67ae313196137542beb61d8dd9c767ba.ppt
- Количество слайдов: 64
“Proteomics & Bioinformatics” MBI, Master's Degree Program in Helsinki, Finland Lecture 5 11 May, 2007 Sophia Kossida, BRF, Academy of Athens, Greece Esa Pitkänen, Univeristy of Helsinki, Finland Juho Rousu, University of Helsinki, Finland
Mining proteomes To identify as many components of the proteome as possible Mapping of proteomes of various organisms and tissues Comparison of protein expression levels for the detection of disease biomarkers
How to select proteome? A proteome is defined by the state of the organism, tissue, or cell that produces it. Because these states are constantly changing, so are the proteomes. Example of proteomes: different kind of cells; liver, … extracellular fluids; blood plasma, urine, CSF…
Applications Systems biology - understand cell-pathways, network, and complex interacting. Biological processes - characterize sub-proteomes such as protein complexes, cellular machines, organelles Biomarkers - discovery of disease (serological, urine, other biological fluids) - diagnostics, treat patients, monitor therapies Drug targets - evaluate toxicity & other biological or pharmaceutical parameters associated with drug treatment
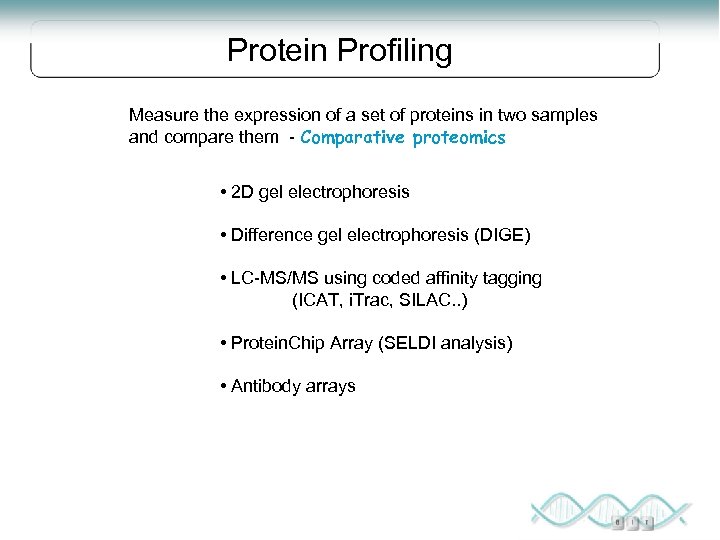
Protein Profiling Measure the expression of a set of proteins in two samples and compare them - Comparative proteomics • 2 D gel electrophoresis • Difference gel electrophoresis (DIGE) • LC-MS/MS using coded affinity tagging (ICAT, i. Trac, SILAC. . ) • Protein. Chip Array (SELDI analysis) • Antibody arrays
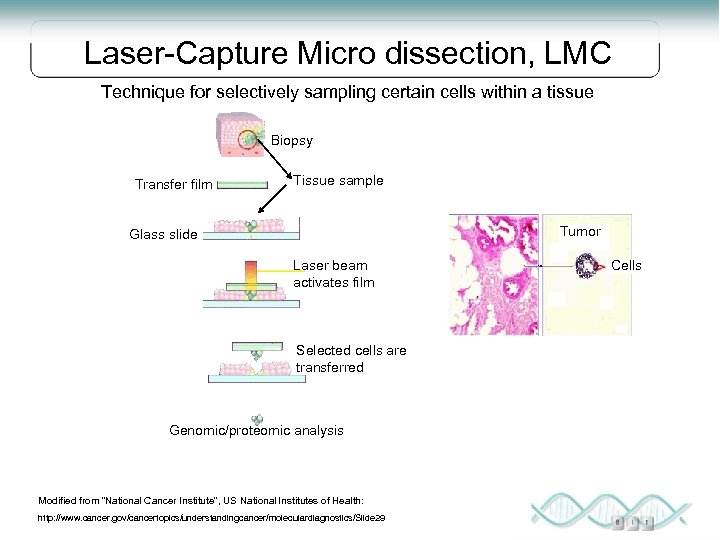
Laser-Capture Micro dissection, LMC Technique for selectively sampling certain cells within a tissue Biopsy Transfer film Tissue sample Tumor Glass slide Laser beam activates film Selected cells are transferred Genomic/proteomic analysis Modified from “National Cancer Institute”, US National Institutes of Health: http: //www. cancer. gov/cancertopics/understandingcancer/moleculardiagnostics/Slide 29 Cells
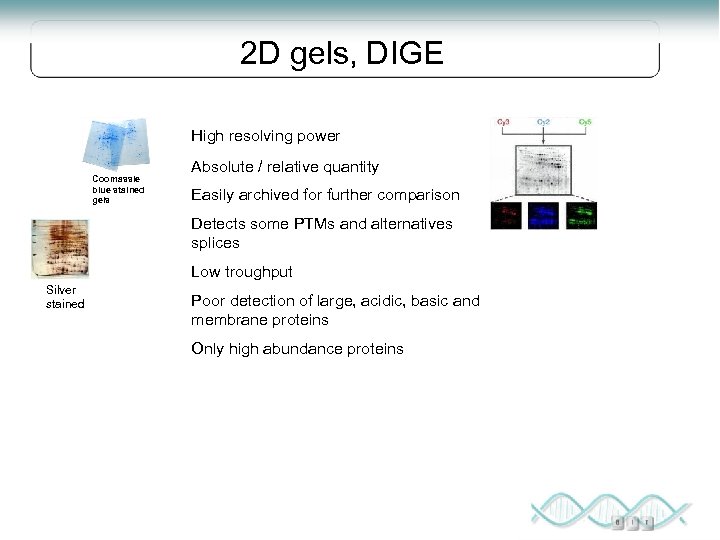
2 D gels, DIGE High resolving power Coomassie blue stained gels Absolute / relative quantity Easily archived for further comparison Detects some PTMs and alternatives splices Low troughput Silver stained Poor detection of large, acidic, basic and membrane proteins Only high abundance proteins
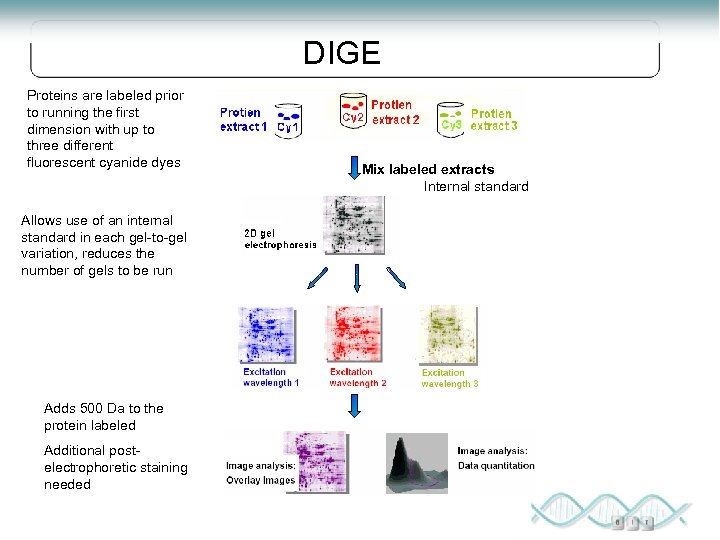
