Proteins Structure and functions

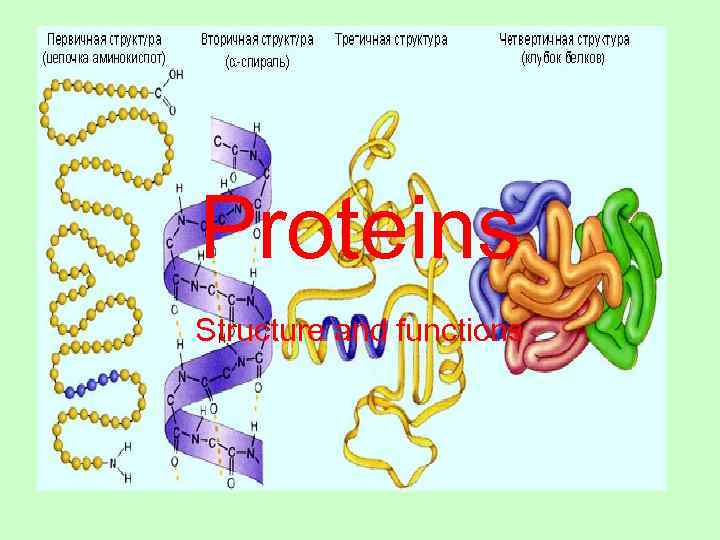

 Proteins Structure and functions
Proteins Structure and functions
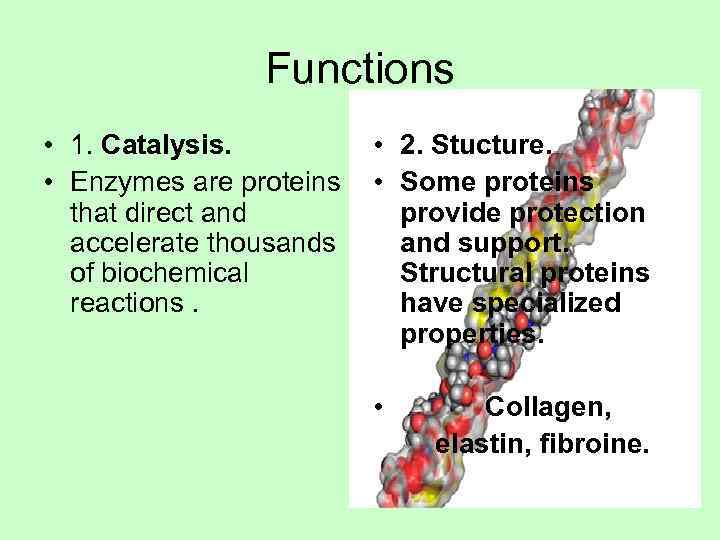
 Functions • 1. Catalysis. • 2. Stucture. • Enzymes are proteins • Some proteins that direct and provide protection accelerate thousands and support. of biochemical Structural proteins reactions. have specialized properties. • Collagen, elastin, fibroine.
Functions • 1. Catalysis. • 2. Stucture. • Enzymes are proteins • Some proteins that direct and provide protection accelerate thousands and support. of biochemical Structural proteins reactions. have specialized properties. • Collagen, elastin, fibroine.
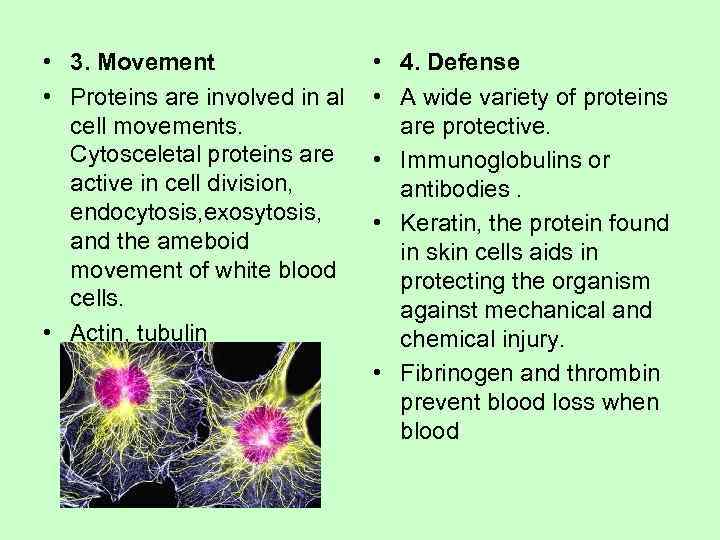
 • 3. Movement • 4. Defense • Proteins are involved in al • A wide variety of proteins cell movements. are protective. Cytosceletal proteins are • Immunoglobulins or active in cell division, antibodies. endocytosis, exosytosis, • Keratin, the protein found and the ameboid in skin cells aids in movement of white blood protecting the organism cells. against mechanical and • Actin, tubulin chemical injury. • Fibrinogen and thrombin prevent blood loss when blood
• 3. Movement • 4. Defense • Proteins are involved in al • A wide variety of proteins cell movements. are protective. Cytosceletal proteins are • Immunoglobulins or active in cell division, antibodies. endocytosis, exosytosis, • Keratin, the protein found and the ameboid in skin cells aids in movement of white blood protecting the organism cells. against mechanical and • Actin, tubulin chemical injury. • Fibrinogen and thrombin prevent blood loss when blood
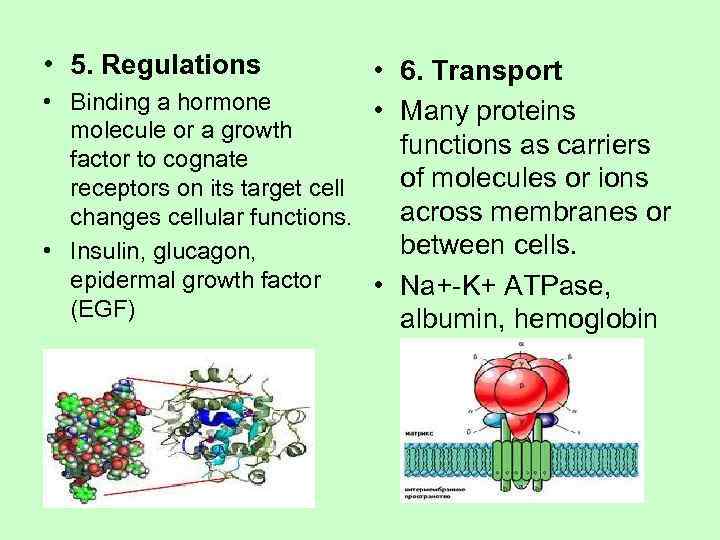
