32f5b7e153bb5902b347b8fee3e2ccc5.ppt
- Количество слайдов: 36
 Protein Sequence Analysis - Overview NIH Proteomics Workshop 2007 Raja Mazumder Scientific Coordinator, PIR Research Assistant Professor, Department of Biochemistry and Molecular Biology Georgetown University Medical Center
Protein Sequence Analysis - Overview NIH Proteomics Workshop 2007 Raja Mazumder Scientific Coordinator, PIR Research Assistant Professor, Department of Biochemistry and Molecular Biology Georgetown University Medical Center
 Topics l l l Proteomics and protein bioinformatics (protein sequence analysis) Why do protein sequence analysis? Searching sequence databases Post-processing search results Detecting remote homologs
Topics l l l Proteomics and protein bioinformatics (protein sequence analysis) Why do protein sequence analysis? Searching sequence databases Post-processing search results Detecting remote homologs
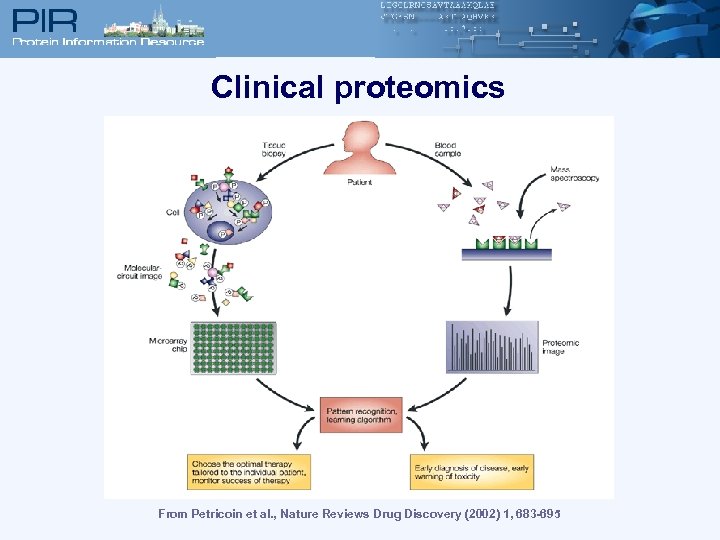 Clinical proteomics From Petricoin et al. , Nature Reviews Drug Discovery (2002) 1, 683 -695
Clinical proteomics From Petricoin et al. , Nature Reviews Drug Discovery (2002) 1, 683 -695
