47188acb6969bb8569d0d4ee33ae8f5b.ppt
- Количество слайдов: 22
 Proteomics: A Challenge for Technology and Information Science CBCB Seminar, November 21, 2005 Tim Griffin Dept. Biochemistry, Molecular Biology and Biophysics tgriffin@umn. edu
Proteomics: A Challenge for Technology and Information Science CBCB Seminar, November 21, 2005 Tim Griffin Dept. Biochemistry, Molecular Biology and Biophysics tgriffin@umn. edu
 What is proteomics? “Proteomics includes not only the identification and quantification of proteins, but also the determination of their localization, modifications, interactions, activities, and, ultimately, their function. ” -Stan Fields in Science, 2001.
What is proteomics? “Proteomics includes not only the identification and quantification of proteins, but also the determination of their localization, modifications, interactions, activities, and, ultimately, their function. ” -Stan Fields in Science, 2001.
 Genomics vs. Proteomics Similarities: Large datasets, tools needed for annotation and interpretation of results Differences: Genomics – generally mature technologies, data processing methods, questions asked usually involve quantitative changes in RNA transcripts (microarrays) Proteomics – still evolving, complexity of protein biochemical properties: expression changes, modifications, interactions, activities – many questions to ask and data to interpret, methods changing, different approaches (mass spec, arrays etc. ),