 • 5. Regulations • 6. Transport • Binding a hormone • Many proteins molecule or a growth factor to cognate functions as carriers receptors on its target cell of molecules or ions changes cellular functions. across membranes or • Insulin, glucagon, between cells. epidermal growth factor • Na+-K+ ATPase, (EGF) albumin, hemoglobin
• 5. Regulations • 6. Transport • Binding a hormone • Many proteins molecule or a growth factor to cognate functions as carriers receptors on its target cell of molecules or ions changes cellular functions. across membranes or • Insulin, glucagon, between cells. epidermal growth factor • Na+-K+ ATPase, (EGF) albumin, hemoglobin
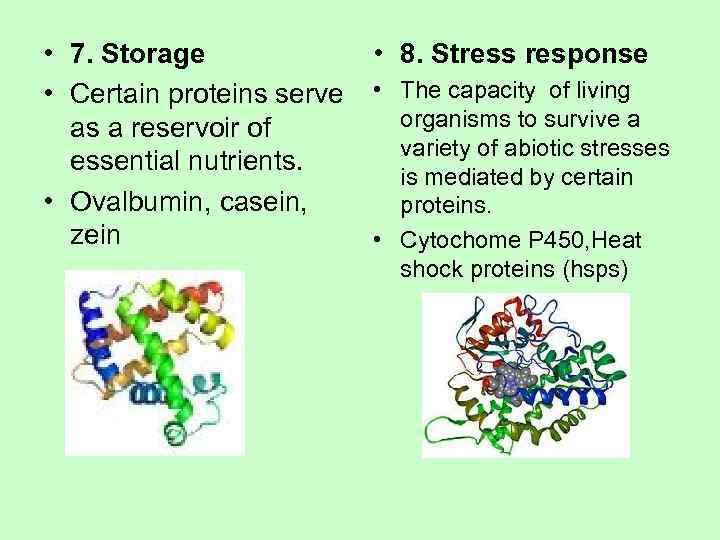
 • 7. Storage • 8. Stress response • Certain proteins serve • The capacity of living as a reservoir of organisms to survive a variety of abiotic stresses essential nutrients. is mediated by certain • Ovalbumin, casein, proteins. zein • Cytochome P 450, Heat shock proteins (hsps)
• 7. Storage • 8. Stress response • Certain proteins serve • The capacity of living as a reservoir of organisms to survive a variety of abiotic stresses essential nutrients. is mediated by certain • Ovalbumin, casein, proteins. zein • Cytochome P 450, Heat shock proteins (hsps)
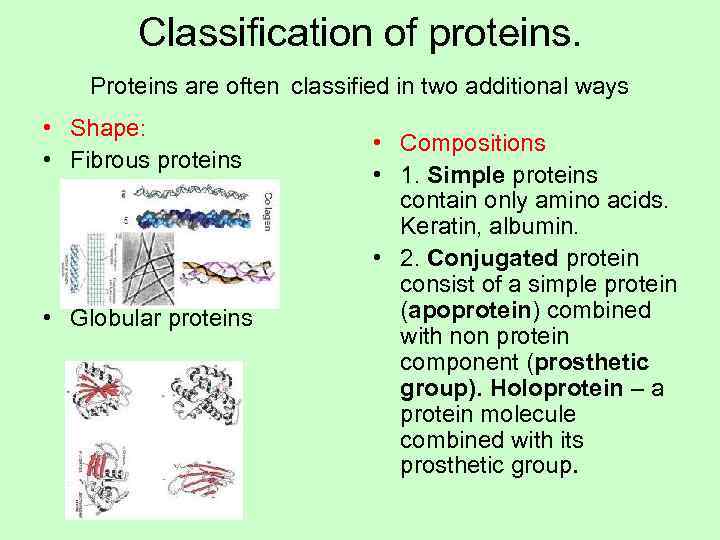
 Classification of proteins. Proteins are often classified in two additional ways • Shape: • Compositions • Fibrous proteins • 1. Simple proteins contain only amino acids. Keratin, albumin. • 2. Conjugated protein consist of a simple protein • Globular proteins (apoprotein) combined with non protein component (prosthetic group). Holoprotein – a protein molecule combined with its prosthetic group.