DIGE Proteins are labeled prior to running the first dimension with up to three different fluorescent cyanide dyes Allows use of an internal standard in each gel-to-gel variation, reduces the number of gels to be run Adds 500 Da to the protein labeled Additional postelectrophoretic staining needed Mix labeled extracts Internal standard
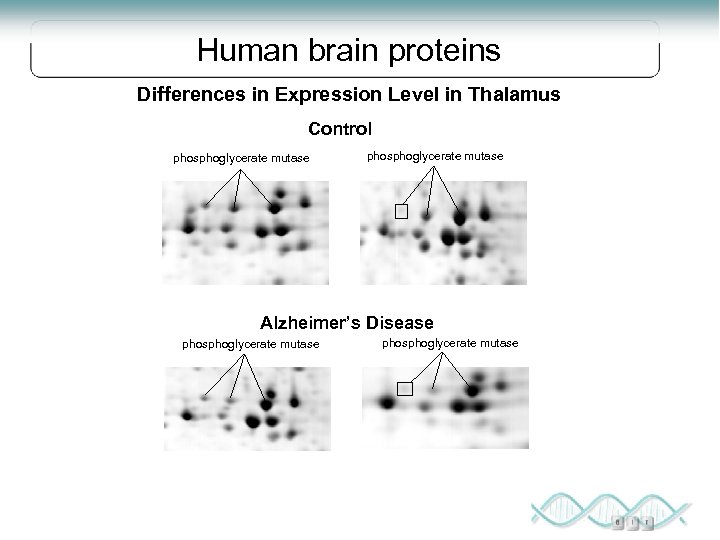
Human brain proteins Differences in Expression Level in Thalamus Control phosphoglycerate mutase Alzheimer’s Disease phosphoglycerate mutase
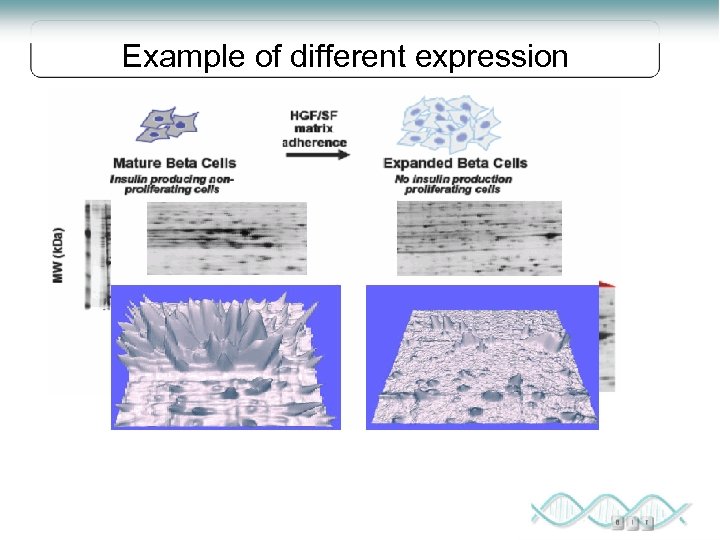
Example of different expression
LC-MS/MS using coded affinity tagging Moderate throughput, but can be automated Detects some low abundance proteins Most isotope label experiments limited to two versions –heavy and light isotope, i. e. binary comparisons only Poor detection of alternative splices and PTMs
Labeling Chemical, ICAT, ITRAQ Chemical modifications to amino acids generally after digestion Most labels differ by 3 -10 Da in mass (not complete / interferences) Compares only 2 -8 samples SILAC Stable isotopes incorporated during cell growth Must be able to grow cells Compares 2 or 3 samples Lys (+8 Da) and Arg (+10 Da) Ion Current No labeling of any kind, See everything in the sample not just what gets labeled Normalization issues, (2 separate runs are compared) Standards needed Robust and many samples and experimental conditions can be compared
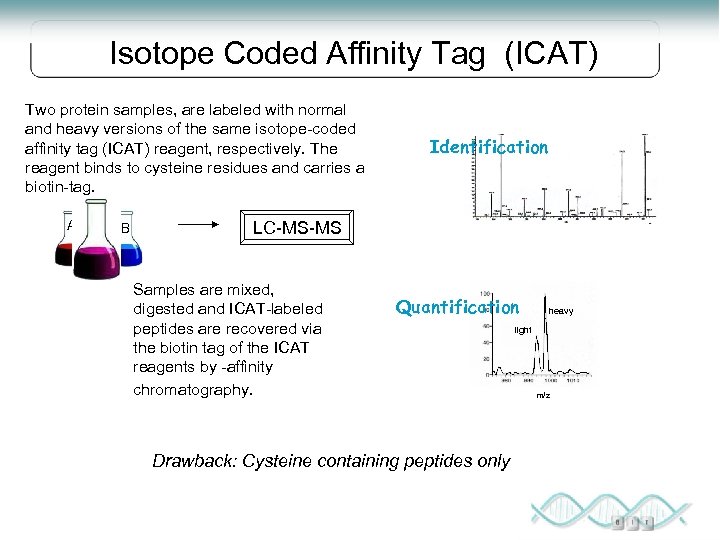
Isotope Coded Affinity Tag (ICAT) Two protein samples, are labeled with normal and heavy versions of the same isotope-coded affinity tag (ICAT) reagent, respectively. The reagent binds to cysteine residues and carries a biotin-tag. A B Identification LC-MS-MS Samples are mixed, digested and ICAT-labeled peptides are recovered via the biotin tag of the ICAT reagents by -affinity chromatography. Quantification Drawback: Cysteine containing peptides only heavy light m/z
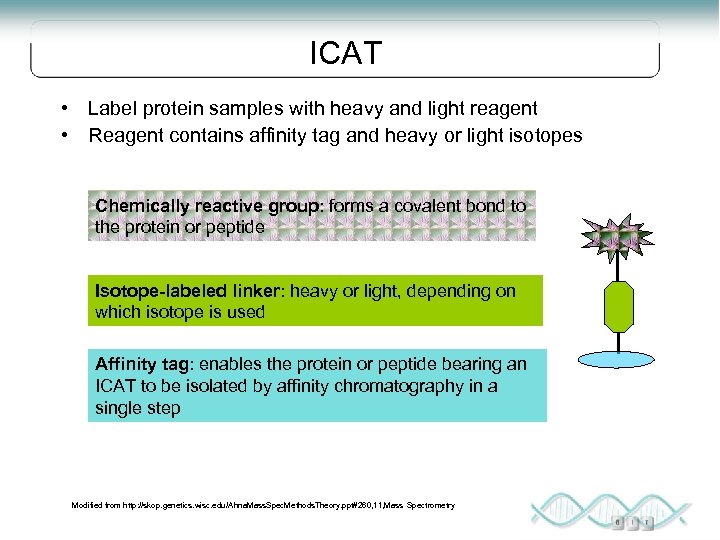
ICAT • Label protein samples with heavy and light reagent • Reagent contains affinity tag and heavy or light isotopes Chemically reactive group: forms a covalent bond to the protein or peptide Isotope-labeled linker: heavy or light, depending on which isotope is used Affinity tag: enables the protein or peptide bearing an ICAT to be isolated by affinity chromatography in a single step Modified from http: //skop. genetics. wisc. edu/Ahna. Mass. Spec. Methods. Theory. ppt#260, 11, Mass Spectrometry
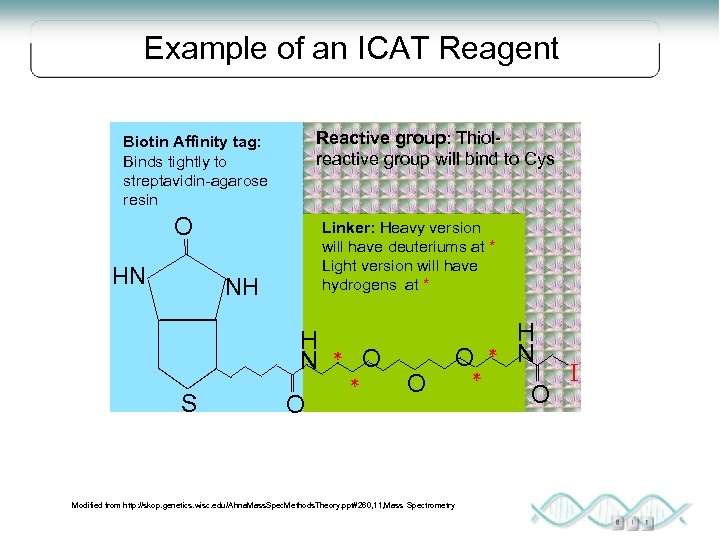