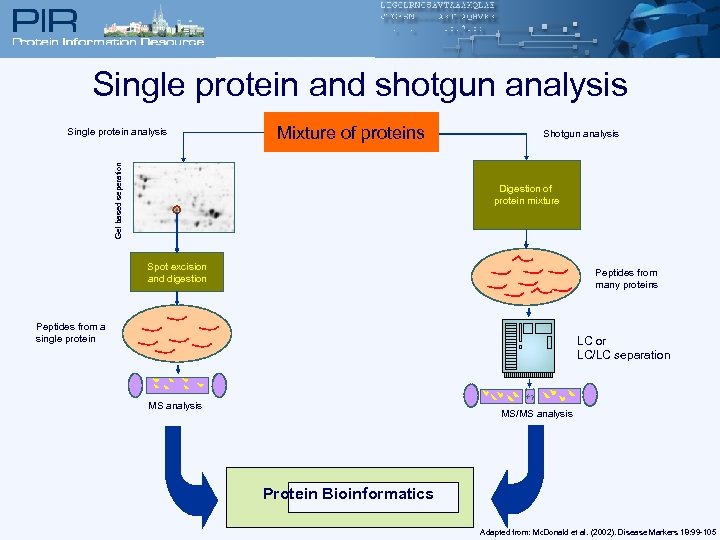 Single protein and shotgun analysis Mixture of proteins Gel based seperation Single protein analysis Shotgun analysis Digestion of protein mixture Spot excision and digestion Peptides from many proteins Peptides from a single protein LC or LC/LC separation MS analysis MS/MS analysis Protein Bioinformatics Adapted from: Mc. Donald et al. (2002). Disease Markers 18: 99 -105
Single protein and shotgun analysis Mixture of proteins Gel based seperation Single protein analysis Shotgun analysis Digestion of protein mixture Spot excision and digestion Peptides from many proteins Peptides from a single protein LC or LC/LC separation MS analysis MS/MS analysis Protein Bioinformatics Adapted from: Mc. Donald et al. (2002). Disease Markers 18: 99 -105
 Protein bioinformatics: protein sequence analysis l Helps characterize protein sequences in silico and allows prediction of protein structure and function l Statistically significant BLAST hits usually signifies sequence homology l Homologous sequences may or may not have the same function but would always (very few exceptions) have the same structural fold l Protein sequence analysis allows protein classification
Protein bioinformatics: protein sequence analysis l Helps characterize protein sequences in silico and allows prediction of protein structure and function l Statistically significant BLAST hits usually signifies sequence homology l Homologous sequences may or may not have the same function but would always (very few exceptions) have the same structural fold l Protein sequence analysis allows protein classification
