS/MS of 525 -550 m/z MS/MS of 550 -575 m/z
DIA on Q Exactive MS MS 3. 6 sec 32 consecutive MS/MS events cover 400 – 1200 m/z precursor range MS/MS of 500 -525 m/z 34 MS/MS of 525 -550 m/z MS/MS of 550 -575 m/z
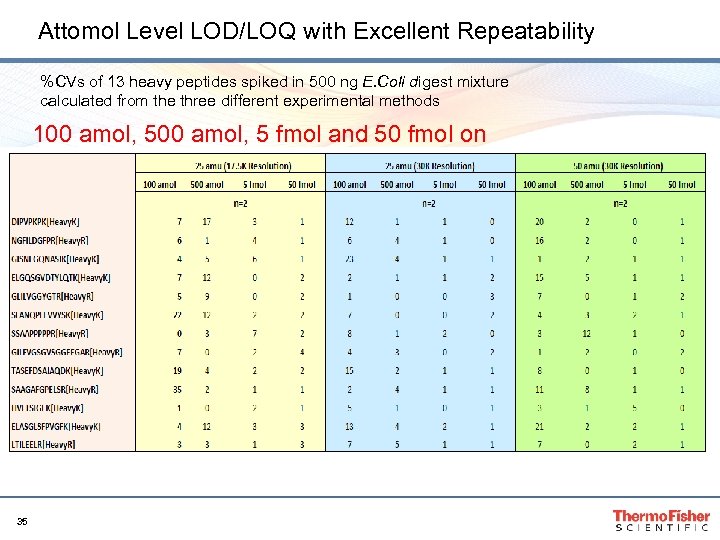 Attomol Level LOD/LOQ with Excellent Repeatability %CVs of 13 heavy peptides spiked in 500 ng E. Coli digest mixture calculated from the three different experimental methods 100 amol, 5 fmol and 50 fmol on column 35
Attomol Level LOD/LOQ with Excellent Repeatability %CVs of 13 heavy peptides spiked in 500 ng E. Coli digest mixture calculated from the three different experimental methods 100 amol, 5 fmol and 50 fmol on column 35
 Conclusions • Q Exactive is a very powerful discovery instrument • Q Exactive is a one-solution platform with equally strong capabilities for discovery as well as targeted quantification experiments • Q Exactive supports the generation of high quality, high resolution, accurate mass spectral libraries that are necessary to succeed with DIA strategies. • With new instrument control software 2. 2*, Q Exactive enables dataindependent data acquisition with its unique capability to use higher resolution set up for more accurate quantitative results and the flexibility to use different combinations of mass range and resolution, to adapt to different research needs. • Most importantly, the Q Exactive already provides equal or better targeted protein quantification performance compared to SRM with its three powerful quantification methods (MS 1, t-SIM, t-HCD, including multiplexing). 36
Conclusions • Q Exactive is a very powerful discovery instrument • Q Exactive is a one-solution platform with equally strong capabilities for discovery as well as targeted quantification experiments • Q Exactive supports the generation of high quality, high resolution, accurate mass spectral libraries that are necessary to succeed with DIA strategies. • With new instrument control software 2. 2*, Q Exactive enables dataindependent data acquisition with its unique capability to use higher resolution set up for more accurate quantitative results and the flexibility to use different combinations of mass range and resolution, to adapt to different research needs. • Most importantly, the Q Exactive already provides equal or better targeted protein quantification performance compared to SRM with its three powerful quantification methods (MS 1, t-SIM, t-HCD, including multiplexing). 36