Example of an ICAT Reagent Reactive group: Thiolreactive group will bind to Cys Biotin Affinity tag: Binds tightly to streptavidin-agarose resin O Linker: Heavy version will have deuteriums at * Light version will have hydrogens at * NH NH H N S O O * * O Modified from http: //skop. genetics. wisc. edu/Ahna. Mass. Spec. Methods. Theory. ppt#260, 11, Mass Spectrometry O * * H N O I
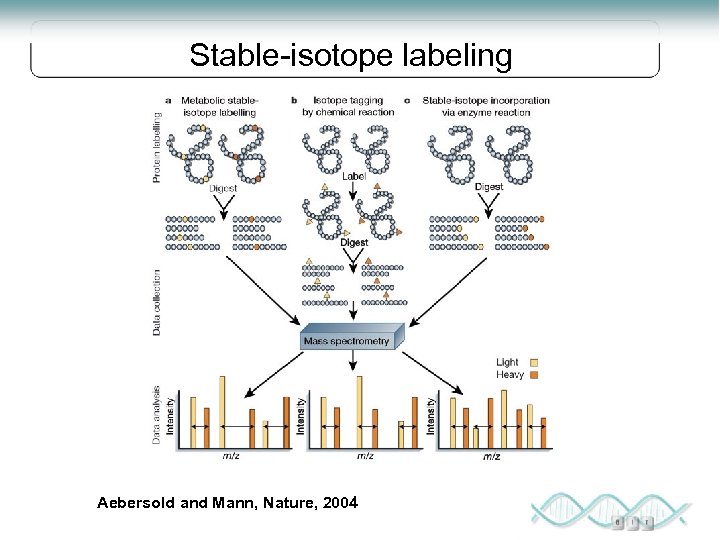
Stable-isotope labeling Aebersold and Mann, Nature, 2004
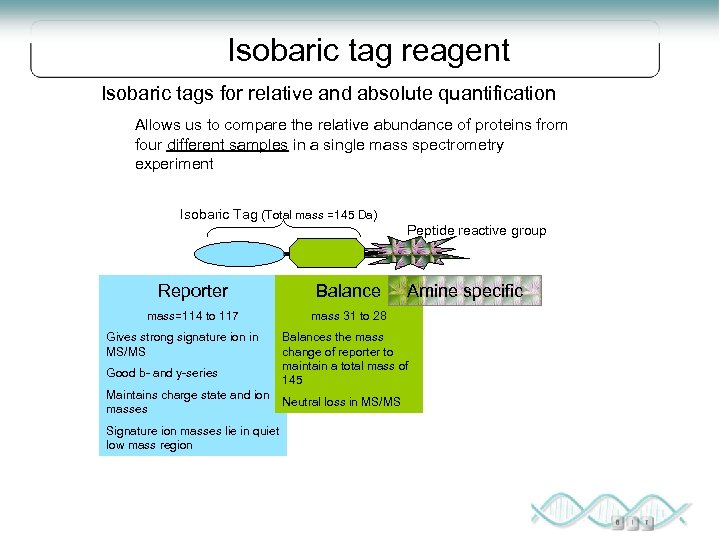
Isobaric tag reagent Isobaric tags for relative and absolute quantification Allows us to compare the relative abundance of proteins from four different samples in a single mass spectrometry experiment Isobaric Tag (Total mass =145 Da) Peptide reactive group Reporter Balance mass=114 to 117 mass 31 to 28 Gives strong signature ion in MS/MS Good b- and y-series Maintains charge state and ion masses Signature ion masses lie in quiet low mass region Amine specific Balances the mass change of reporter to maintain a total mass of 145 Neutral loss in MS/MS
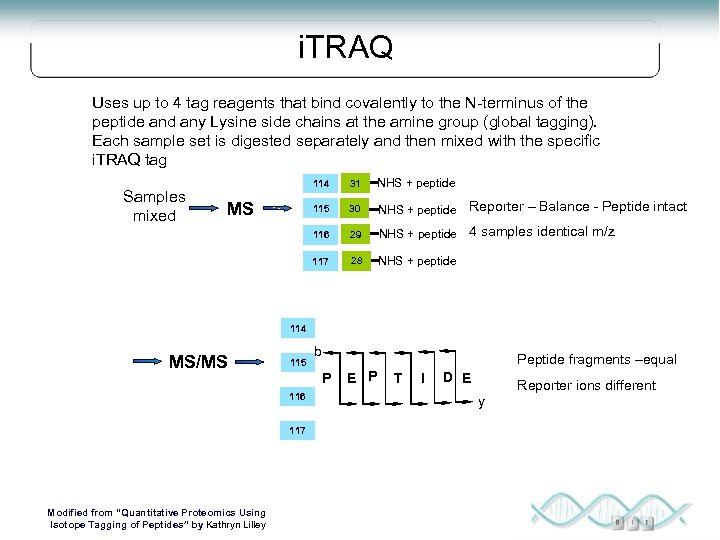
i. TRAQ Uses up to 4 tag reagents that bind covalently to the N-terminus of the peptide and any Lysine side chains at the amine group (global tagging). Each sample set is digested separately and then mixed with the specific i. TRAQ tag NHS + peptide 115 30 NHS + peptide Reporter – Balance - Peptide intact 29 NHS + peptide 4 samples identical m/z 117 MS 31 116 Samples mixed 114 28 NHS + peptide 114 MS/MS 115 Peptide fragments –equal P 116 117 Modified from “Quantitative Proteomics Using Isotope Tagging of Peptides” by Kathryn Lilley b E P T I D E Reporter ions different y
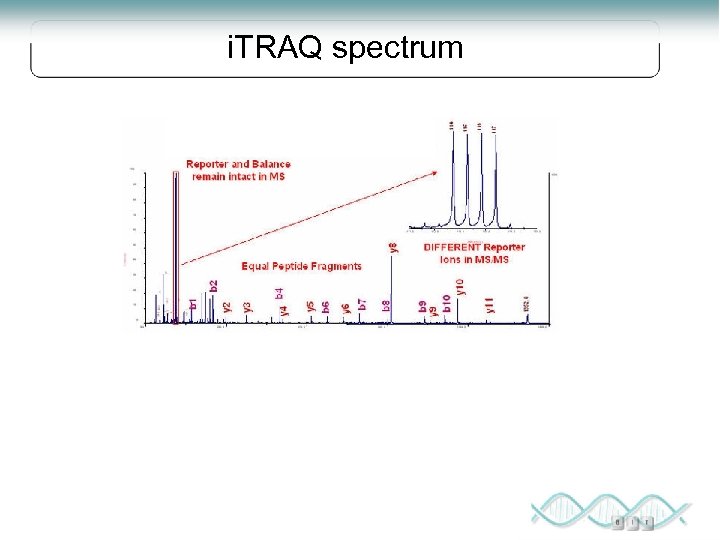
i. TRAQ spectrum
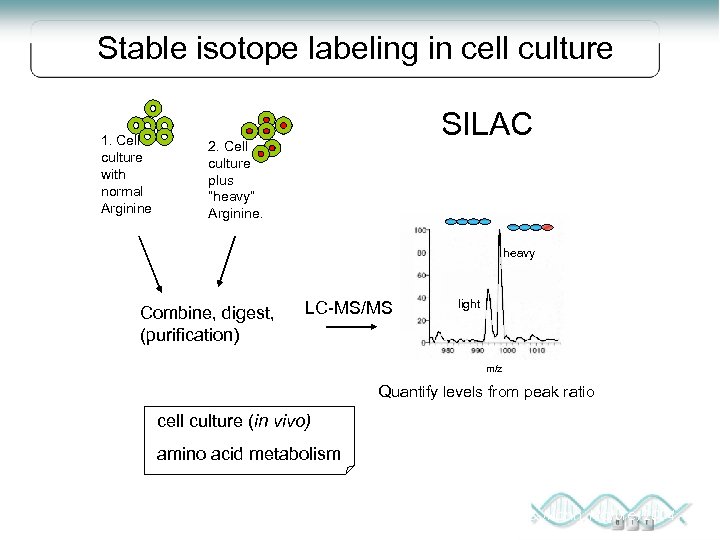
Stable isotope labeling in cell culture 1. Cell culture with normal Arginine SILAC 2. Cell culture plus “heavy” Arginine. heavy Combine, digest, (purification) LC-MS/MS light m/z Quantify levels from peak ratio cell culture (in vivo) amino acid metabolism Steen & Mann, Nature, 2004
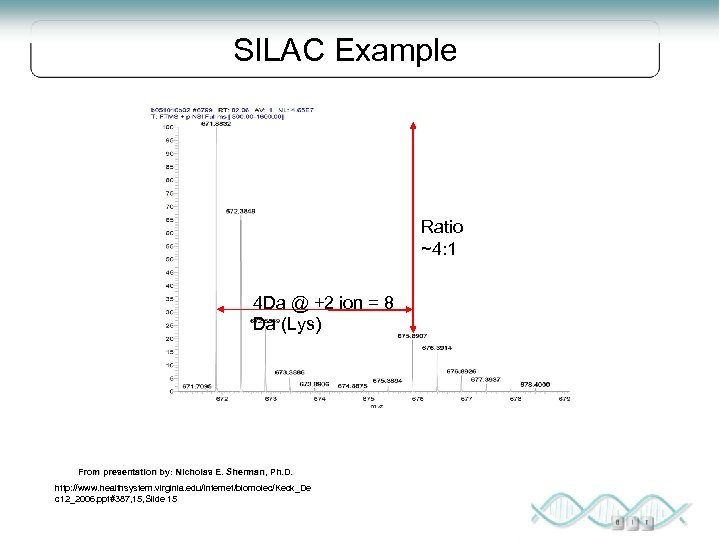
SILAC Example Ratio ~4: 1 4 Da @ +2 ion = 8 Da (Lys) From presentation by: Nicholas E. Sherman, Ph. D. http: //www. healthsystem. virginia. edu/internet/biomolec/Keck_De c 12_2006. ppt#387, 15, Slide 15
SELDI Surface Enhanced Laser Desorption Ionization Ionized proteins are detected and their mass accurately determined by Time-of-Flight Mass Spectrometry High throughput Small amounts of sample More reproducible than 2 DE, but lower resolving power Applied for the analysis of crude samples Process is not standardized