Classification of proteins. Proteins are often classified in two additional ways • Shape: • Compositions • Fibrous proteins • 1. Simple proteins contain only amino acids. Keratin, albumin. • 2. Conjugated protein consist of a simple protein • Globular proteins (apoprotein) combined with non protein component (prosthetic group). Holoprotein – a protein molecule combined with its prosthetic group.
 Conjugated proteins are classified according to the nature of their prosthetic group • 1. Glycoproteins • 3. Metalloproteins contain carbohydrate contain metal ions. component. • 4. Phosphoproteins • 2. Lipoproteins contain phosphate contain lipids groups. molecules • 5. Hemoproteins contain heme groups
Conjugated proteins are classified according to the nature of their prosthetic group • 1. Glycoproteins • 3. Metalloproteins contain carbohydrate contain metal ions. component. • 4. Phosphoproteins • 2. Lipoproteins contain phosphate contain lipids groups. molecules • 5. Hemoproteins contain heme groups
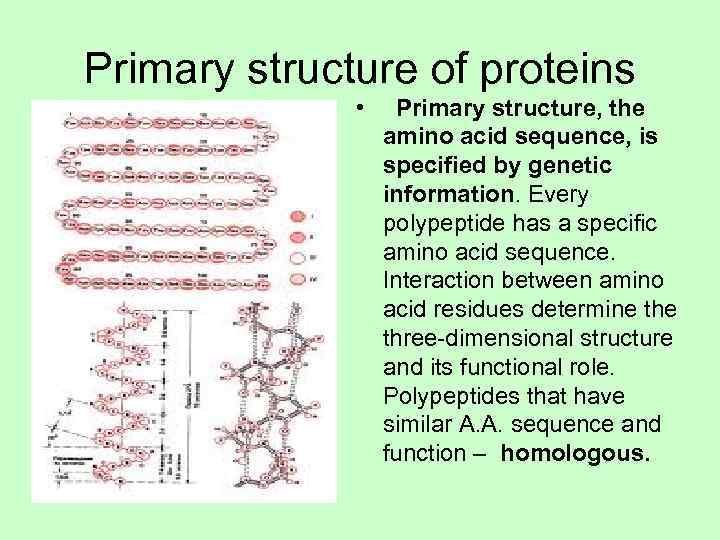
 Primary structure of proteins • Primary structure, the amino acid sequence, is specified by genetic information. Every polypeptide has a specific amino acid sequence. Interaction between amino acid residues determine the three-dimensional structure and its functional role. Polypeptides that have similar A. A. sequence and function – homologous.
Primary structure of proteins • Primary structure, the amino acid sequence, is specified by genetic information. Every polypeptide has a specific amino acid sequence. Interaction between amino acid residues determine the three-dimensional structure and its functional role. Polypeptides that have similar A. A. sequence and function – homologous.
 Molecular diseases • molecular disease any disease in which the pathogenesis can be traced to a single molecule, usually a protein, which is either abnormal in structure or present in reduced amounts.
Molecular diseases • molecular disease any disease in which the pathogenesis can be traced to a single molecule, usually a protein, which is either abnormal in structure or present in reduced amounts.