Genomics vs. Proteomics Similarities: Large datasets, tools needed for annotation and interpretation of results Differences: Genomics – generally mature technologies, data processing methods, questions asked usually involve quantitative changes in RNA transcripts (microarrays) Proteomics – still evolving, complexity of protein biochemical properties: expression changes, modifications, interactions, activities – many questions to ask and data to interpret, methods changing, different approaches (mass spec, arrays etc. ),
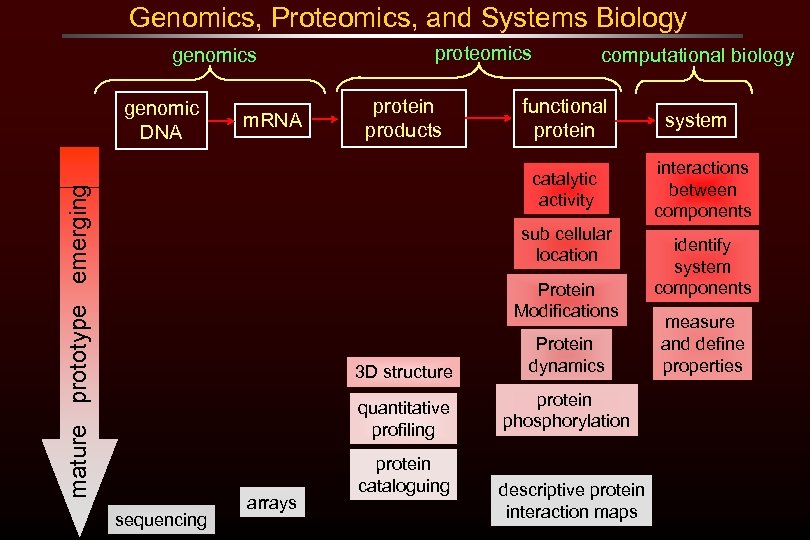 Genomics, Proteomics, and Systems Biology genomics genomic DNA m. RNA proteomics protein products computational biology functional protein mature prototype emerging catalytic activity sub cellular location Protein Modifications 3 D structure quantitative profiling sequencing arrays Protein dynamics protein phosphorylation protein cataloguing descriptive protein interaction maps system interactions between components identify system components measure and define properties
Genomics, Proteomics, and Systems Biology genomics genomic DNA m. RNA proteomics protein products computational biology functional protein mature prototype emerging catalytic activity sub cellular location Protein Modifications 3 D structure quantitative profiling sequencing arrays Protein dynamics protein phosphorylation protein cataloguing descriptive protein interaction maps system interactions between components identify system components measure and define properties