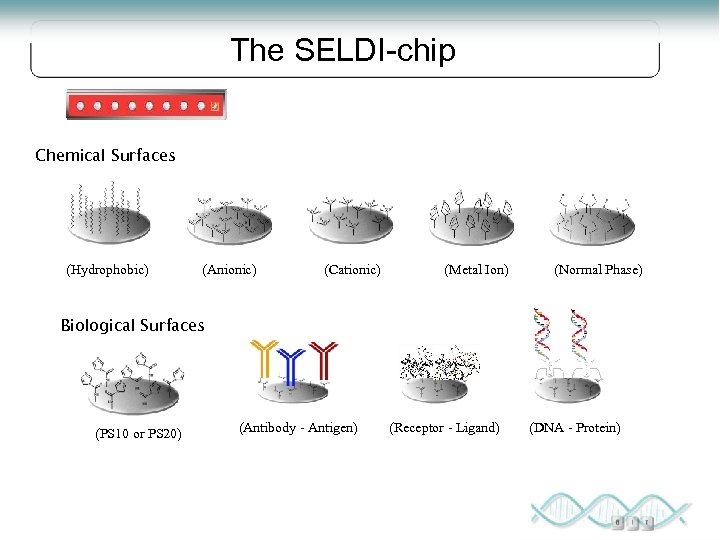
The SELDI-chip Chemical Surfaces (Hydrophobic) (Anionic) (Cationic) (Metal Ion) (Normal Phase) Biological Surfaces (PS 10 or PS 20) (Antibody - Antigen) (Receptor - Ligand) (DNA - Protein)
Antibody arrays Not discovery based Must have 1 or 2 specific high affinity antibodies Very high throughput Can be highly quantitative - relative and absolute Can design reagents to detect PTMs, splice forms
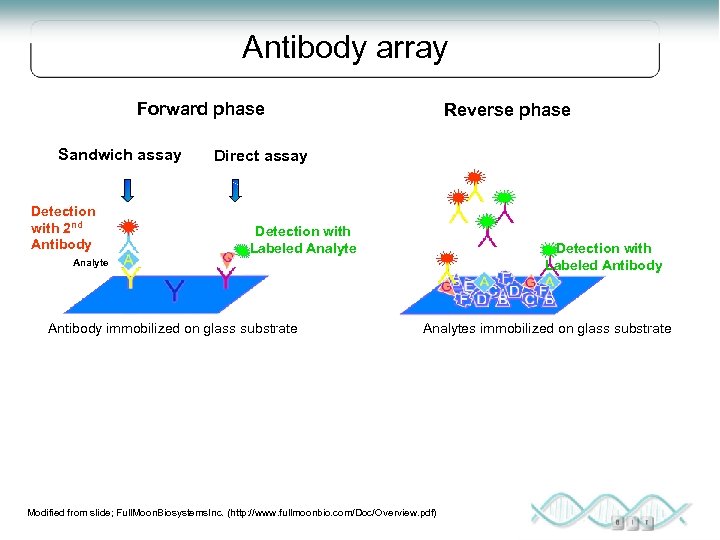
Antibody array Forward phase Sandwich assay Detection with 2 nd Antibody Reverse phase Direct assay Detection with Labeled Analyte Detection with Labeled Antibody Analyte Antibody immobilized on glass substrate Analytes immobilized on glass substrate Modified from slide; Full. Moon. Biosystems. Inc. (http: //www. fullmoonbio. com/Doc/Overview. pdf)
Protein Interactions From single proteins to systems biology
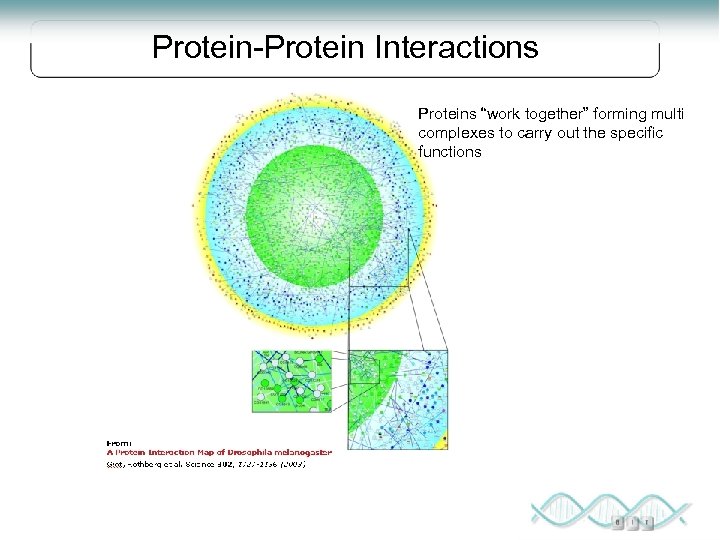
Protein-Protein Interactions Proteins “work together” forming multi complexes to carry out the specific functions
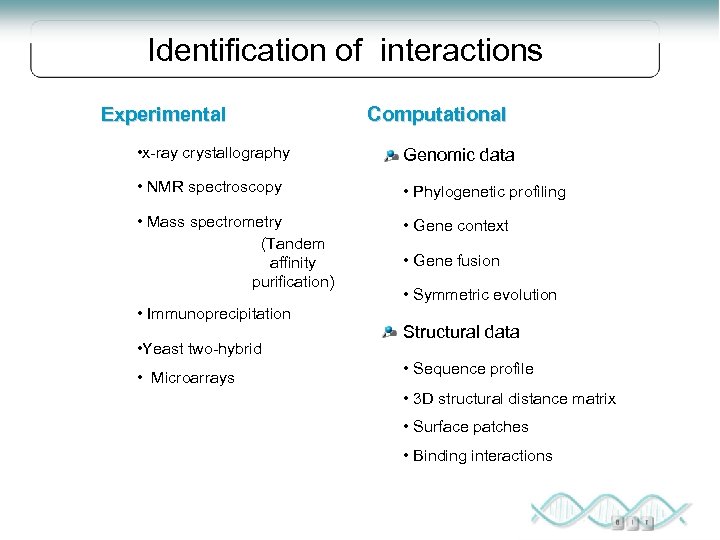
Identification of interactions Experimental Computational • x-ray crystallography Genomic data • NMR spectroscopy • Phylogenetic profiling • Mass spectrometry (Tandem affinity purification) • Gene context • Immunoprecipitation • Yeast two-hybrid • Microarrays • Gene fusion • Symmetric evolution Structural data • Sequence profile • 3 D structural distance matrix • Surface patches • Binding interactions
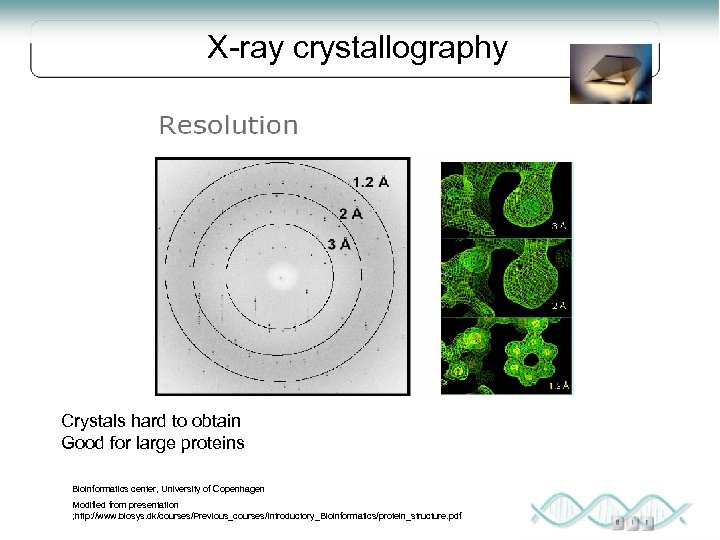
X-ray crystallography Crystals hard to obtain Good for large proteins Bioinformatics center, University of Copenhagen Modified from presentation ; http: //www. biosys. dk/courses/Previous_courses/Introductory_Bioinformatics/protein_structure. pdf
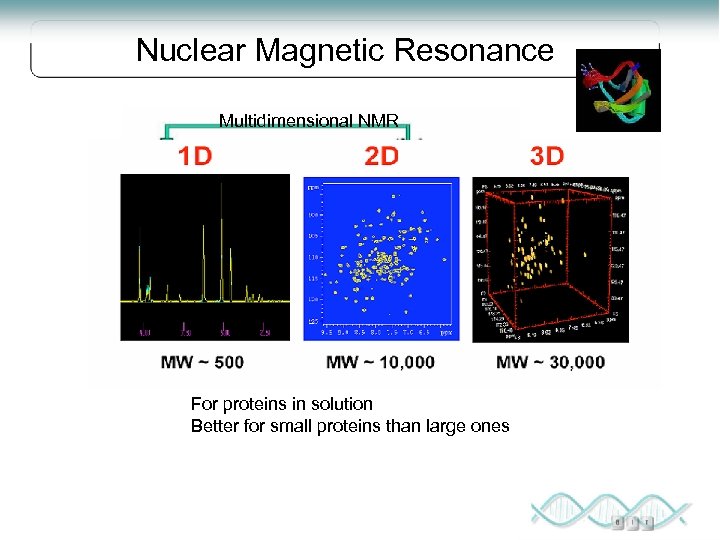
Nuclear Magnetic Resonance Multidimensional NMR Spectroscopy For proteins in solution Better for small proteins than large ones
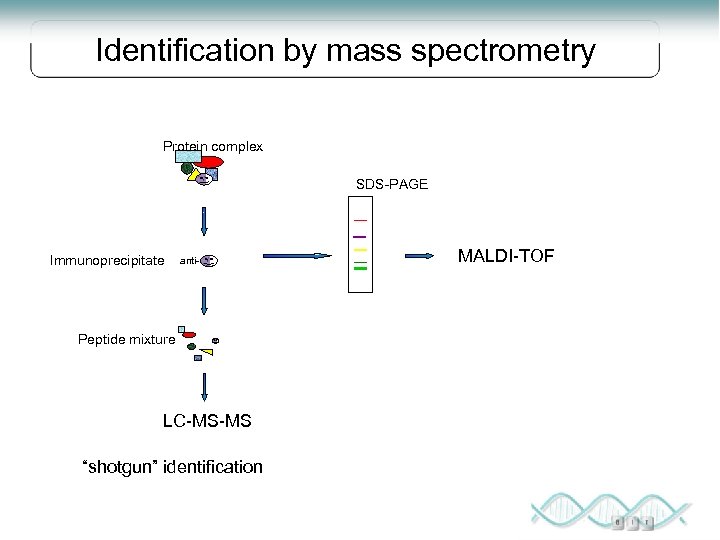
Identification by mass spectrometry Protein complex SDS-PAGE Immunoprecipitate anti- Peptide mixture LC-MS-MS “shotgun” identification MALDI-TOF
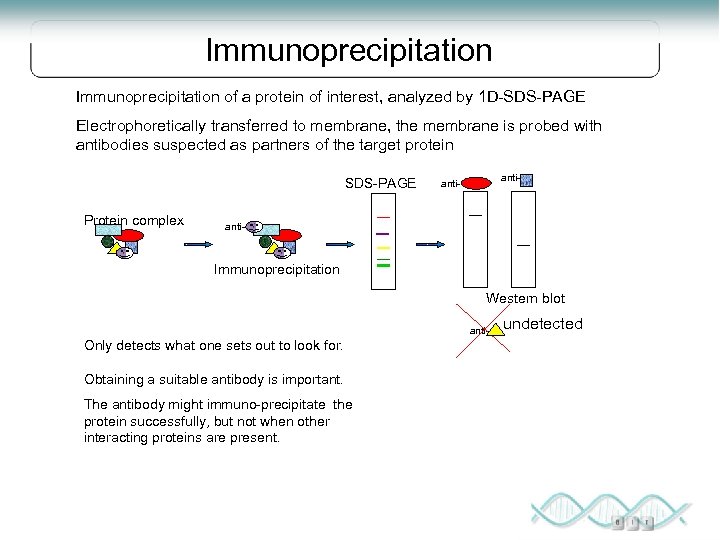
Immunoprecipitation of a protein of interest, analyzed by 1 D-SDS-PAGE Electrophoretically transferred to membrane, the membrane is probed with antibodies suspected as partners of the target protein SDS-PAGE Protein complex anti- Immunoprecipitation Western blot anti- Only detects what one sets out to look for. Obtaining a suitable antibody is important. The antibody might immuno-precipitate the protein successfully, but not when other interacting proteins are present. undetected