 Development of protein sequence databases l Atlas of protein sequence and structure – Dayhoff (1966) first sequence database (pre-bioinformatics). Currently known as Protein Information Resource (PIR) l Protein data bank (PDB) – structural database (1972) remains most widely used database of structures l Uni. Prot – The Universal Protein Resource (2003) is a central database of protein sequence and function created by joining the forces of the Swiss-Prot, Tr. EMBL and PIR protein database activities
Development of protein sequence databases l Atlas of protein sequence and structure – Dayhoff (1966) first sequence database (pre-bioinformatics). Currently known as Protein Information Resource (PIR) l Protein data bank (PDB) – structural database (1972) remains most widely used database of structures l Uni. Prot – The Universal Protein Resource (2003) is a central database of protein sequence and function created by joining the forces of the Swiss-Prot, Tr. EMBL and PIR protein database activities
 Comparative protein sequence analysis and evolution l Patterns of conservation in sequences allows us to determine which residues are under selective constraint (and thus likely important for protein function) l Comparative analysis of proteins is more sensitive than comparing DNA l Homologous proteins have a common ancestor l Different proteins evolve at different rates l Protein classification systems based on evolution: PIRSF and COG
Comparative protein sequence analysis and evolution l Patterns of conservation in sequences allows us to determine which residues are under selective constraint (and thus likely important for protein function) l Comparative analysis of proteins is more sensitive than comparing DNA l Homologous proteins have a common ancestor l Different proteins evolve at different rates l Protein classification systems based on evolution: PIRSF and COG
 PIRSF and large-scale annotation of proteins l PIRSF is a protein classification system based on the evolutionary relationships of whole proteins l As part of the Uni. Prot project, PIR has developed this classification strategy to assist in the propagation and standardization of protein annotation
PIRSF and large-scale annotation of proteins l PIRSF is a protein classification system based on the evolutionary relationships of whole proteins l As part of the Uni. Prot project, PIR has developed this classification strategy to assist in the propagation and standardization of protein annotation