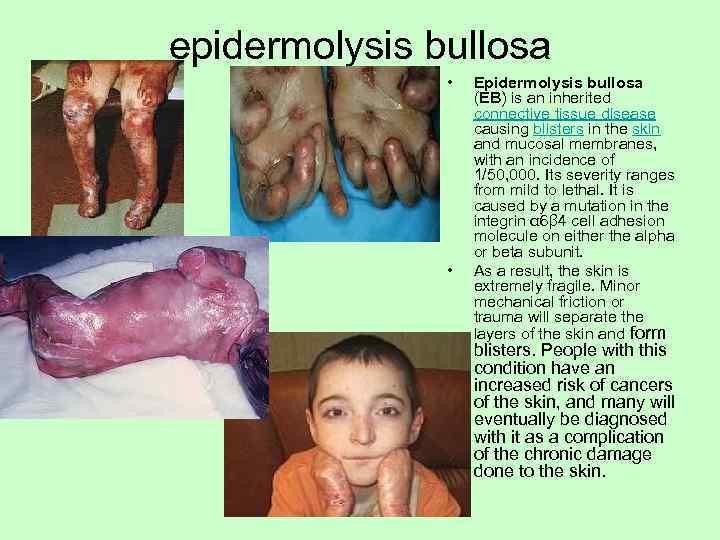
 epidermolysis bullosa • Epidermolysis bullosa (EB) is an inherited connective tissue disease causing blisters in the skin and mucosal membranes, with an incidence of 1/50, 000. Its severity ranges from mild to lethal. It is caused by a mutation in the integrin α 6β 4 cell adhesion molecule on either the alpha or beta subunit. • As a result, the skin is extremely fragile. Minor mechanical friction or trauma will separate the layers of the skin and form blisters. People with this condition have an increased risk of cancers of the skin, and many will eventually be diagnosed with it as a complication of the chronic damage done to the skin.
epidermolysis bullosa • Epidermolysis bullosa (EB) is an inherited connective tissue disease causing blisters in the skin and mucosal membranes, with an incidence of 1/50, 000. Its severity ranges from mild to lethal. It is caused by a mutation in the integrin α 6β 4 cell adhesion molecule on either the alpha or beta subunit. • As a result, the skin is extremely fragile. Minor mechanical friction or trauma will separate the layers of the skin and form blisters. People with this condition have an increased risk of cancers of the skin, and many will eventually be diagnosed with it as a complication of the chronic damage done to the skin.
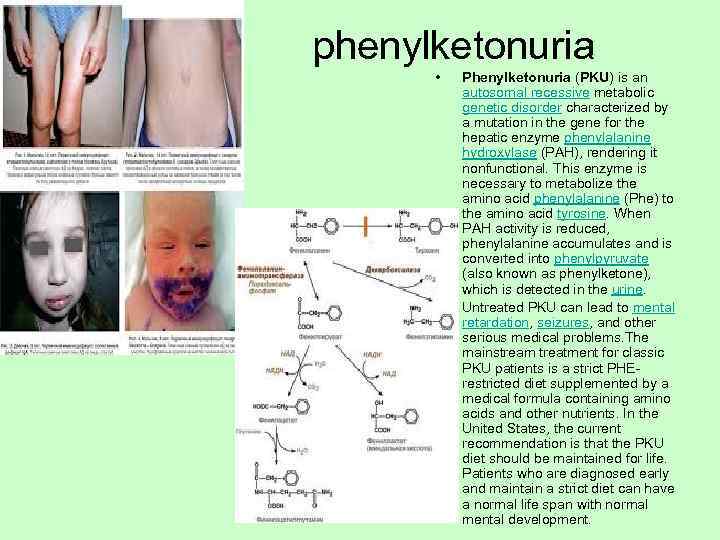
 phenylketonuria • Phenylketonuria (PKU) is an autosomal recessive metabolic genetic disorder characterized by a mutation in the gene for the hepatic enzyme phenylalanine hydroxylase (PAH), rendering it nonfunctional. This enzyme is necessary to metabolize the amino acid phenylalanine (Phe) to the amino acid tyrosine. When PAH activity is reduced, phenylalanine accumulates and is converted into phenylpyruvate (also known as phenylketone), which is detected in the urine. • Untreated PKU can lead to mental retardation, seizures, and other serious medical problems. The mainstream treatment for classic PKU patients is a strict PHE- restricted diet supplemented by a medical formula containing amino acids and other nutrients. In the United States, the current recommendation is that the PKU diet should be maintained for life. Patients who are diagnosed early and maintain a strict diet can have a normal life span with normal mental development.
phenylketonuria • Phenylketonuria (PKU) is an autosomal recessive metabolic genetic disorder characterized by a mutation in the gene for the hepatic enzyme phenylalanine hydroxylase (PAH), rendering it nonfunctional. This enzyme is necessary to metabolize the amino acid phenylalanine (Phe) to the amino acid tyrosine. When PAH activity is reduced, phenylalanine accumulates and is converted into phenylpyruvate (also known as phenylketone), which is detected in the urine. • Untreated PKU can lead to mental retardation, seizures, and other serious medical problems. The mainstream treatment for classic PKU patients is a strict PHE- restricted diet supplemented by a medical formula containing amino acids and other nutrients. In the United States, the current recommendation is that the PKU diet should be maintained for life. Patients who are diagnosed early and maintain a strict diet can have a normal life span with normal mental development.