 Conclusions on Targeted Peptide Quantitation • The greatest challenge to targeted peptide quantitation is background signal • Reduction in background signal through increased chromatographic or sample preparation methods can significantly extend quantitative capabilities. • HR/AM shows tremendous promise to complement Qq. Q experiments • HR/AM-driven selectivity is an essential tool to cope with abundant matrix signal • Q Exactive hardware facilitates a wide range of data acquisition schemes – all of which incorporate HR/AM • Continued development of better chromatography enables more robust targeted quantitation • Qual aspects are automatically built into data acquisition and processing methods to help determine background interference • Pinpoint software ties all aspects of targeted peptide quantitation together 37
Conclusions on Targeted Peptide Quantitation • The greatest challenge to targeted peptide quantitation is background signal • Reduction in background signal through increased chromatographic or sample preparation methods can significantly extend quantitative capabilities. • HR/AM shows tremendous promise to complement Qq. Q experiments • HR/AM-driven selectivity is an essential tool to cope with abundant matrix signal • Q Exactive hardware facilitates a wide range of data acquisition schemes – all of which incorporate HR/AM • Continued development of better chromatography enables more robust targeted quantitation • Qual aspects are automatically built into data acquisition and processing methods to help determine background interference • Pinpoint software ties all aspects of targeted peptide quantitation together 37
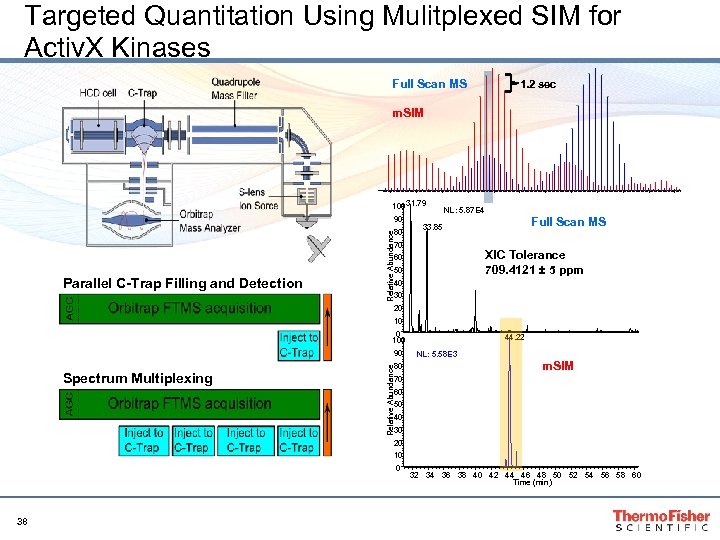 Targeted Quantitation Using Mulitplexed SIM for Activ. X Kinases Full Scan MS 1. 2 sec m. SIM 100 31. 79 90 Relative Abundance 80 NL: 5. 87 E 4 70 XIC Tolerance 709. 4121 ± 5 ppm 60 50 Parallel C-Trap Filling and Detection Full Scan MS 33. 85 40 30 20 100 90 Relative Abundance Spectrum Multiplexing 80 44. 22 NL: 5. 58 E 3 m. SIM 70 60 50 40 30 20 10 0 38 32 34 36 38 40 42 44 46 48 50 52 54 56 58 60 Time (min)