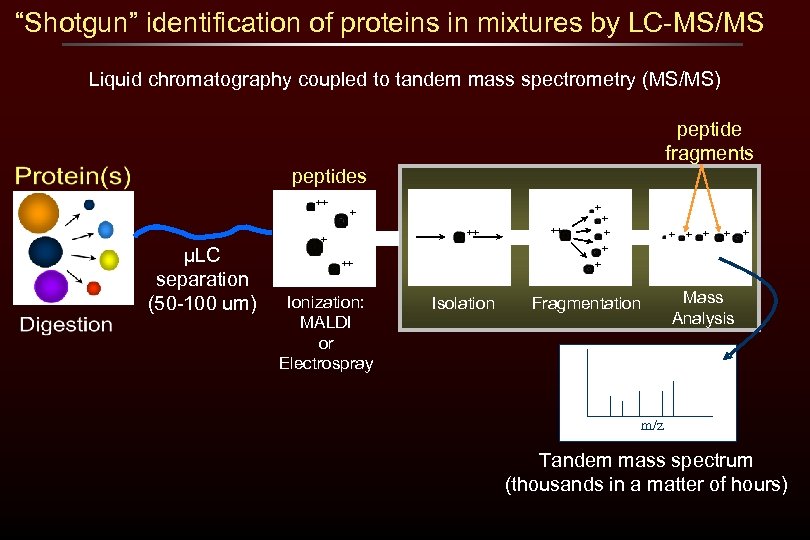 “Shotgun” identification of proteins in mixtures by LC-MS/MS Liquid chromatography coupled to tandem mass spectrometry (MS/MS) peptide fragments peptides ++ µLC separation (50 -100 um) + + ++ + ++ Ionization: MALDI or Electrospray + Isolation Mass Analysis Fragmentation m/z Tandem mass spectrum (thousands in a matter of hours)
“Shotgun” identification of proteins in mixtures by LC-MS/MS Liquid chromatography coupled to tandem mass spectrometry (MS/MS) peptide fragments peptides ++ µLC separation (50 -100 um) + + ++ + ++ Ionization: MALDI or Electrospray + Isolation Mass Analysis Fragmentation m/z Tandem mass spectrum (thousands in a matter of hours)
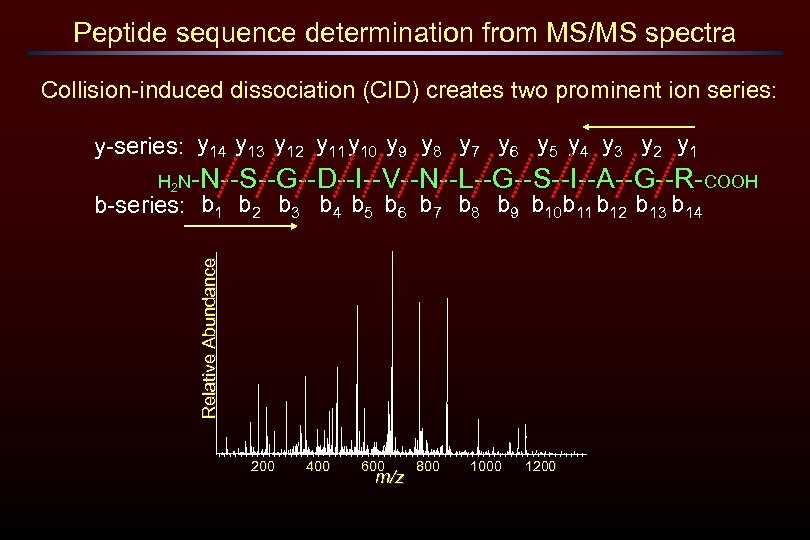 Peptide sequence determination from MS/MS spectra Collision-induced dissociation (CID) creates two prominent ion series: y-series: y 14 y 13 y 12 y 11 y 10 y 9 y 8 y 7 y 6 y 5 y 4 y 3 y 2 y 1 H 2 N-N--S--G--D--I--V--N--L--G--S--I--A--G--R-COOH Relative Abundance b-series: b 1 b 2 b 3 b 4 b 5 b 6 b 7 b 8 b 9 b 10 b 11 b 12 b 13 b 14 200 400 600 m/z 800 1000 1200
Peptide sequence determination from MS/MS spectra Collision-induced dissociation (CID) creates two prominent ion series: y-series: y 14 y 13 y 12 y 11 y 10 y 9 y 8 y 7 y 6 y 5 y 4 y 3 y 2 y 1 H 2 N-N--S--G--D--I--V--N--L--G--S--I--A--G--R-COOH Relative Abundance b-series: b 1 b 2 b 3 b 4 b 5 b 6 b 7 b 8 b 9 b 10 b 11 b 12 b 13 b 14 200 400 600 m/z 800 1000 1200
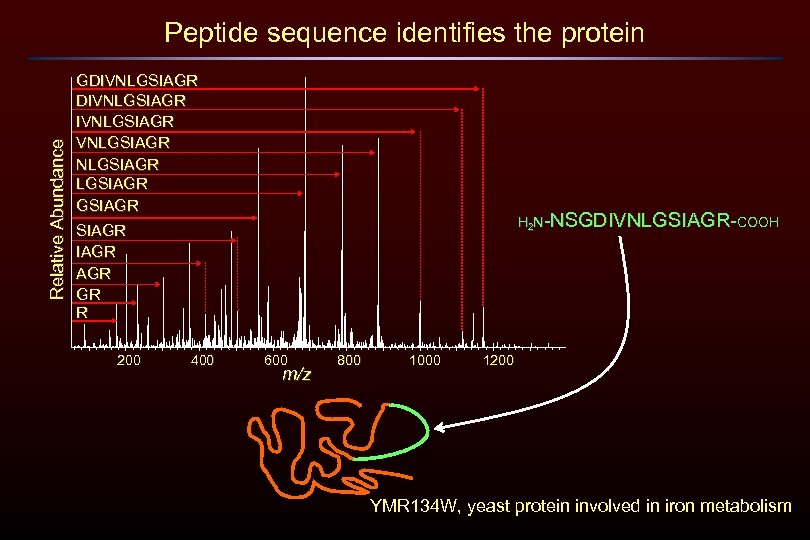 Relative Abundance Peptide sequence identifies the protein GDIVNLGSIAGR NLGSIAGR IAGR GR R 200 400 H 2 N-NSGDIVNLGSIAGR-COOH 600 m/z 800 1000 1200 YMR 134 W, yeast protein involved in iron metabolism