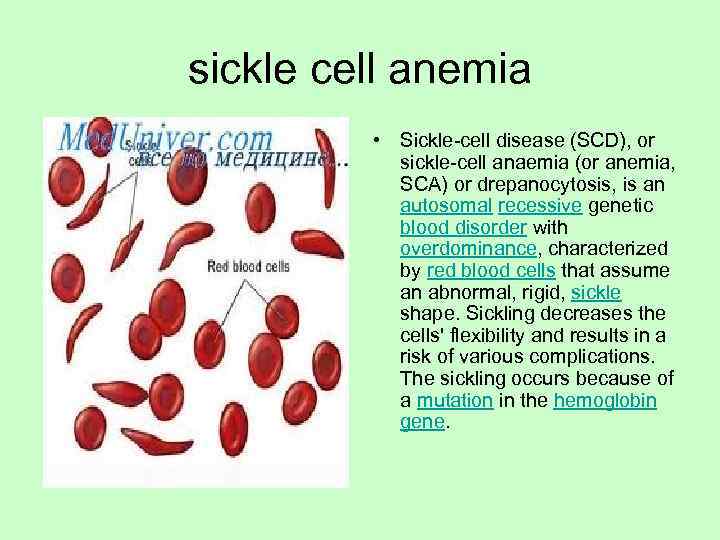
 sickle cell anemia • Sickle-cell disease (SCD), or sickle-cell anaemia (or anemia, SCA) or drepanocytosis, is an autosomal recessive genetic blood disorder with overdominance, characterized by red blood cells that assume an abnormal, rigid, sickle shape. Sickling decreases the cells' flexibility and results in a risk of various complications. The sickling occurs because of a mutation in the hemoglobin gene.
sickle cell anemia • Sickle-cell disease (SCD), or sickle-cell anaemia (or anemia, SCA) or drepanocytosis, is an autosomal recessive genetic blood disorder with overdominance, characterized by red blood cells that assume an abnormal, rigid, sickle shape. Sickling decreases the cells' flexibility and results in a risk of various complications. The sickling occurs because of a mutation in the hemoglobin gene.
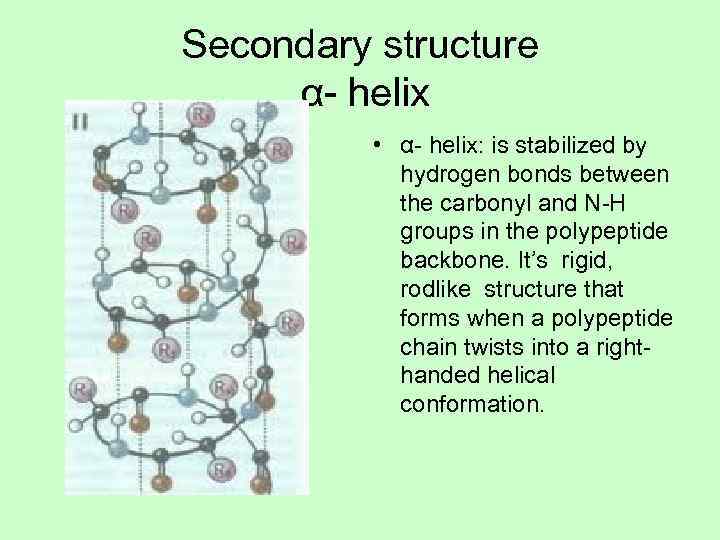
 Secondary structure α- helix • α- helix: is stabilized by hydrogen bonds between the carbonyl and N-H groups in the polypeptide backbone. It’s rigid, rodlike structure that forms when a polypeptide chain twists into a right- handed helical conformation.
Secondary structure α- helix • α- helix: is stabilized by hydrogen bonds between the carbonyl and N-H groups in the polypeptide backbone. It’s rigid, rodlike structure that forms when a polypeptide chain twists into a right- handed helical conformation.
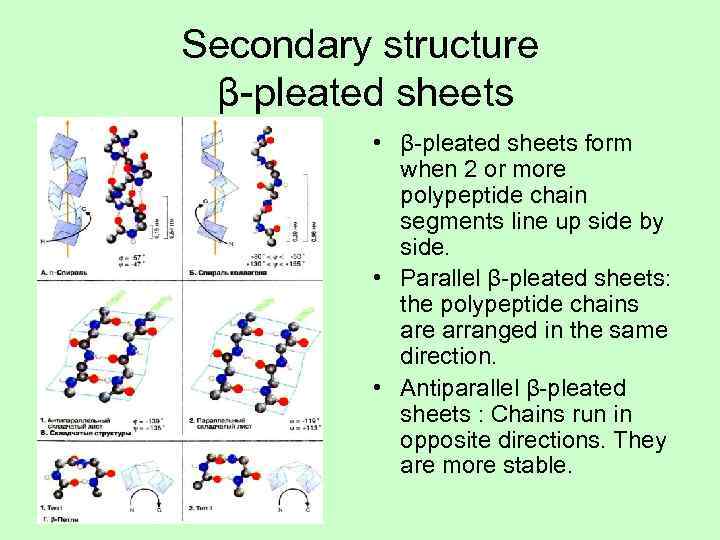
 Secondary structure β-pleated sheets • β-pleated sheets form when 2 or more polypeptide chain segments line up side by side. • Parallel β-pleated sheets: the polypeptide chains are arranged in the same direction. • Antiparallel β-pleated sheets : Chains run in opposite directions. They are more stable.