Targeted Quantitation Using Mulitplexed SIM for Activ. X Kinases Full Scan MS 1. 2 sec m. SIM 100 31. 79 90 Relative Abundance 80 NL: 5. 87 E 4 70 XIC Tolerance 709. 4121 ± 5 ppm 60 50 Parallel C-Trap Filling and Detection Full Scan MS 33. 85 40 30 20 100 90 Relative Abundance Spectrum Multiplexing 80 44. 22 NL: 5. 58 E 3 m. SIM 70 60 50 40 30 20 10 0 38 32 34 36 38 40 42 44 46 48 50 52 54 56 58 60 Time (min)
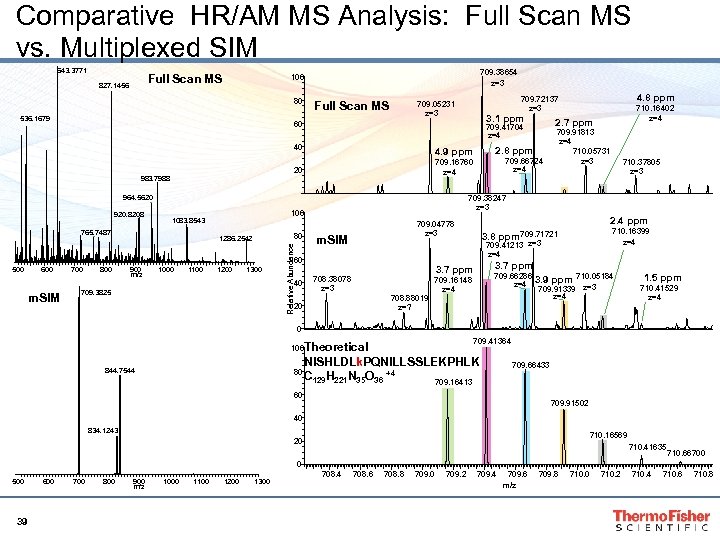 Comparative HR/AM MS Analysis: Full Scan MS vs. Multiplexed SIM 643. 3771 Full Scan MS 827. 1456 709. 38654 z=3 100 80 536. 1679 Full Scan MS 100 1083. 8543 1286. 2542 Relative Abundance 80 m. SIM 700 800 900 m/z 1000 1100 1200 1300 40 709. 3825 710. 37805 z=3 2. 4 ppm 709. 04778 z=3 m. SIM 60 600 709. 91813 z=4 710. 05731 z=3 709. 38247 z=3 964. 5620 765. 7487 709. 66724 z=4 709. 16760 z=4 983. 7988 920. 8208 2. 8 ppm 4. 9 ppm 710. 16402 z=4 2. 7 ppm 709. 41704 z=4 20 500 3. 1 ppm 60 40 4. 8 ppm 709. 72137 z=3 709. 05231 z=3 3. 7 ppm 708. 38078 z=3 708. 88019 z=? 20 710. 16399 z=4 3. 8 ppm 709. 71721 709. 41213 z=4 709. 66286 z=4 709. 16148 z=4 710. 05184 709. 91339 z=3 z=4 3. 9 ppm 1. 5 ppm 710. 41529 z=4 0 709. 41364 100 Theoretical 844. 7544 80 NISHLDLk. PQNILLSSLEKPHLK C 129 H 221 N 35 O 36 +4 709. 16413 709. 66433 60 709. 91502 40 834. 1243 710. 16569 20 710. 41635 710. 66700 0 500 39 600 700 800 900 m/z 1000 1100 1200 1300 708. 4 708. 6 708. 8 709. 0 709. 2 709. 4 709. 6 m/z 709. 8 710. 0 710. 2 710. 4 710. 6 710. 8
Comparative HR/AM MS Analysis: Full Scan MS vs. Multiplexed SIM 643. 3771 Full Scan MS 827. 1456 709. 38654 z=3 100 80 536. 1679 Full Scan MS 100 1083. 8543 1286. 2542 Relative Abundance 80 m. SIM 700 800 900 m/z 1000 1100 1200 1300 40 709. 3825 710. 37805 z=3 2. 4 ppm 709. 04778 z=3 m. SIM 60 600 709. 91813 z=4 710. 05731 z=3 709. 38247 z=3 964. 5620 765. 7487 709. 66724 z=4 709. 16760 z=4 983. 7988 920. 8208 2. 8 ppm 4. 9 ppm 710. 16402 z=4 2. 7 ppm 709. 41704 z=4 20 500 3. 1 ppm 60 40 4. 8 ppm 709. 72137 z=3 709. 05231 z=3 3. 7 ppm 708. 38078 z=3 708. 88019 z=? 20 710. 16399 z=4 3. 8 ppm 709. 71721 709. 41213 z=4 709. 66286 z=4 709. 16148 z=4 710. 05184 709. 91339 z=3 z=4 3. 9 ppm 1. 5 ppm 710. 41529 z=4 0 709. 41364 100 Theoretical 844. 7544 80 NISHLDLk. PQNILLSSLEKPHLK C 129 H 221 N 35 O 36 +4 709. 16413 709. 66433 60 709. 91502 40 834. 1243 710. 16569 20 710. 41635 710. 66700 0 500 39 600 700 800 900 m/z 1000 1100 1200 1300 708. 4 708. 6 708. 8 709. 0 709. 2 709. 4 709. 6 m/z 709. 8 710. 0 710. 2 710. 4 710. 6 710. 8