Relative Abundance Peptide sequence identifies the protein GDIVNLGSIAGR NLGSIAGR IAGR GR R 200 400 H 2 N-NSGDIVNLGSIAGR-COOH 600 m/z 800 1000 1200 YMR 134 W, yeast protein involved in iron metabolism
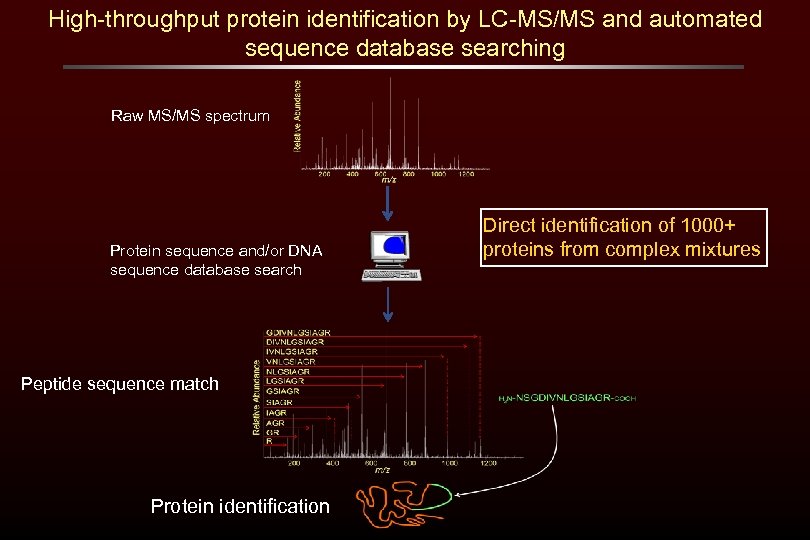 High-throughput protein identification by LC-MS/MS and automated sequence database searching Raw MS/MS spectrum Protein sequence and/or DNA sequence database search Peptide sequence match Protein identification Direct identification of 1000+ proteins from complex mixtures
High-throughput protein identification by LC-MS/MS and automated sequence database searching Raw MS/MS spectrum Protein sequence and/or DNA sequence database search Peptide sequence match Protein identification Direct identification of 1000+ proteins from complex mixtures
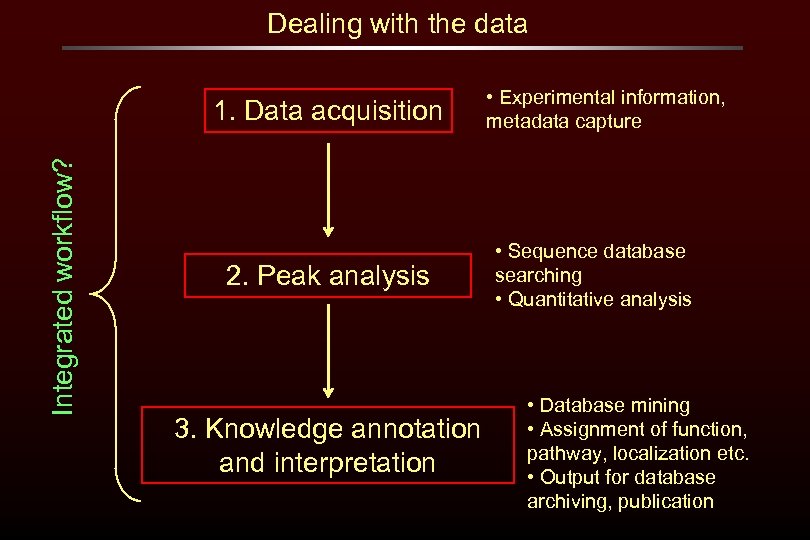 Dealing with the data Integrated workflow? 1. Data acquisition 2. Peak analysis 3. Knowledge annotation and interpretation • Experimental information, metadata capture • Sequence database searching • Quantitative analysis • Database mining • Assignment of function, pathway, localization etc. • Output for database archiving, publication
Dealing with the data Integrated workflow? 1. Data acquisition 2. Peak analysis 3. Knowledge annotation and interpretation • Experimental information, metadata capture • Sequence database searching • Quantitative analysis • Database mining • Assignment of function, pathway, localization etc. • Output for database archiving, publication