Secondary structure β-pleated sheets • β-pleated sheets form when 2 or more polypeptide chain segments line up side by side. • Parallel β-pleated sheets: the polypeptide chains are arranged in the same direction. • Antiparallel β-pleated sheets : Chains run in opposite directions. They are more stable.
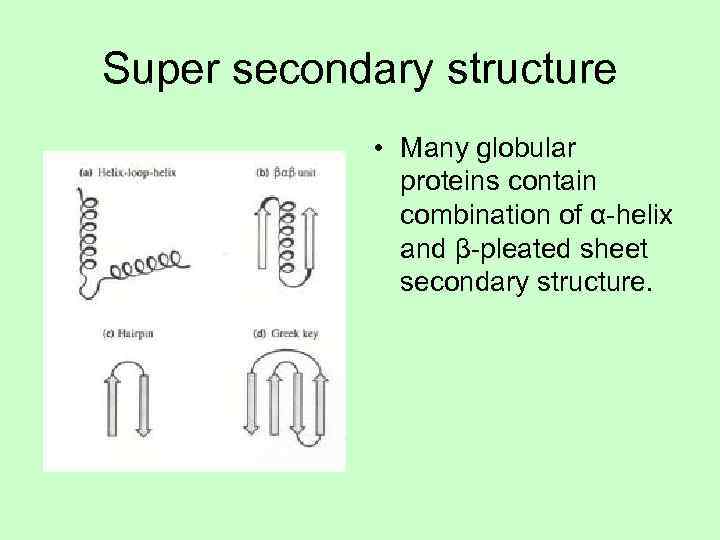
 Super secondary structure • Many globular proteins contain combination of α-helix and β-pleated sheet secondary structure.
Super secondary structure • Many globular proteins contain combination of α-helix and β-pleated sheet secondary structure.
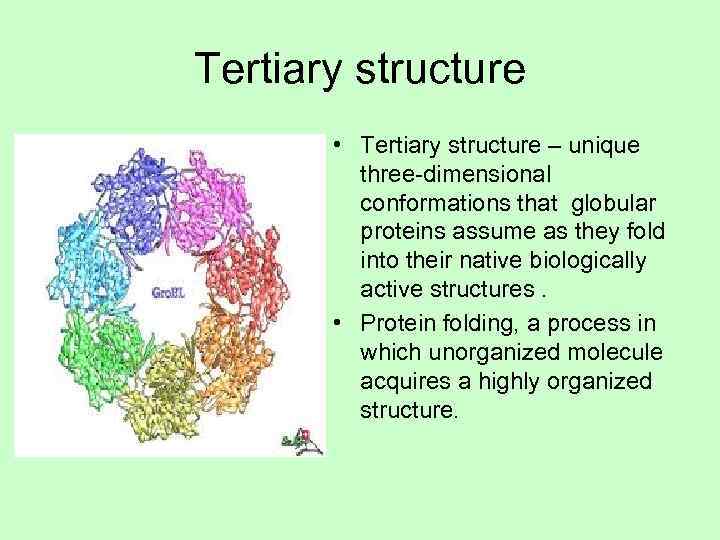
 Tertiary structure • Tertiary structure – unique three-dimensional conformations that globular proteins assume as they fold into their native biologically active structures. • Protein folding, a process in which unorganized molecule acquires a highly organized structure.
Tertiary structure • Tertiary structure – unique three-dimensional conformations that globular proteins assume as they fold into their native biologically active structures. • Protein folding, a process in which unorganized molecule acquires a highly organized structure.
 Tertiary structure has several important features: • 1. Many polypeptides fold in such a fashion that amino acid • 3. Large globular proteins residues that are distant from (more than 200 amino acid each other in the primary residues) often contain several structure come into close compact units called – proximity. DOMAINS. Domains are typically structurally • 2. Because of efficient packing as independent segments that the polypeptide chain folds, have specific functions globular proteins are compact. During this process, most water molecules are excluded from the protein’s interior making interactions between both polar and nonpolar groups possible.
Tertiary structure has several important features: • 1. Many polypeptides fold in such a fashion that amino acid • 3. Large globular proteins residues that are distant from (more than 200 amino acid each other in the primary residues) often contain several structure come into close compact units called – proximity. DOMAINS. Domains are typically structurally • 2. Because of efficient packing as independent segments that the polypeptide chain folds, have specific functions globular proteins are compact. During this process, most water molecules are excluded from the protein’s interior making interactions between both polar and nonpolar groups possible.