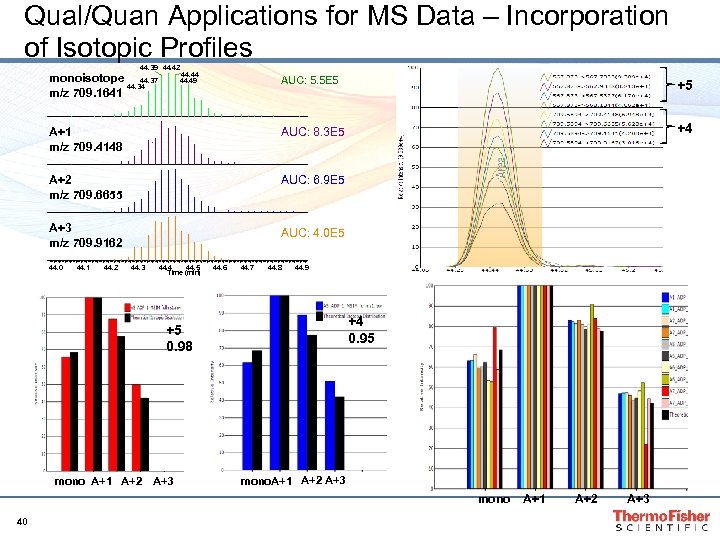 Qual/Quan Applications for MS Data – Incorporation of Isotopic Profiles 44. 39 44. 42 44. 44 44. 49 monoisotope 44. 37 44. 34 m/z 709. 1641 AUC: 5. 5 E 5 +5 A+1 m/z 709. 4148 AUC: 8. 3 E 5 +4 A+2 m/z 709. 6655 AUC: 6. 9 E 5 A+3 m/z 709. 9162 AUC: 4. 0 E 5 44. 0 44. 1 44. 2 44. 3 44. 4 44. 5 Time (min) 44. 6 44. 7 44. 8 44. 9 +4 0. 95 +5 0. 98 mono A+1 A+2 A+3 mono 40 A+1 A+2 A+3
Qual/Quan Applications for MS Data – Incorporation of Isotopic Profiles 44. 39 44. 42 44. 44 44. 49 monoisotope 44. 37 44. 34 m/z 709. 1641 AUC: 5. 5 E 5 +5 A+1 m/z 709. 4148 AUC: 8. 3 E 5 +4 A+2 m/z 709. 6655 AUC: 6. 9 E 5 A+3 m/z 709. 9162 AUC: 4. 0 E 5 44. 0 44. 1 44. 2 44. 3 44. 4 44. 5 Time (min) 44. 6 44. 7 44. 8 44. 9 +4 0. 95 +5 0. 98 mono A+1 A+2 A+3 mono 40 A+1 A+2 A+3
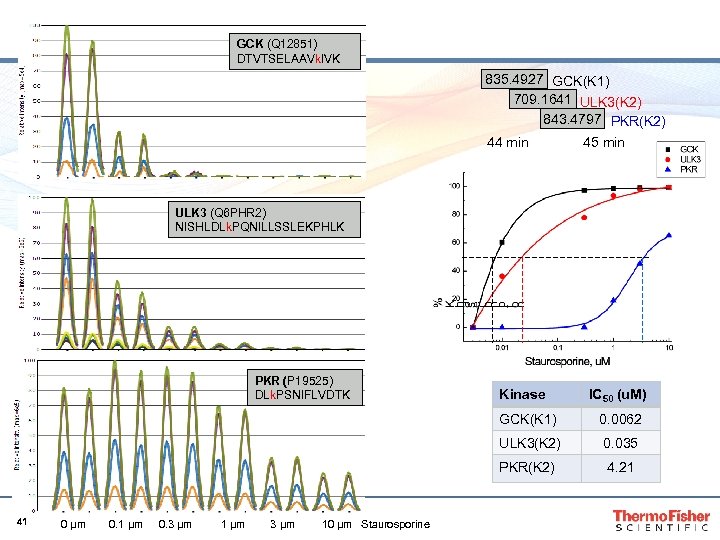 GCK (Q 12851) DTVTSELAAVk. IVK 835. 4927 GCK(K 1) 709. 1641 ULK 3(K 2) 843. 4797 PKR(K 2) 44 min 45 min ULK 3 (Q 6 PHR 2) NISHLDLk. PQNILLSSLEKPHLK PKR (P 19525) DLk. PSNIFLVDTK Kinase IC 50 (u. M) GCK(K 1) ULK 3(K 2) 0 µm 0. 1 µm 0. 3 µm 1 µm 3 µm 10 µm Staurosporine 0. 035 PKR(K 2) 41 0. 0062 4. 21
GCK (Q 12851) DTVTSELAAVk. IVK 835. 4927 GCK(K 1) 709. 1641 ULK 3(K 2) 843. 4797 PKR(K 2) 44 min 45 min ULK 3 (Q 6 PHR 2) NISHLDLk. PQNILLSSLEKPHLK PKR (P 19525) DLk. PSNIFLVDTK Kinase IC 50 (u. M) GCK(K 1) ULK 3(K 2) 0 µm 0. 1 µm 0. 3 µm 1 µm 3 µm 10 µm Staurosporine 0. 035 PKR(K 2) 41 0. 0062 4. 21
 Acknowledgements • Reiko Kiyonami, Yi Zhang, Andreas Huhmer, Yue Xuan, Markus Kellmann, Amol Prakash, Scott Peterman, Gene Ciccimaro, Martin Hornshaw, Alan Atkins, Jenny Ho, Myriam Demant , Michaela Scigelova, Sigrid Baumgarten, Madalina Oppermann and probably many more colleagues in Thermo • Our collaborators and users in EU and World-Wide 42
Acknowledgements • Reiko Kiyonami, Yi Zhang, Andreas Huhmer, Yue Xuan, Markus Kellmann, Amol Prakash, Scott Peterman, Gene Ciccimaro, Martin Hornshaw, Alan Atkins, Jenny Ho, Myriam Demant , Michaela Scigelova, Sigrid Baumgarten, Madalina Oppermann and probably many more colleagues in Thermo • Our collaborators and users in EU and World-Wide 42