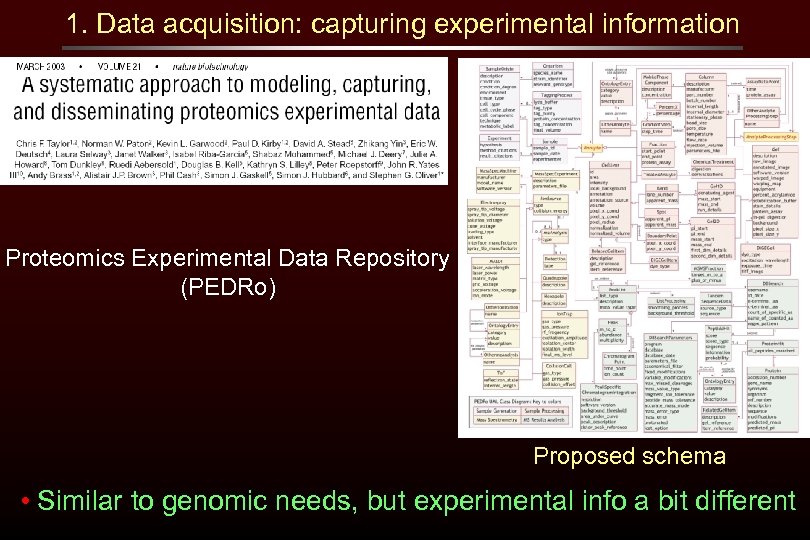 1. Data acquisition: capturing experimental information Proteomics Experimental Data Repository (PEDRo) Proposed schema • Similar to genomic needs, but experimental info a bit different
1. Data acquisition: capturing experimental information Proteomics Experimental Data Repository (PEDRo) Proposed schema • Similar to genomic needs, but experimental info a bit different
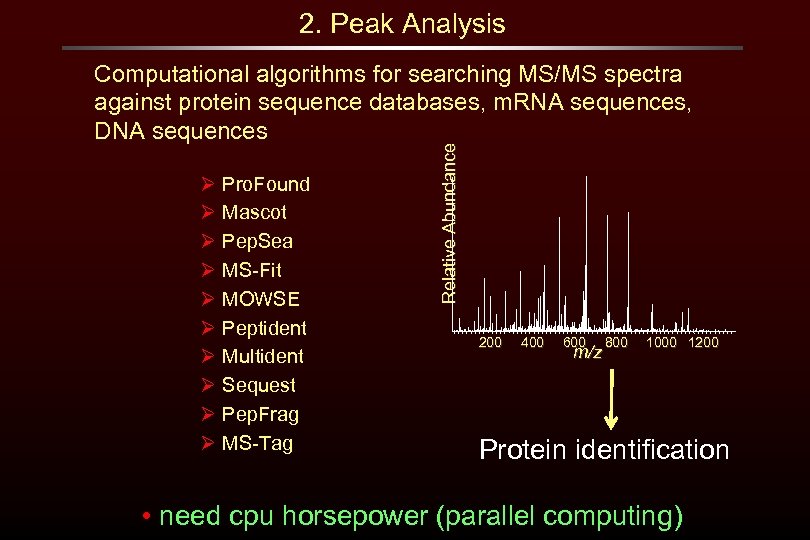 2. Peak Analysis Ø Pro. Found Ø Mascot Ø Pep. Sea Ø MS-Fit Ø MOWSE Ø Peptident Ø Multident Ø Sequest Ø Pep. Frag Ø MS-Tag Relative Abundance Computational algorithms for searching MS/MS spectra against protein sequence databases, m. RNA sequences, DNA sequences 200 400 600 m/z 800 1000 1200 Protein identification • need cpu horsepower (parallel computing)
2. Peak Analysis Ø Pro. Found Ø Mascot Ø Pep. Sea Ø MS-Fit Ø MOWSE Ø Peptident Ø Multident Ø Sequest Ø Pep. Frag Ø MS-Tag Relative Abundance Computational algorithms for searching MS/MS spectra against protein sequence databases, m. RNA sequences, DNA sequences 200 400 600 m/z 800 1000 1200 Protein identification • need cpu horsepower (parallel computing)
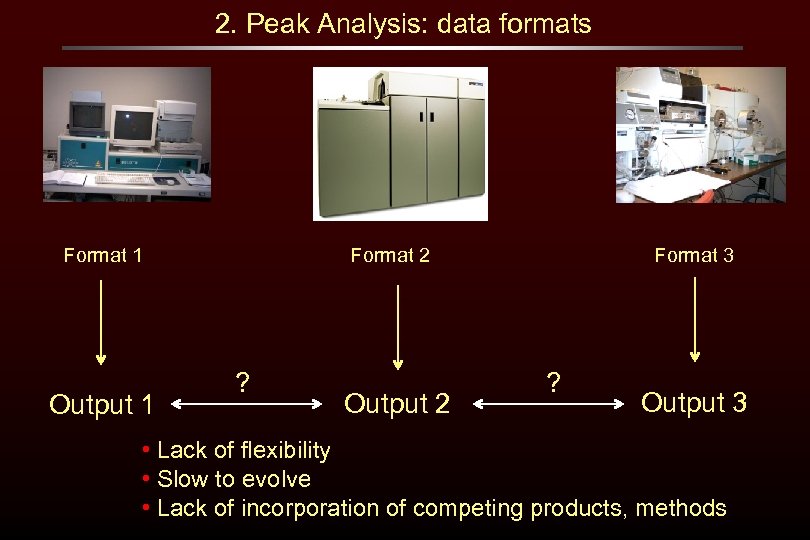 2. Peak Analysis: data formats Format 1 Output 1 Format 2 ? Output 2 Format 3 ? Output 3 • Lack of flexibility • Slow to evolve • Lack of incorporation of competing products, methods