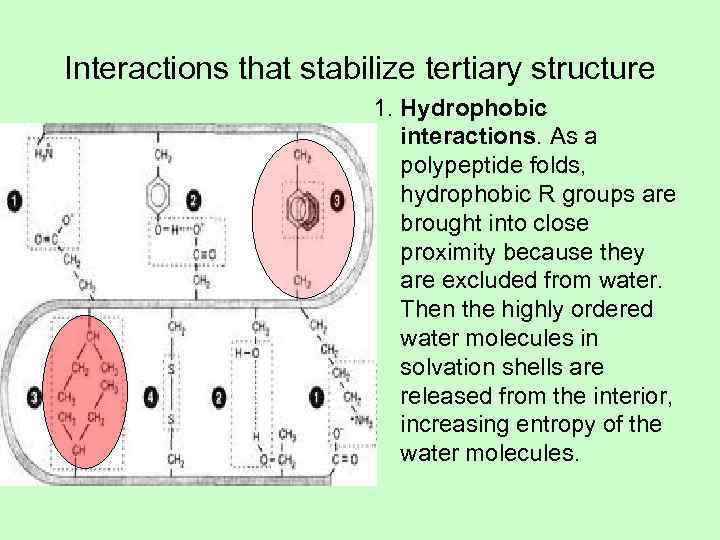
 Interactions that stabilize tertiary structure 1. Hydrophobic interactions. As a polypeptide folds, hydrophobic R groups are brought into close proximity because they are excluded from water. Then the highly ordered water molecules in solvation shells are released from the interior, increasing entropy of the water molecules.
Interactions that stabilize tertiary structure 1. Hydrophobic interactions. As a polypeptide folds, hydrophobic R groups are brought into close proximity because they are excluded from water. Then the highly ordered water molecules in solvation shells are released from the interior, increasing entropy of the water molecules.
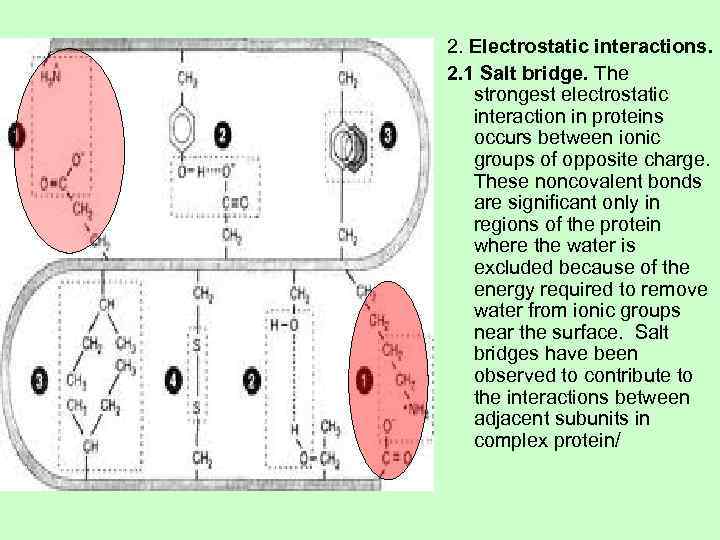
 2. Electrostatic interactions. 2. 1 Salt bridge. The strongest electrostatic interaction in proteins occurs between ionic groups of opposite charge. These noncovalent bonds are significant only in regions of the protein where the water is excluded because of the energy required to remove water from ionic groups near the surface. Salt bridges have been observed to contribute to the interactions between adjacent subunits in complex protein/
2. Electrostatic interactions. 2. 1 Salt bridge. The strongest electrostatic interaction in proteins occurs between ionic groups of opposite charge. These noncovalent bonds are significant only in regions of the protein where the water is excluded because of the energy required to remove water from ionic groups near the surface. Salt bridges have been observed to contribute to the interactions between adjacent subunits in complex protein/
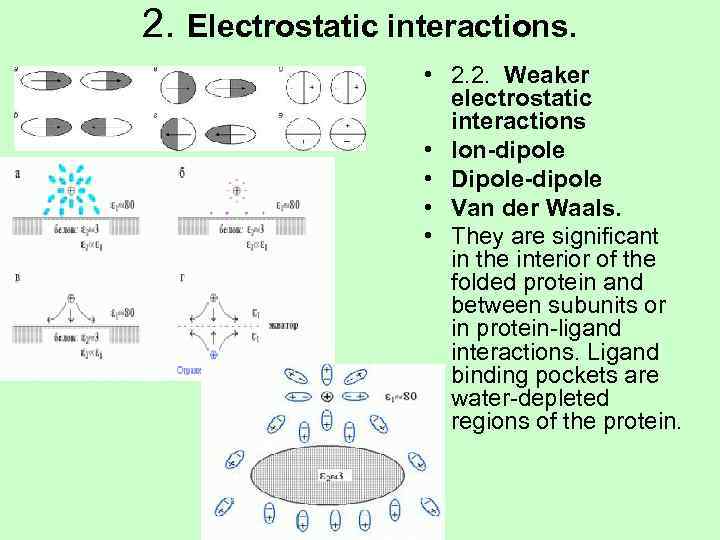
 2. Electrostatic interactions. • 2. 2. Weaker electrostatic interactions • Ion-dipole • Dipole-dipole • Van der Waals. • They are significant in the interior of the folded protein and between subunits or in protein-ligand interactions. Ligand binding pockets are water-depleted regions of the protein.