 Publications • 1: Gallien S, Duriez E, Crone C, Kellmann M, Moehring T, Domon B. Targeted Proteomic Quantification on Quadrupole-Orbitrap Mass Spectrometer. Mol Cell Proteomics. 2012 Sep 7. [Epub ahead of print] Pub. Med PMID: 22962056. • 2: Peterson AC, Russell JD, Bailey DJ, Westphall MS, Coon JJ. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol Cell Proteomics. 2012 Aug 3. [Epub ahead of print] Pub. Med PMID: 22865924. • 3: Bailey DJ, Rose CM, Mc. Alister GC, Brumbaugh J, Yu P, Wenger CD, Westphall MS, Thomson JA, Coon JJ. Instant spectral assignment for advanced decision tree-driven mass spectrometry. Proc Natl Acad Sci U S A. 2012 May 29; 109(22): 8411 -6. Epub 2012 May 14. Pub. Med PMID: 22586074; Pub. Med Central PMCID: PMC 3365209. • 4: Kelstrup CD, Young C, Lavallee R, Nielsen ML, Olsen JV. Optimized Fast and Sensitive Acquisition Methods for Shotgun Proteomics on a Quadrupole Orbitrap Mass Spectrometer. J Proteome Res. 2012 May 10. [Epub ahead of print] Pub. Med PMID: 22537090. • 5: Deeb SJ, D'Souza RC, Cox J, Schmidt-Supprian M, Mann M. Super-SILAC allows classification of diffuse large B-cell lymphoma subtypes by their protein expression profiles. Mol Cell Proteomics. 2012 May; 11(5): 77 -89. Epub 2012 Mar 21. Pub. Med PMID: 22442255; Pub. Med Central PMCID: PMC 3418848. • 6: Huang X, Tian C, Liu M, Wang Y, Tolmachev AV, Sharma S, Yu F, Fu K, Zheng J, Ding SJ. Quantitative proteomic analysis of mouse embryonic fibroblasts and induced pluripotent stem cells using 16 O/18 O labeling. J Proteome Res. 2012 Apr 6; 11(4): 2091 -102. Epub 2012 Mar 15. Pub. Med PMID: 22375802. • 7: Nagaraj N, Kulak NA, Cox J, Neuhauser N, Mayr K, Hoerning O, Vorm O, Mann M. System-wide perturbation analysis with nearly complete coverage of the yeast proteome by single-shot ultra HPLC runs on a bench top Orbitrap. Mol Cell Proteomics. 2012 Mar; 11(3): M 111. 013722. Epub 2011 Oct 20. Pub. Med PMID: 22021278; Pub. Med Central PMCID: PMC 3316726. • 8: Michalski A, Damoc E, Hauschild JP, Lange O, Wieghaus A, Makarov A, Nagaraj N, Cox J, Mann M, Horning S. Mass spectrometrybased proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol Cell Proteomics. 2011 Sep; 10(9): M 111. 011015. Epub 2011 Jun 3. Pub. Med PMID: 21642640; Pub. Med Central PMCID: PMC 3284220. 43