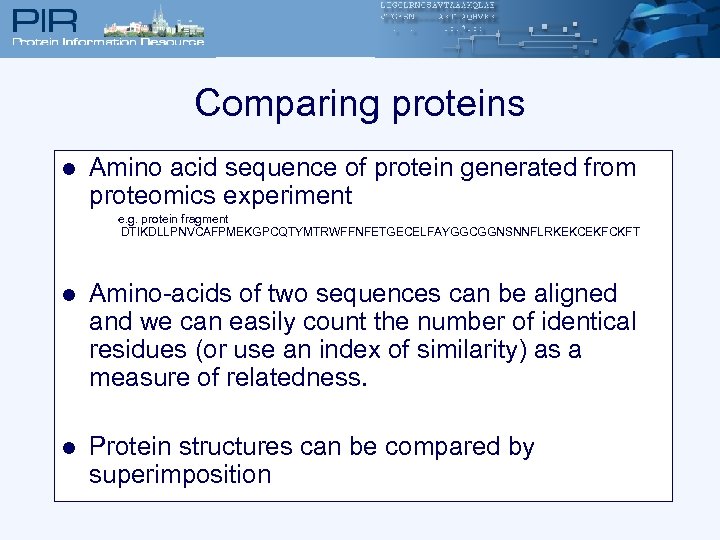 Comparing proteins l Amino acid sequence of protein generated from proteomics experiment e. g. protein fragment DTIKDLLPNVCAFPMEKGPCQTYMTRWFFNFETGECELFAYGGCGGNSNNFLRKEKCEKFCKFT l Amino-acids of two sequences can be aligned and we can easily count the number of identical residues (or use an index of similarity) as a measure of relatedness. l Protein structures can be compared by superimposition
Comparing proteins l Amino acid sequence of protein generated from proteomics experiment e. g. protein fragment DTIKDLLPNVCAFPMEKGPCQTYMTRWFFNFETGECELFAYGGCGGNSNNFLRKEKCEKFCKFT l Amino-acids of two sequences can be aligned and we can easily count the number of identical residues (or use an index of similarity) as a measure of relatedness. l Protein structures can be compared by superimposition
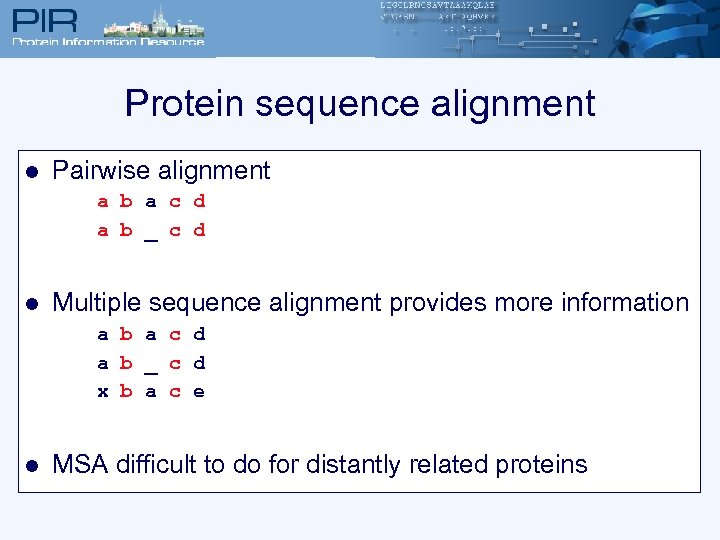 Protein sequence alignment l Pairwise alignment a b a c d a b _ c d l Multiple sequence alignment provides more information a b a c d a b _ c d x b a c e l MSA difficult to do for distantly related proteins
Protein sequence alignment l Pairwise alignment a b a c d a b _ c d l Multiple sequence alignment provides more information a b a c d a b _ c d x b a c e l MSA difficult to do for distantly related proteins