2. Electrostatic interactions. • 2. 2. Weaker electrostatic interactions • Ion-dipole • Dipole-dipole • Van der Waals. • They are significant in the interior of the folded protein and between subunits or in protein-ligand interactions. Ligand binding pockets are water-depleted regions of the protein.
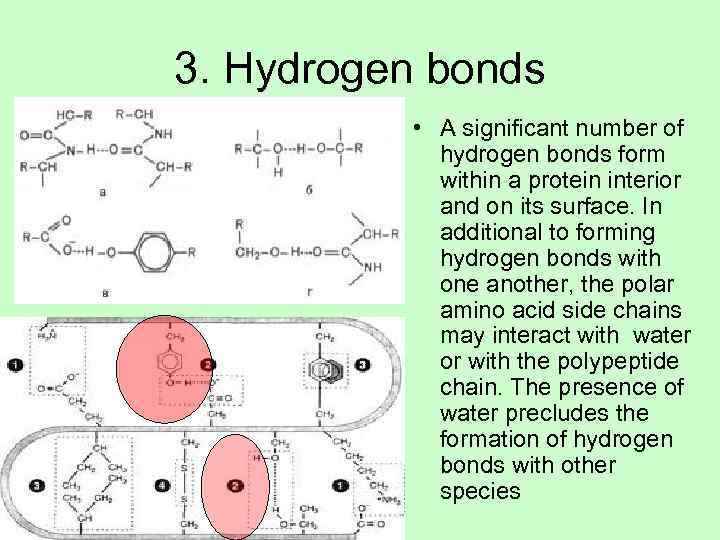
 3. Hydrogen bonds • A significant number of hydrogen bonds form within a protein interior and on its surface. In additional to forming hydrogen bonds with one another, the polar amino acid side chains may interact with water or with the polypeptide chain. The presence of water precludes the formation of hydrogen bonds with other species
3. Hydrogen bonds • A significant number of hydrogen bonds form within a protein interior and on its surface. In additional to forming hydrogen bonds with one another, the polar amino acid side chains may interact with water or with the polypeptide chain. The presence of water precludes the formation of hydrogen bonds with other species
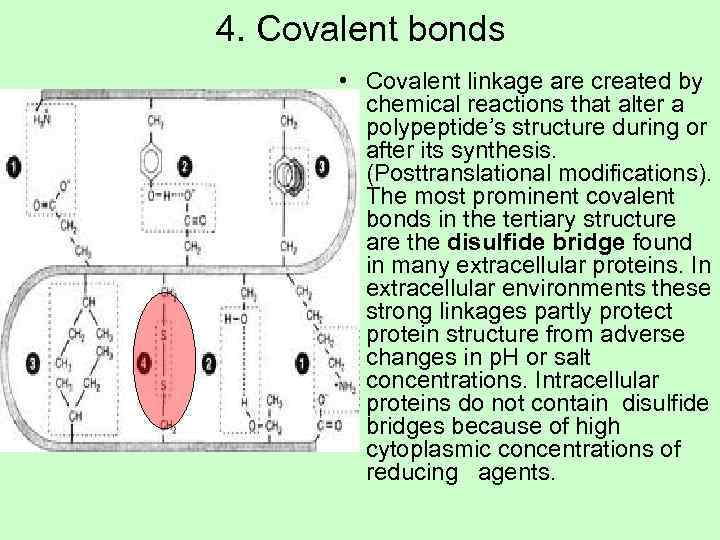
 4. Covalent bonds • Covalent linkage are created by chemical reactions that alter a polypeptide’s structure during or after its synthesis. (Posttranslational modifications). The most prominent covalent bonds in the tertiary structure are the disulfide bridge found in many extracellular proteins. In extracellular environments these strong linkages partly protect protein structure from adverse changes in p. H or salt concentrations. Intracellular proteins do not contain disulfide bridges because of high cytoplasmic concentrations of reducing agents.
4. Covalent bonds • Covalent linkage are created by chemical reactions that alter a polypeptide’s structure during or after its synthesis. (Posttranslational modifications). The most prominent covalent bonds in the tertiary structure are the disulfide bridge found in many extracellular proteins. In extracellular environments these strong linkages partly protect protein structure from adverse changes in p. H or salt concentrations. Intracellular proteins do not contain disulfide bridges because of high cytoplasmic concentrations of reducing agents.