2. Peak Analysis: data formats Format 1 Output 1 Format 2 ? Output 2 Format 3 ? Output 3 • Lack of flexibility • Slow to evolve • Lack of incorporation of competing products, methods
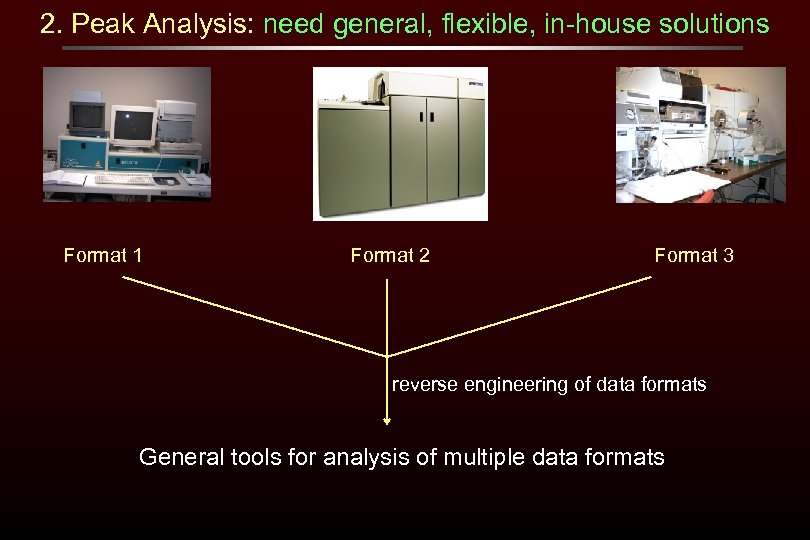 2. Peak Analysis: need general, flexible, in-house solutions Format 1 Format 2 Format 3 reverse engineering of data formats General tools for analysis of multiple data formats
2. Peak Analysis: need general, flexible, in-house solutions Format 1 Format 2 Format 3 reverse engineering of data formats General tools for analysis of multiple data formats
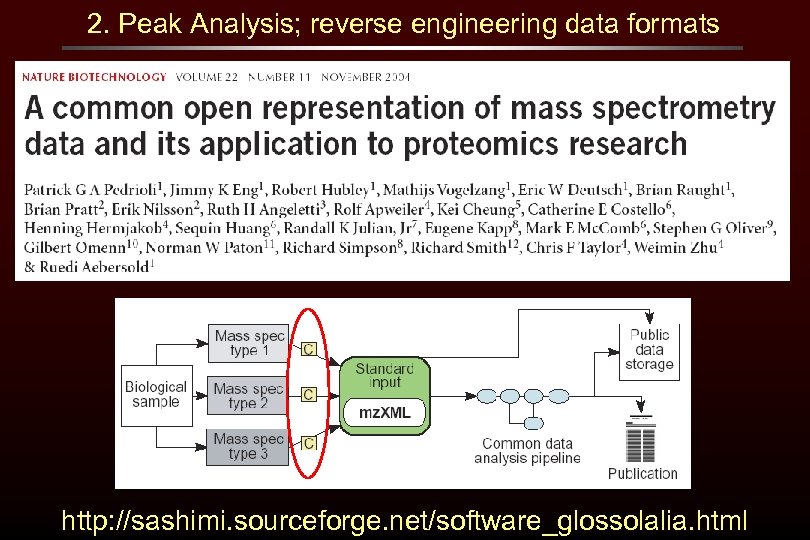 2. Peak Analysis; reverse engineering data formats http: //sashimi. sourceforge. net/software_glossolalia. html
2. Peak Analysis; reverse engineering data formats http: //sashimi. sourceforge. net/software_glossolalia. html
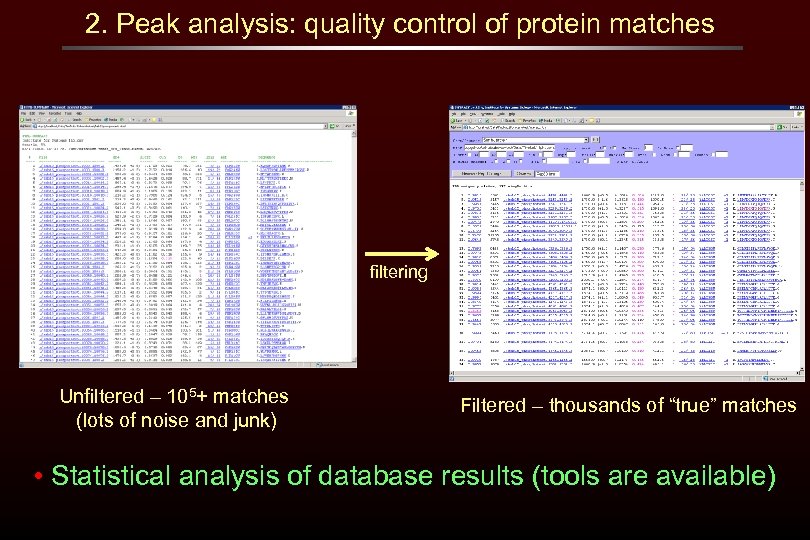 2. Peak analysis: quality control of protein matches filtering Unfiltered – 105+ matches (lots of noise and junk) Filtered – thousands of “true” matches • Statistical analysis of database results (tools are available)
2. Peak analysis: quality control of protein matches filtering Unfiltered – 105+ matches (lots of noise and junk) Filtered – thousands of “true” matches • Statistical analysis of database results (tools are available)
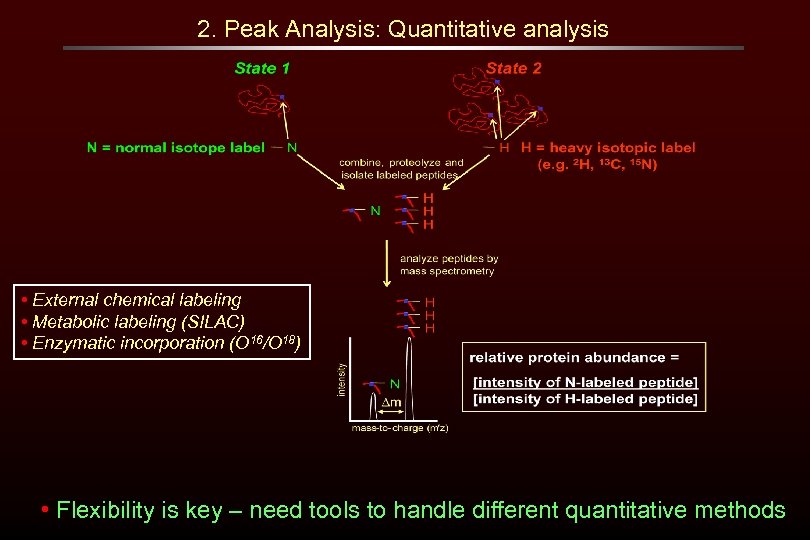 2. Peak Analysis: Quantitative analysis • External chemical labeling • Metabolic labeling (SILAC) • Enzymatic incorporation (O 16/O 18) • Flexibility is key – need tools to handle different quantitative methods