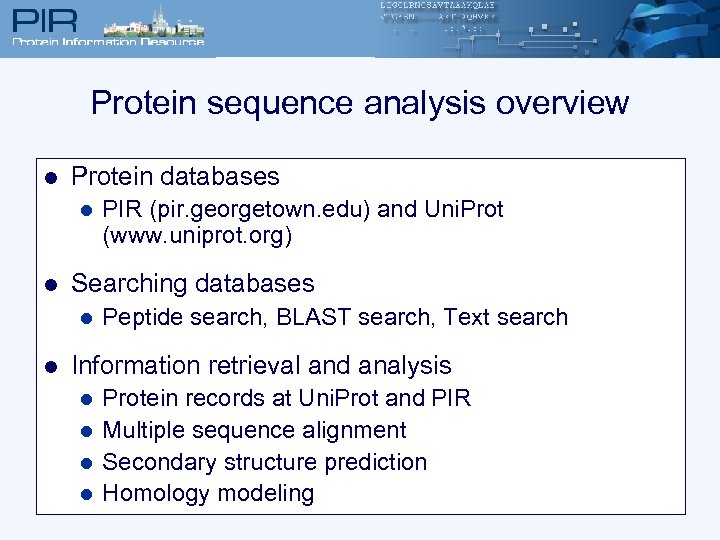 Protein sequence analysis overview l Protein databases l l Searching databases l l PIR (pir. georgetown. edu) and Uni. Prot (www. uniprot. org) Peptide search, BLAST search, Text search Information retrieval and analysis l l Protein records at Uni. Prot and PIR Multiple sequence alignment Secondary structure prediction Homology modeling
Protein sequence analysis overview l Protein databases l l Searching databases l l PIR (pir. georgetown. edu) and Uni. Prot (www. uniprot. org) Peptide search, BLAST search, Text search Information retrieval and analysis l l Protein records at Uni. Prot and PIR Multiple sequence alignment Secondary structure prediction Homology modeling
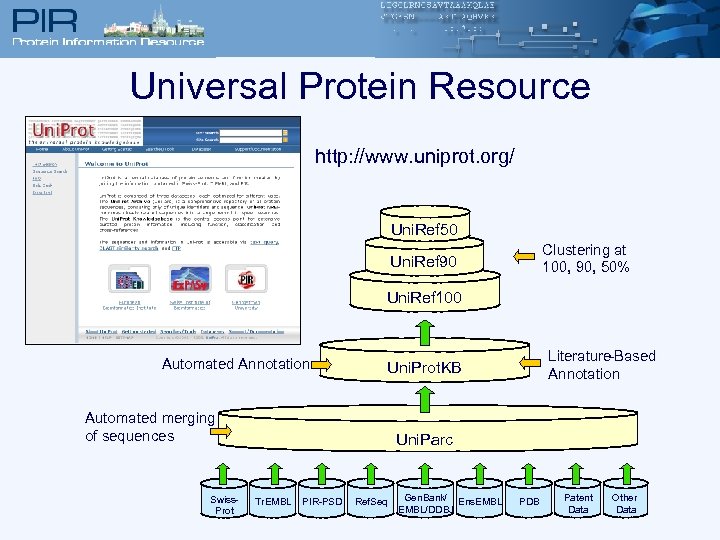 Universal Protein Resource http: //www. uniprot. org/ Uni. Ref 50 Clustering at 100, 90, 50% Uni. Ref 90 Uni. Prot NREF Uni. Ref 100 Automated Annotation Automated merging of sequences Swiss. Prot Literature-Based Annotation Uni. Prot Knowledgebase Uni. Prot. KB Uni. Prot Archive Uni. Parc Tr. EMBL PIR-PSD Ref. Seq Gen. Bank/ Ens. EMBL/DDBJ PDB Patent Data Other Data
Universal Protein Resource http: //www. uniprot. org/ Uni. Ref 50 Clustering at 100, 90, 50% Uni. Ref 90 Uni. Prot NREF Uni. Ref 100 Automated Annotation Automated merging of sequences Swiss. Prot Literature-Based Annotation Uni. Prot Knowledgebase Uni. Prot. KB Uni. Prot Archive Uni. Parc Tr. EMBL PIR-PSD Ref. Seq Gen. Bank/ Ens. EMBL/DDBJ PDB Patent Data Other Data