Publications • 1: Gallien S, Duriez E, Crone C, Kellmann M, Moehring T, Domon B. Targeted Proteomic Quantification on Quadrupole-Orbitrap Mass Spectrometer. Mol Cell Proteomics. 2012 Sep 7. [Epub ahead of print] Pub. Med PMID: 22962056. • 2: Peterson AC, Russell JD, Bailey DJ, Westphall MS, Coon JJ. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol Cell Proteomics. 2012 Aug 3. [Epub ahead of print] Pub. Med PMID: 22865924. • 3: Bailey DJ, Rose CM, Mc. Alister GC, Brumbaugh J, Yu P, Wenger CD, Westphall MS, Thomson JA, Coon JJ. Instant spectral assignment for advanced decision tree-driven mass spectrometry. Proc Natl Acad Sci U S A. 2012 May 29; 109(22): 8411 -6. Epub 2012 May 14. Pub. Med PMID: 22586074; Pub. Med Central PMCID: PMC 3365209. • 4: Kelstrup CD, Young C, Lavallee R, Nielsen ML, Olsen JV. Optimized Fast and Sensitive Acquisition Methods for Shotgun Proteomics on a Quadrupole Orbitrap Mass Spectrometer. J Proteome Res. 2012 May 10. [Epub ahead of print] Pub. Med PMID: 22537090. • 5: Deeb SJ, D'Souza RC, Cox J, Schmidt-Supprian M, Mann M. Super-SILAC allows classification of diffuse large B-cell lymphoma subtypes by their protein expression profiles. Mol Cell Proteomics. 2012 May; 11(5): 77 -89. Epub 2012 Mar 21. Pub. Med PMID: 22442255; Pub. Med Central PMCID: PMC 3418848. • 6: Huang X, Tian C, Liu M, Wang Y, Tolmachev AV, Sharma S, Yu F, Fu K, Zheng J, Ding SJ. Quantitative proteomic analysis of mouse embryonic fibroblasts and induced pluripotent stem cells using 16 O/18 O labeling. J Proteome Res. 2012 Apr 6; 11(4): 2091 -102. Epub 2012 Mar 15. Pub. Med PMID: 22375802. • 7: Nagaraj N, Kulak NA, Cox J, Neuhauser N, Mayr K, Hoerning O, Vorm O, Mann M. System-wide perturbation analysis with nearly complete coverage of the yeast proteome by single-shot ultra HPLC runs on a bench top Orbitrap. Mol Cell Proteomics. 2012 Mar; 11(3): M 111. 013722. Epub 2011 Oct 20. Pub. Med PMID: 22021278; Pub. Med Central PMCID: PMC 3316726. • 8: Michalski A, Damoc E, Hauschild JP, Lange O, Wieghaus A, Makarov A, Nagaraj N, Cox J, Mann M, Horning S. Mass spectrometrybased proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol Cell Proteomics. 2011 Sep; 10(9): M 111. 011015. Epub 2011 Jun 3. Pub. Med PMID: 21642640; Pub. Med Central PMCID: PMC 3284220. 43