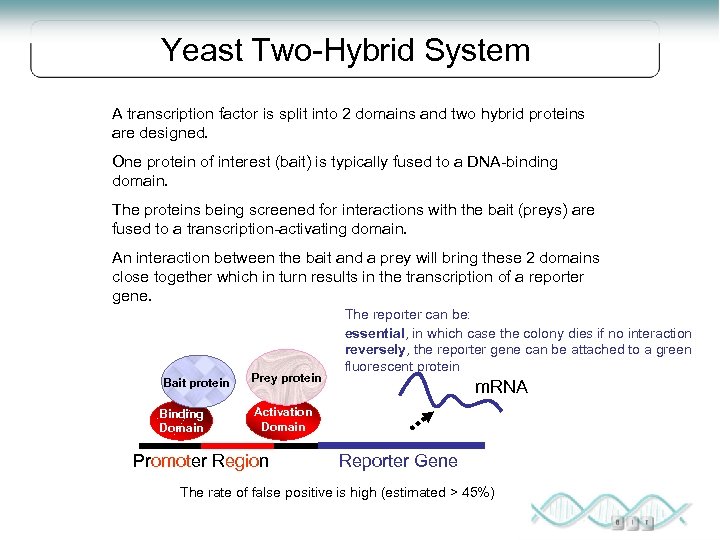
Yeast Two-Hybrid System A transcription factor is split into 2 domains and two hybrid proteins are designed. One protein of interest (bait) is typically fused to a DNA-binding domain. The proteins being screened for interactions with the bait (preys) are fused to a transcription-activating domain. An interaction between the bait and a prey will bring these 2 domains close together which in turn results in the transcription of a reporter gene. Bait protein Binding Domain Prey protein The reporter can be: essential, in which case the colony dies if no interaction reversely, the reporter gene can be attached to a green fluorescent protein m. RNA Activation Domain Promoter Region Reporter Gene The rate of false positive is high (estimated > 45%)
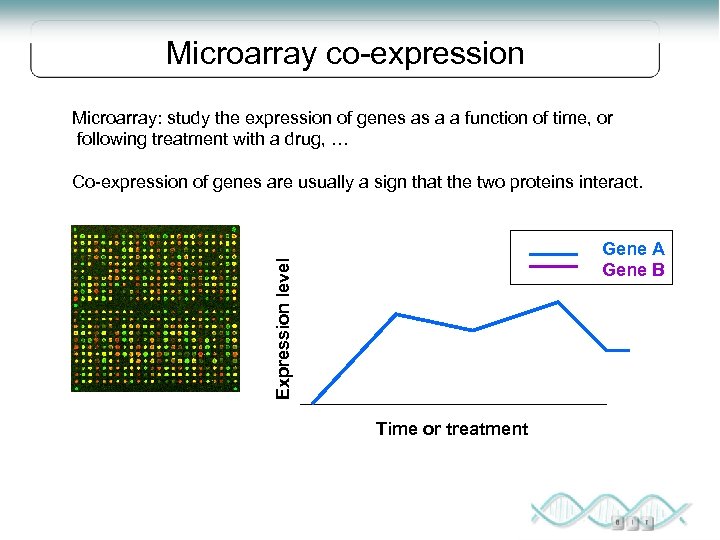
Microarray co-expression Microarray: study the expression of genes as a a function of time, or following treatment with a drug, … Co-expression of genes are usually a sign that the two proteins interact. Expression level Gene A Gene B Time or treatment
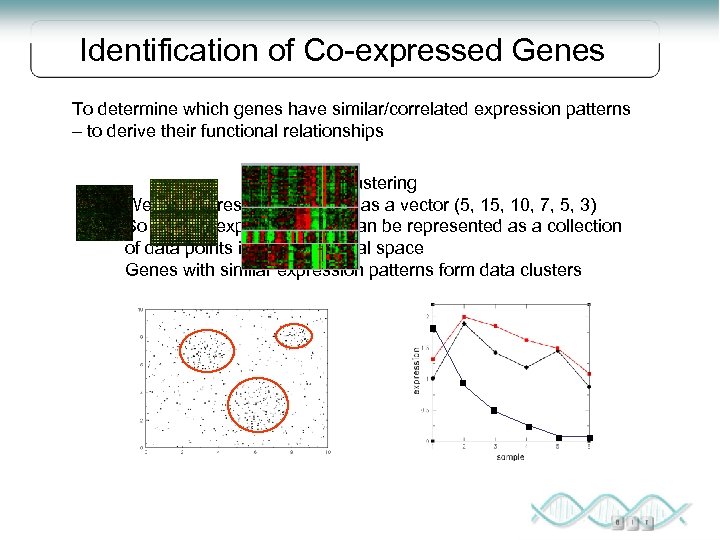
Identification of Co-expressed Genes To determine which genes have similar/correlated expression patterns – to derive their functional relationships Data clustering We can represent each gene as a vector (5, 10, 7, 5, 3) So a set of expression data can be represented as a collection of data points in K-dimensional space Genes with similar expression patterns form data clusters
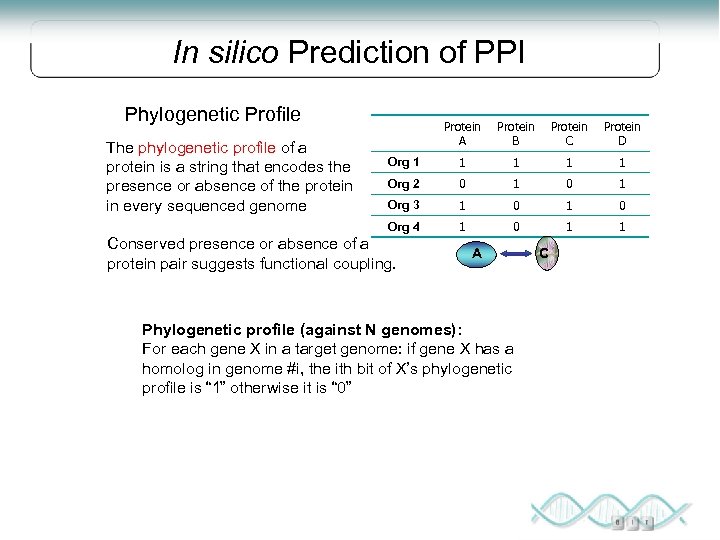
In silico Prediction of PPI Phylogenetic Profile Protein B Protein C Protein D Org 1 1 1 Org 2 0 1 Org 3 1 0 Org 4 The phylogenetic profile of a protein is a string that encodes the presence or absence of the protein in every sequenced genome Protein A 1 0 1 1 Conserved presence or absence of a protein pair suggests functional coupling. A Phylogenetic profile (against N genomes): For each gene X in a target genome: if gene X has a homolog in genome #i, the ith bit of X’s phylogenetic profile is “ 1” otherwise it is “ 0” C
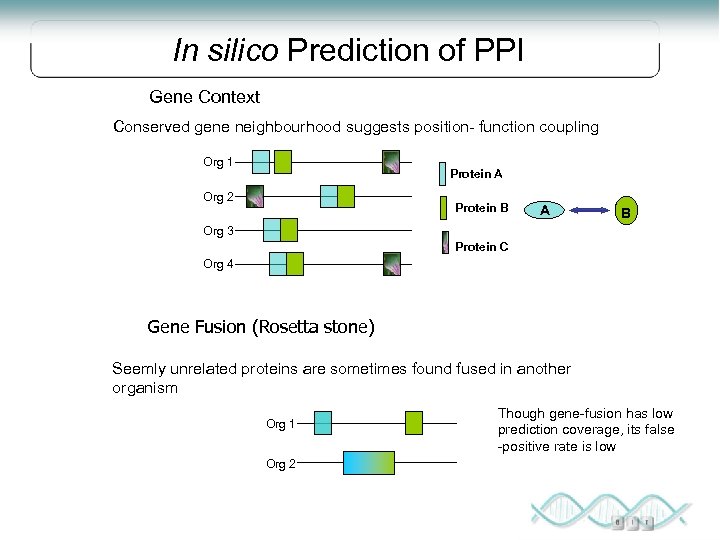