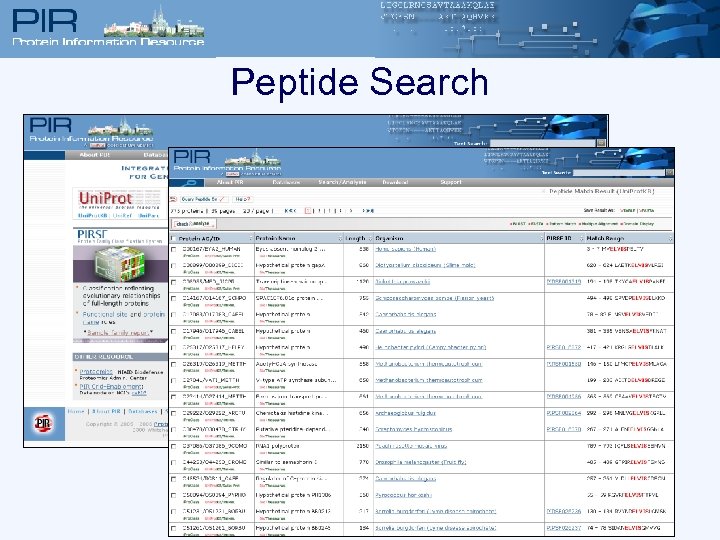 Peptide Search
Peptide Search
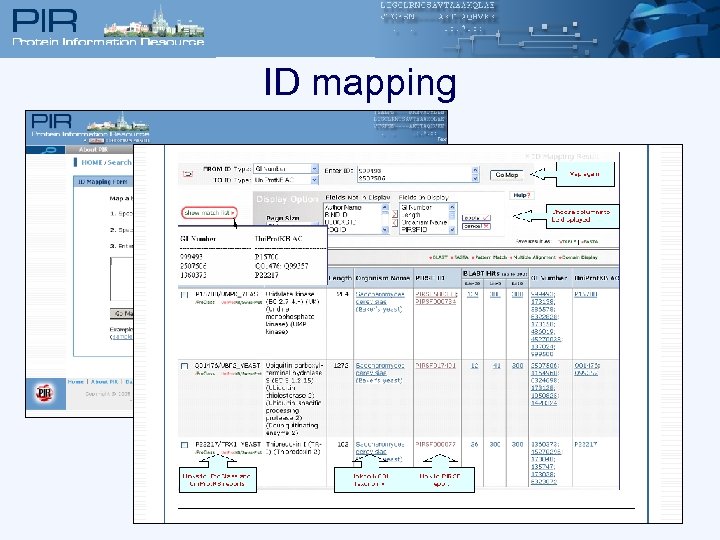 ID mapping
ID mapping
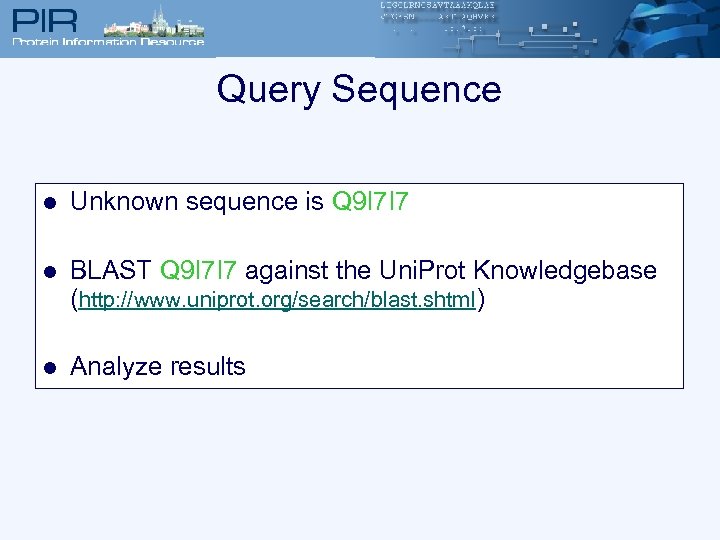 Query Sequence l Unknown sequence is Q 9 I 7 I 7 l BLAST Q 9 I 7 I 7 against the Uni. Prot Knowledgebase (http: //www. uniprot. org/search/blast. shtml) l Analyze results
Query Sequence l Unknown sequence is Q 9 I 7 I 7 l BLAST Q 9 I 7 I 7 against the Uni. Prot Knowledgebase (http: //www. uniprot. org/search/blast. shtml) l Analyze results
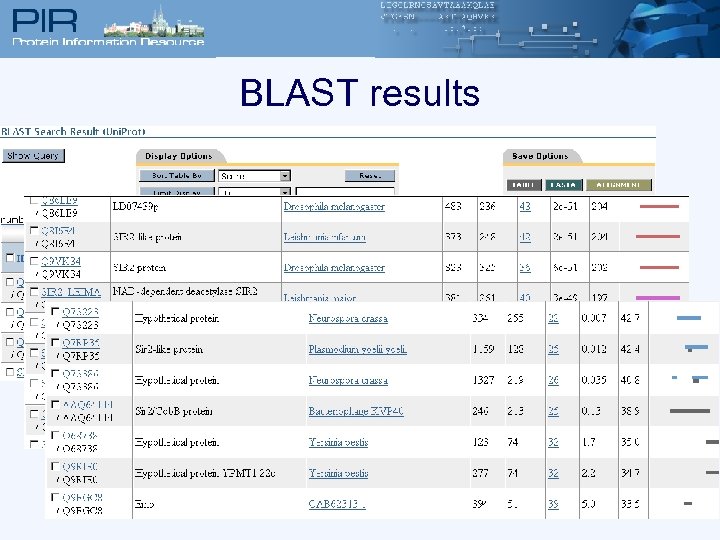 BLAST results
BLAST results
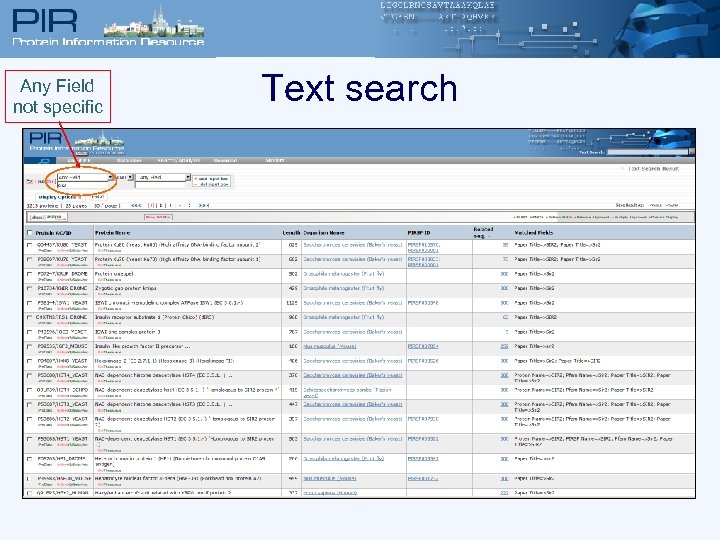 Any Field not specific Text search
Any Field not specific Text search
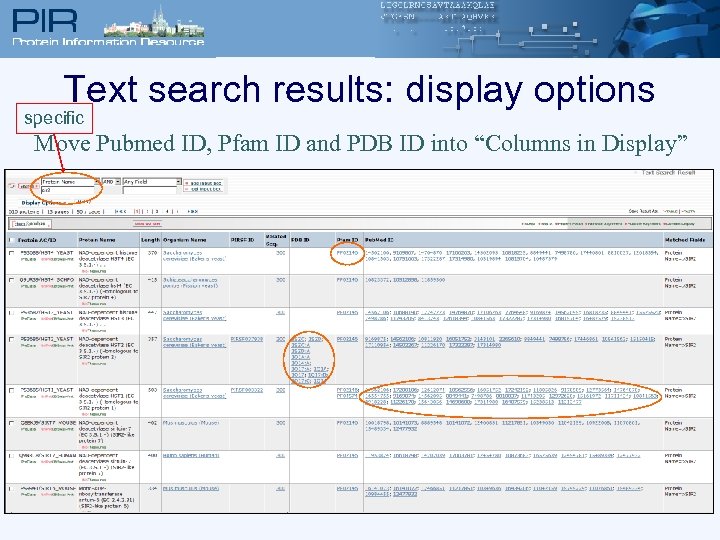 Text search results: display options specific Move Pubmed ID, Pfam ID and PDB ID into “Columns in Display”
Text search results: display options specific Move Pubmed ID, Pfam ID and PDB ID into “Columns in Display”
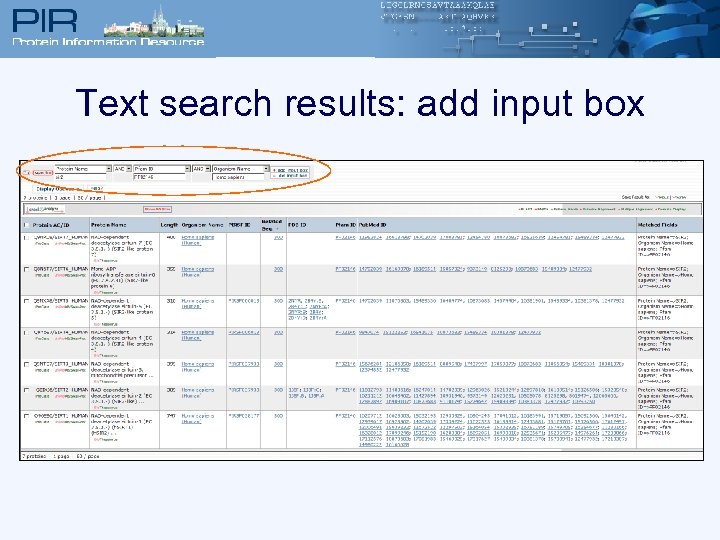 Text search results: add input box
Text search results: add input box
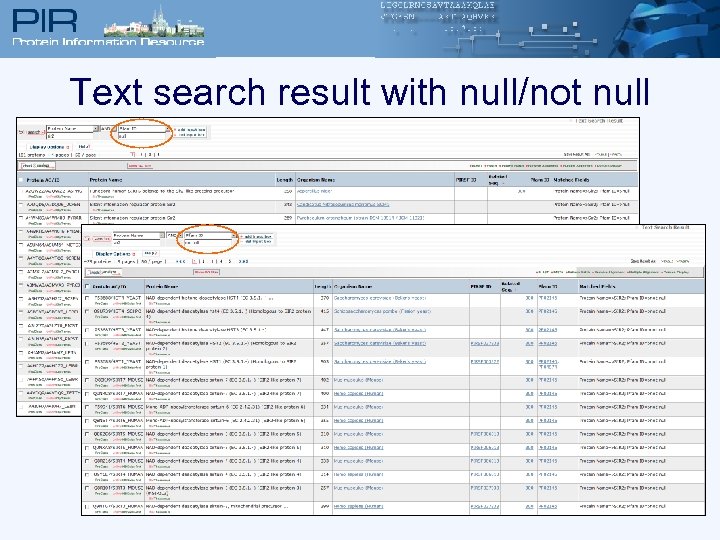 Text search result with null/not null
Text search result with null/not null
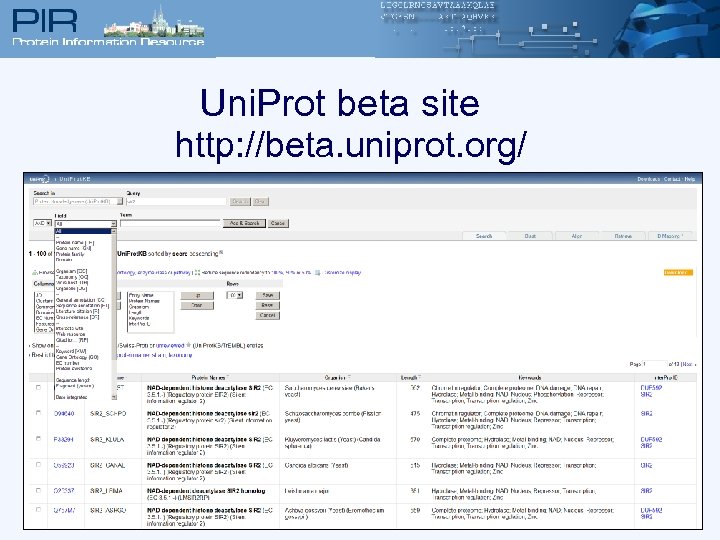 Uni. Prot beta site http: //beta. uniprot. org/