2. Peak Analysis: Quantitative analysis • External chemical labeling • Metabolic labeling (SILAC) • Enzymatic incorporation (O 16/O 18) • Flexibility is key – need tools to handle different quantitative methods
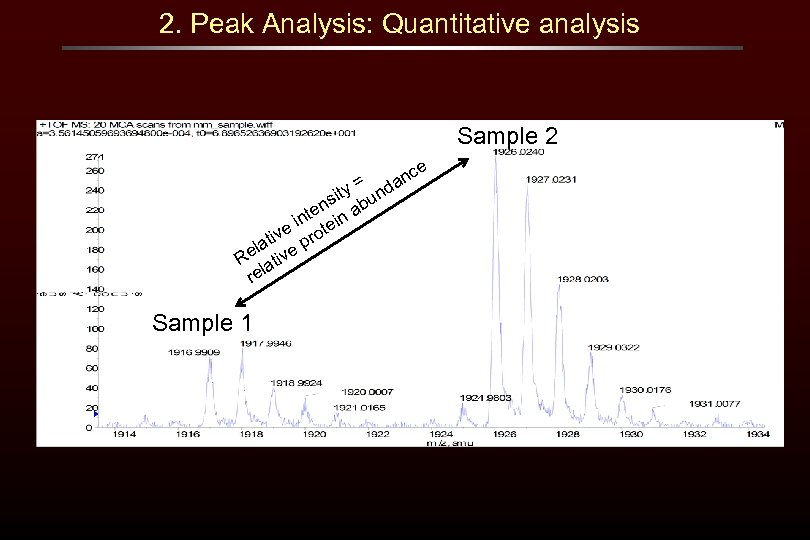 2. Peak Analysis: Quantitative analysis Sample 2 e nc a = ity und s ten in ab n e i rote iv lat e p Re ativ rel Sample 1
2. Peak Analysis: Quantitative analysis Sample 2 e nc a = ity und s ten in ab n e i rote iv lat e p Re ativ rel Sample 1
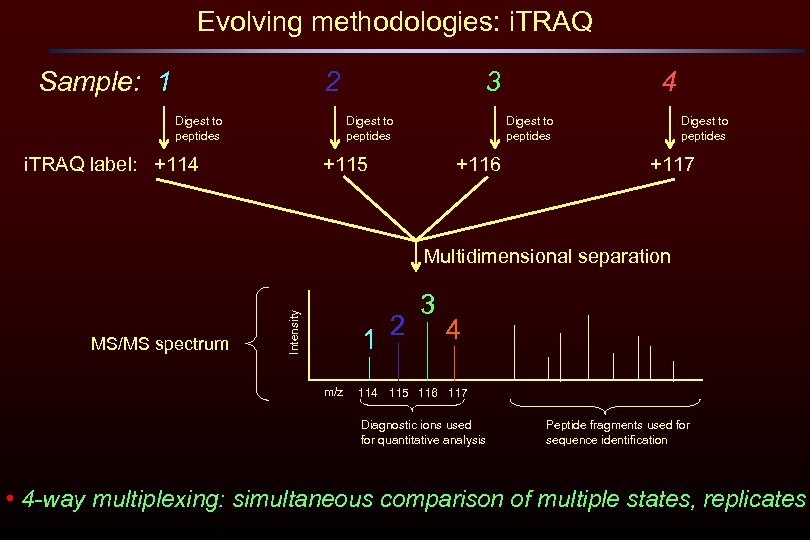 Evolving methodologies: i. TRAQ Sample: 1 2 Digest to peptides 3 Digest to peptides i. TRAQ label: +114 4 Digest to peptides +115 +116 Digest to peptides +117 Multidimensional separation Intensity MS/MS spectrum 2 1 m/z 3 4 115 116 117 Diagnostic ions used for quantitative analysis Peptide fragments used for sequence identification • 4 -way multiplexing: simultaneous comparison of multiple states, replicates
Evolving methodologies: i. TRAQ Sample: 1 2 Digest to peptides 3 Digest to peptides i. TRAQ label: +114 4 Digest to peptides +115 +116 Digest to peptides +117 Multidimensional separation Intensity MS/MS spectrum 2 1 m/z 3 4 115 116 117 Diagnostic ions used for quantitative analysis Peptide fragments used for sequence identification • 4 -way multiplexing: simultaneous comparison of multiple states, replicates
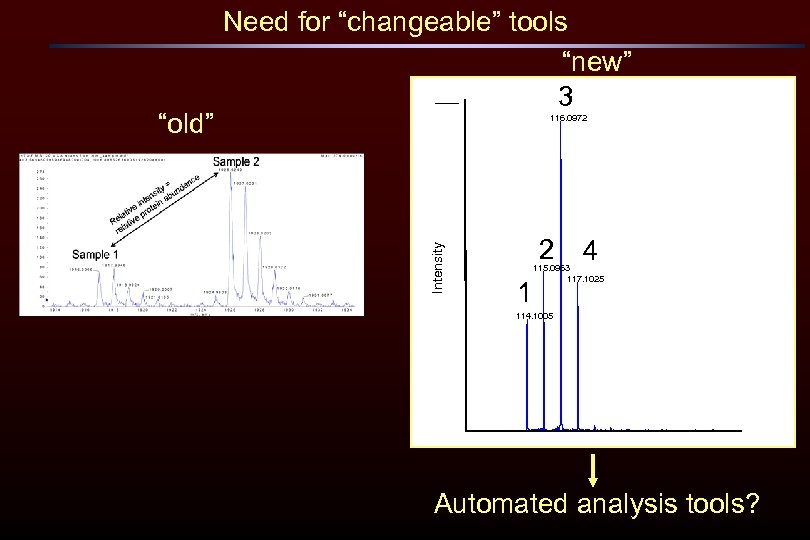 116. 0972 Intensity “old” Need for “changeable” tools “new” 3 2 4 115. 0963 117. 1025 1 114. 1005 Automated analysis tools?