In silico Prediction of PPI Gene Context Conserved gene neighbourhood suggests position- function coupling Org 1 Protein A Org 2 Protein B A B Org 3 Protein C Org 4 Gene Fusion (Rosetta stone) Seemly unrelated proteins are sometimes found fused in another organism Org 1 Org 2 Though gene-fusion has low prediction coverage, its false -positive rate is low
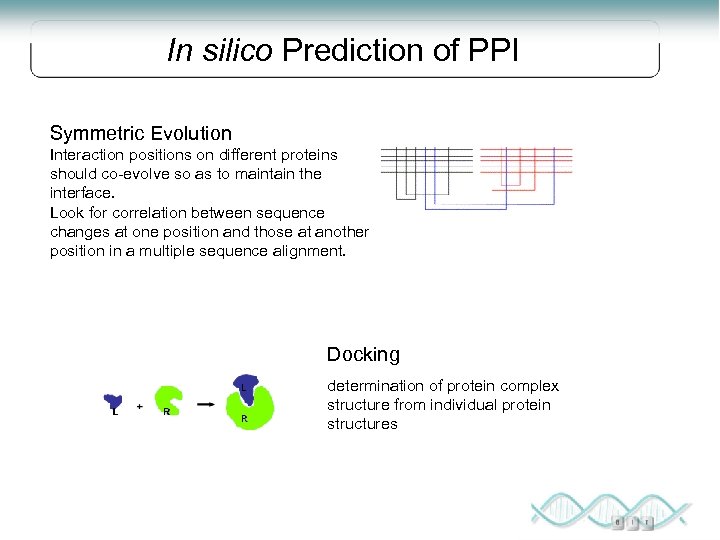
In silico Prediction of PPI Symmetric Evolution Interaction positions on different proteins should co-evolve so as to maintain the interface. Look for correlation between sequence changes at one position and those at another position in a multiple sequence alignment. Docking determination of protein complex structure from individual protein structures
Structure- and interaction databases STRING (EMBL) BOND (Unleashed Informatics) DIP (UCLA) i. HOP
STRING http: //string. embl. de
Biomolecular Object Network Databank BOND http: //bond. unleashedinformatics. com
Database of Interacting Proteins The DIP database catalogs experimentally determined interactions between proteins. It combines information from a variety of sources to create a single, consistent set of protein interactions. http: //dip. doe-mbi. ucla. edu/
ihop http: //www. ihop-net. org/Uni. Pub/i. HOP/
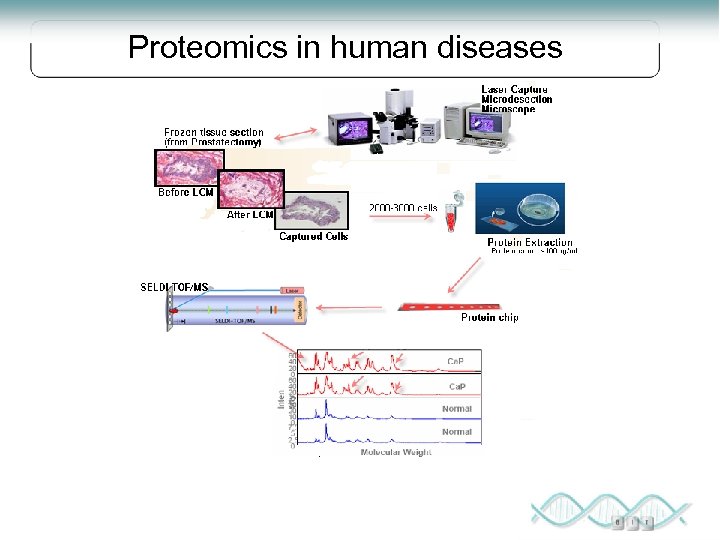
Proteomics in human diseases
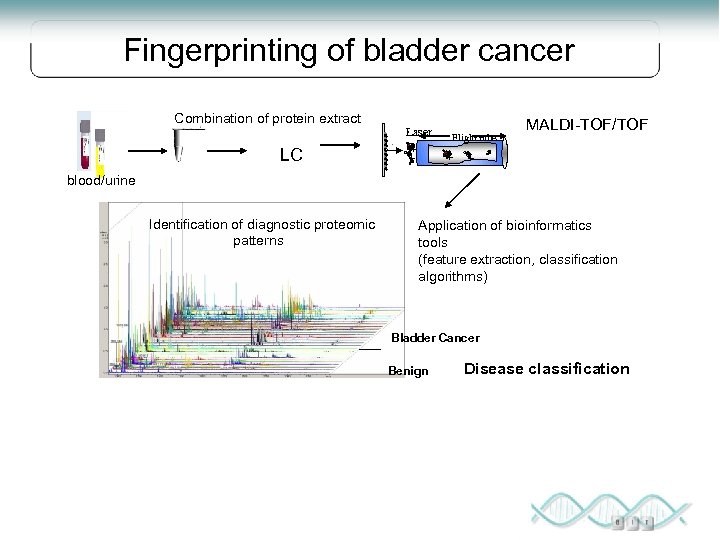
Fingerprinting of bladder cancer Combination of protein extract LC Laser + + Flight tube + MALDI-TOF/TOF + blood/urine Identification of diagnostic proteomic patterns Application of bioinformatics tools (feature extraction, classification algorithms) Bladder Cancer Benign Disease classification
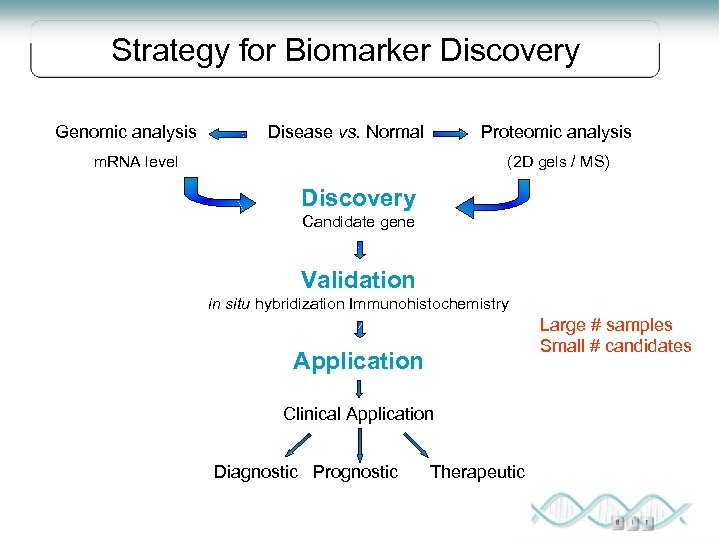
Strategy for Biomarker Discovery Genomic analysis Disease vs. Normal Proteomic analysis m. RNA level (2 D gels / MS) Discovery Candidate gene Validation in situ hybridization Immunohistochemistry Large # samples Small # candidates Application Clinical Application Diagnostic Prognostic Therapeutic
Proteins as biomarkers The protein composition may be associated with disease processes in the organism and thus have potential utility as diagnostic markers. Proteins are closer to the actual disease process, in most cases, than parent genes Proteins are ultimate regulators of cellular function Most cancer markers are proteins The vast majority of drug targets are proteins Individual biomarkers are not sufficient for accurate disease detection Panel of biomarkers should be established
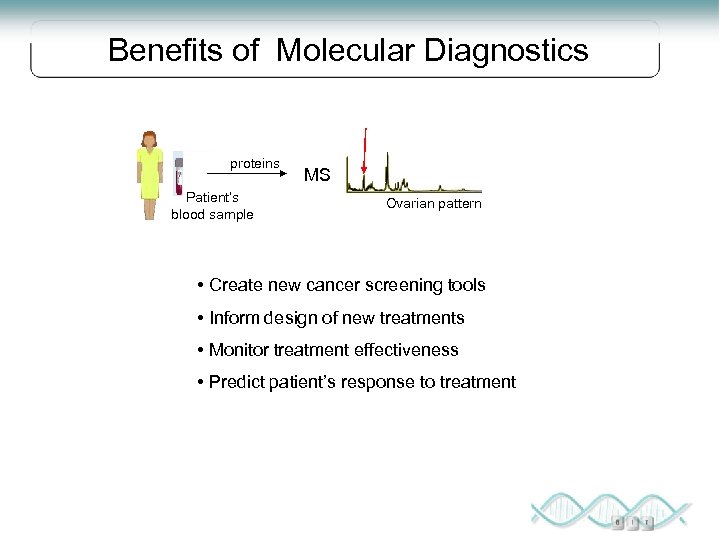
Benefits of Molecular Diagnostics proteins Patient’s blood sample MS Ovarian pattern • Create new cancer screening tools • Inform design of new treatments • Monitor treatment effectiveness • Predict patient’s response to treatment
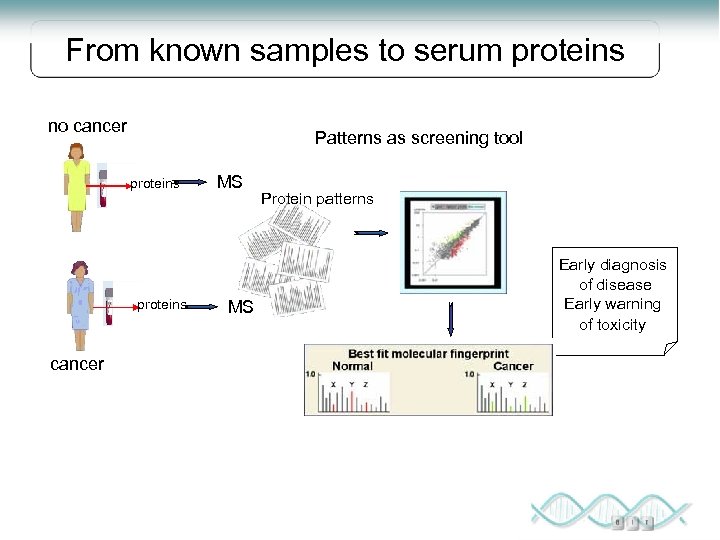
From known samples to serum proteins no cancer Patterns as screening tool proteins cancer MS MS Protein patterns Early diagnosis of disease Early warning of toxicity
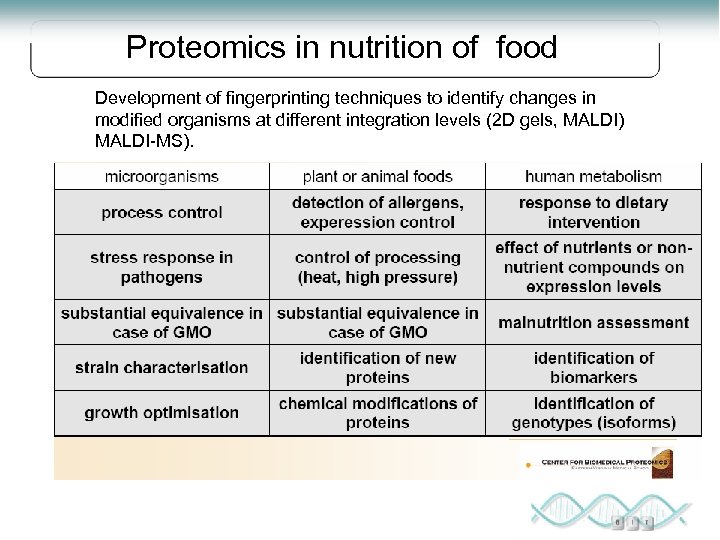
Proteomics in nutrition of food Development of fingerprinting techniques to identify changes in modified organisms at different integration levels (2 D gels, MALDI) MALDI-MS).
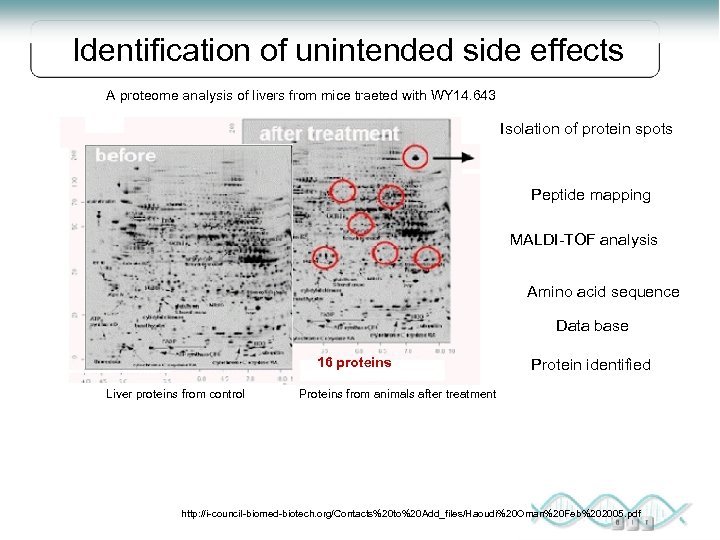
Identification of unintended side effects A proteome analysis of livers from mice traeted with WY 14. 643 Isolation of protein spots Peptide mapping MALDI-TOF analysis Amino acid sequence Data base 16 proteins Liver proteins from control Protein identified Proteins from animals after treatment http: //i-council-biomed-biotech. org/Contacts%20 to%20 Add_files/Haoudi%20 Oman%20 Feb%202005. pdf
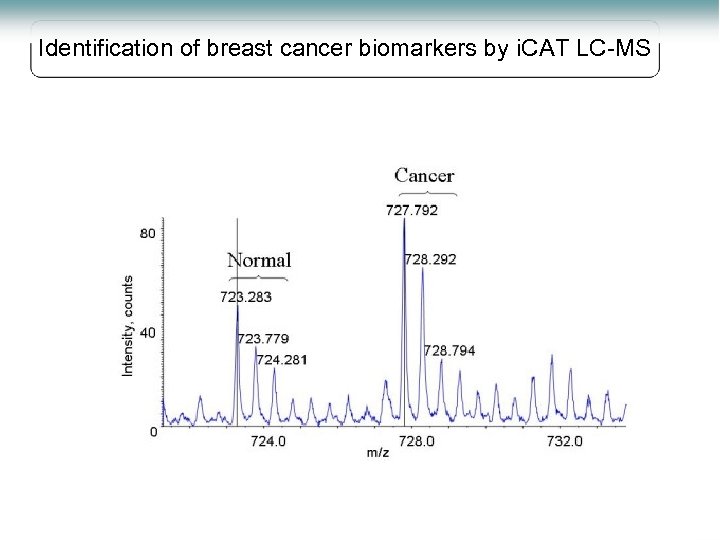
Identification of breast cancer biomarkers by i. CAT LC-MS
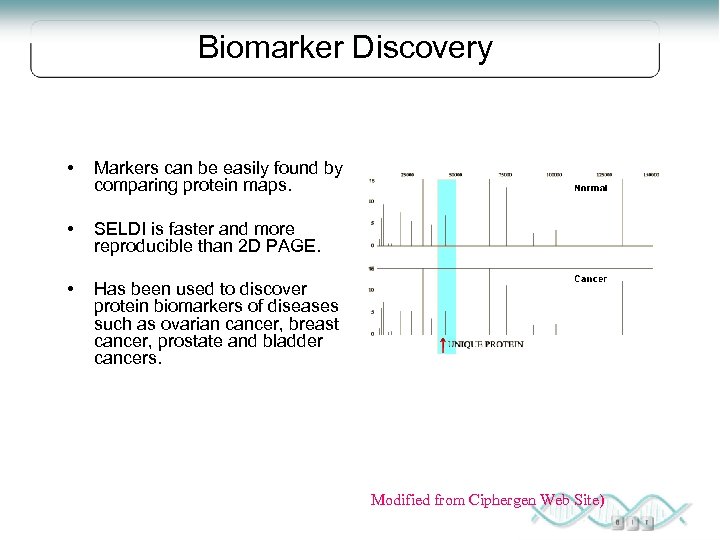
Biomarker Discovery • Markers can be easily found by comparing protein maps. • SELDI is faster and more reproducible than 2 D PAGE. • Has been used to discover protein biomarkers of diseases such as ovarian cancer, breast cancer, prostate and bladder cancers. Modified from Ciphergen Web Site)
Gene Ontology A knowledge representation about the word or some part of it. An ontology is used as a description of the concepts and relationships that exist for a community of agents. Ontology generally describes: • Individuals: the basic or “ground level” objects • Classes: sets, collections, or types of objects • Attributes: properties, features, characteristics, or parameters that objects can have and share • Relations: ways that objects can be related to one another from: wikipedia

Goals Develop a set of controlled, structured vocabularies – gene ontology (GO) to describe aspects of molecular biology Describe gene products using vocabulary terms (annotation) Provide a public resource, allowing access to the GO, annotations and software tools developed for use with the GO data www. geneontology. org
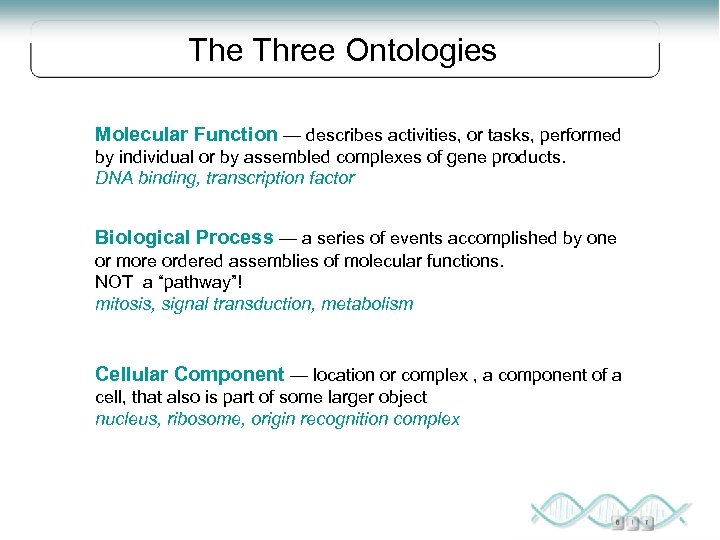
The Three Ontologies Molecular Function — describes activities, or tasks, performed by individual or by assembled complexes of gene products. DNA binding, transcription factor Biological Process — a series of events accomplished by one or more ordered assemblies of molecular functions. NOT a “pathway”! mitosis, signal transduction, metabolism Cellular Component — location or complex , a component of a cell, that also is part of some larger object nucleus, ribosome, origin recognition complex