116. 0972 Intensity “old” Need for “changeable” tools “new” 3 2 4 115. 0963 117. 1025 1 114. 1005 Automated analysis tools?
 3. Knowledge annotation: making sense of lists of data
3. Knowledge annotation: making sense of lists of data
 3. Knowledge annotation: mining proteomic/genomic databases
3. Knowledge annotation: mining proteomic/genomic databases
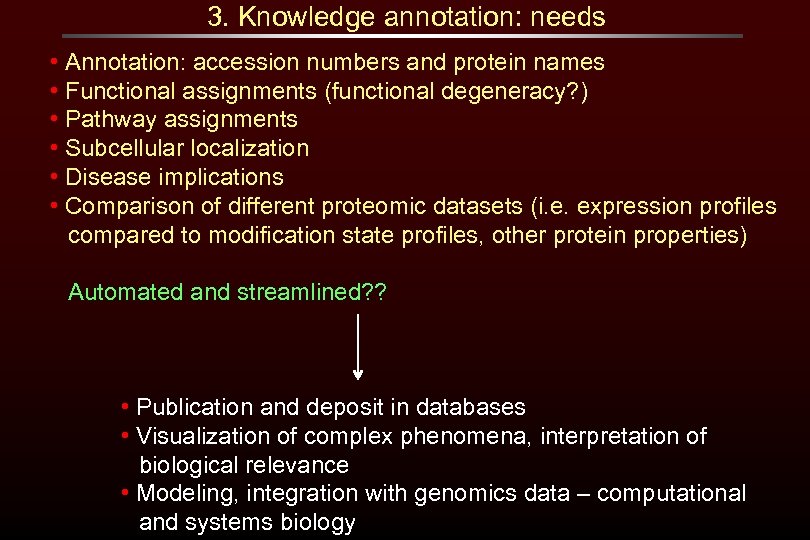 3. Knowledge annotation: needs • Annotation: accession numbers and protein names • Functional assignments (functional degeneracy? ) • Pathway assignments • Subcellular localization • Disease implications • Comparison of different proteomic datasets (i. e. expression profiles compared to modification state profiles, other protein properties) Automated and streamlined? ? • Publication and deposit in databases • Visualization of complex phenomena, interpretation of biological relevance • Modeling, integration with genomics data – computational and systems biology
3. Knowledge annotation: needs • Annotation: accession numbers and protein names • Functional assignments (functional degeneracy? ) • Pathway assignments • Subcellular localization • Disease implications • Comparison of different proteomic datasets (i. e. expression profiles compared to modification state profiles, other protein properties) Automated and streamlined? ? • Publication and deposit in databases • Visualization of complex phenomena, interpretation of biological relevance • Modeling, integration with genomics data – computational and systems biology