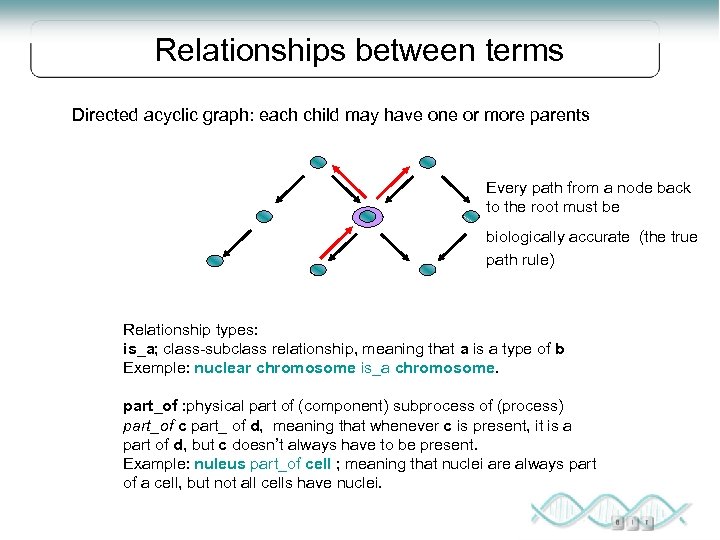
Relationships between terms Directed acyclic graph: each child may have one or more parents Every path from a node back to the root must be biologically accurate (the true path rule) Relationship types: is_a; class-subclass relationship, meaning that a is a type of b Exemple: nuclear chromosome is_a chromosome. part_of : physical part of (component) subprocess of (process) part_of c part_ of d, meaning that whenever c is present, it is a part of d, but c doesn’t always have to be present. Example: nuleus part_of cell ; meaning that nuclei are always part of a cell, but not all cells have nuclei.
Relationships between terms Example: the biological process term hexose biosynthesis has two parents, hexose metabolism and monosaccaride biosynthesis. This is because biosynthesis is a subtype of metabolism, and a hexose is a subtype of monosaccharide. When any gene involved in hexose biosynthesis is annotated to this term, it is automatically annotated to both hexose metabolsim and monosaccharide biosynthesis, because every GO term must obey the “true path rule”, if the child term deescribes the gene product, then all its parent terms must also apply to that gene product. .
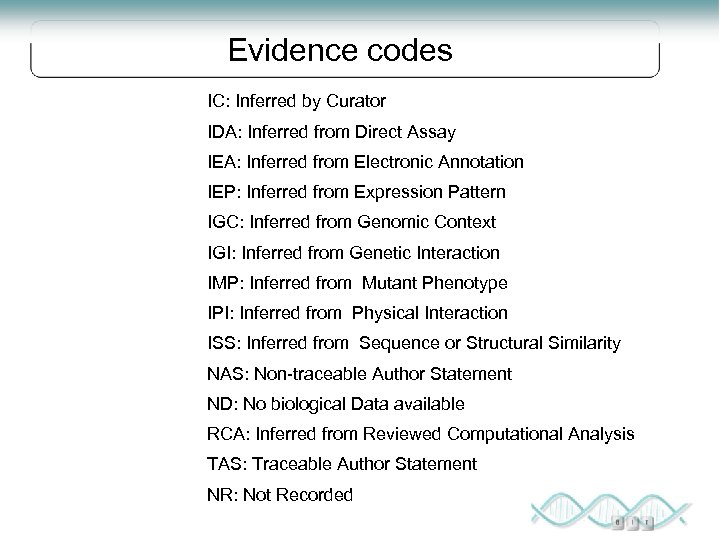
Evidence codes IC: Inferred by Curator IDA: Inferred from Direct Assay IEA: Inferred from Electronic Annotation IEP: Inferred from Expression Pattern IGC: Inferred from Genomic Context IGI: Inferred from Genetic Interaction IMP: Inferred from Mutant Phenotype IPI: Inferred from Physical Interaction ISS: Inferred from Sequence or Structural Similarity NAS: Non-traceable Author Statement ND: No biological Data available RCA: Inferred from Reviewed Computational Analysis TAS: Traceable Author Statement NR: Not Recorded
Gene Ontology Home
GO tools • search for gene products and view the terms with which they are associated; • search or browse the ontology for GO terms of interest and see term details and gene product annotations. • Ami. GO also provides a BLAST search engine, which searches the sequences of genes and gene products that have been annotated to a GO term and submitted to the GO Consortium.
Annotation tools
Re. BIL

Gene expression tools
67ae313196137542beb61d8dd9c767ba.ppt