Uni. Prot beta site http: //beta. uniprot. org/
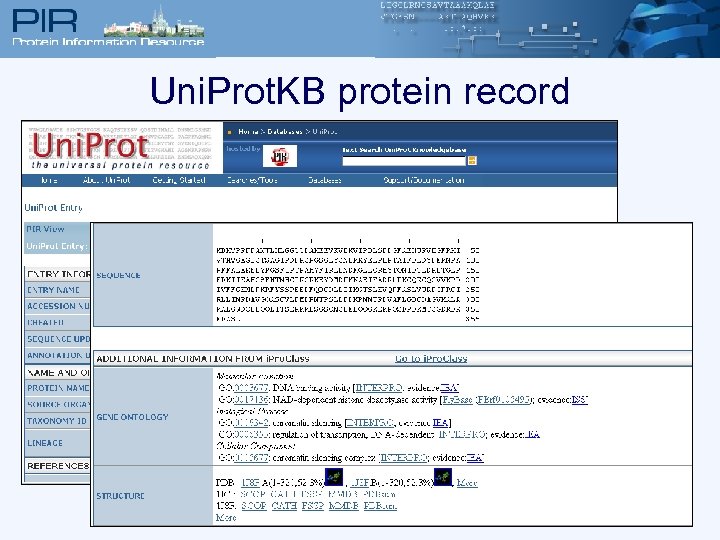 Uni. Prot. KB protein record
Uni. Prot. KB protein record
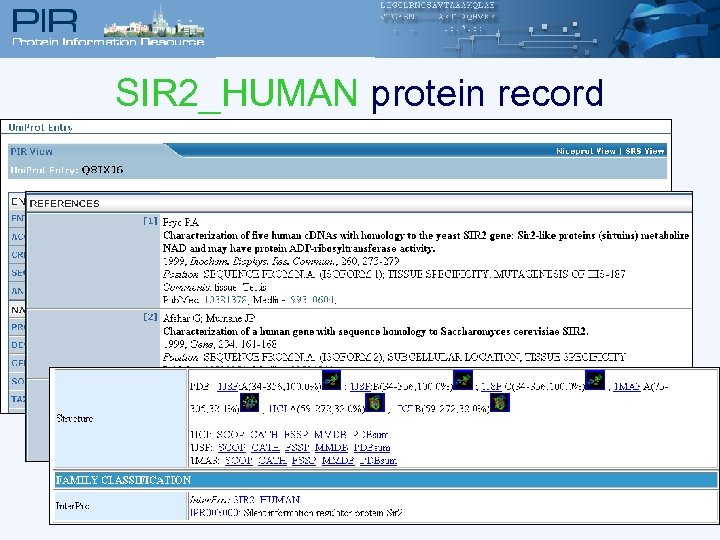 SIR 2_HUMAN protein record
SIR 2_HUMAN protein record
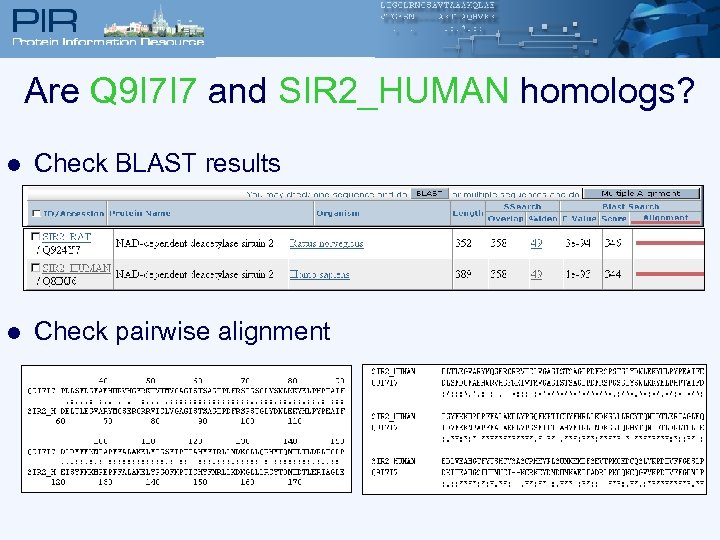 Are Q 9 I 7 I 7 and SIR 2_HUMAN homologs? l Check BLAST results l Check pairwise alignment
Are Q 9 I 7 I 7 and SIR 2_HUMAN homologs? l Check BLAST results l Check pairwise alignment
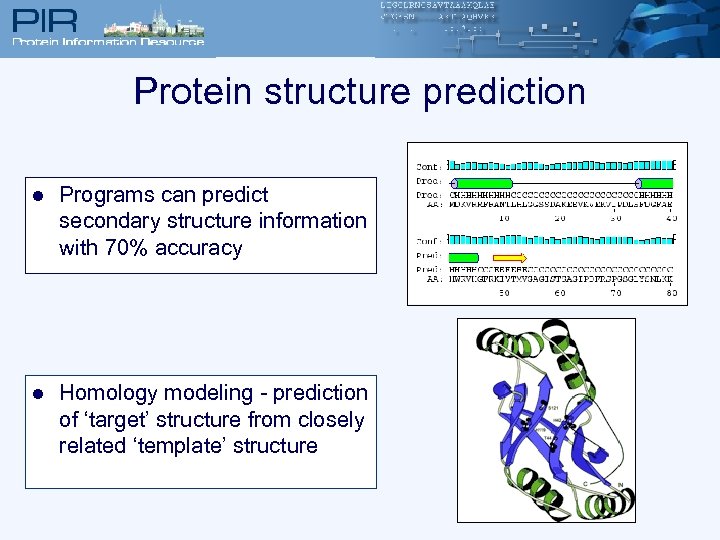 Protein structure prediction l Programs can predict secondary structure information with 70% accuracy l Homology modeling - prediction of ‘target’ structure from closely related ‘template’ structure
Protein structure prediction l Programs can predict secondary structure information with 70% accuracy l Homology modeling - prediction of ‘target’ structure from closely related ‘template’ structure
 Secondary structure prediction http: //bioinf. cs. ucl. ac. uk/psipred/
Secondary structure prediction http: //bioinf. cs. ucl. ac. uk/psipred/
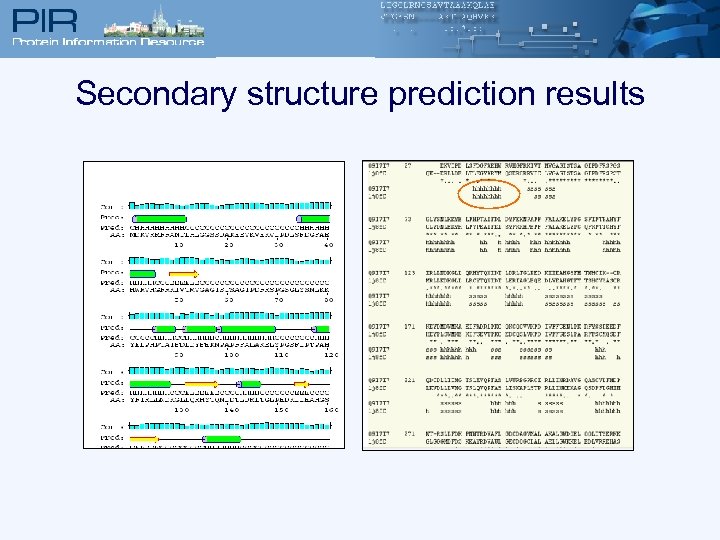 Secondary structure prediction results
Secondary structure prediction results
 Sir 2 structure
Sir 2 structure
 Homology modeling http: //www. expasy. org/swissmod/SWISS-MODEL. html
Homology modeling http: //www. expasy. org/swissmod/SWISS-MODEL. html
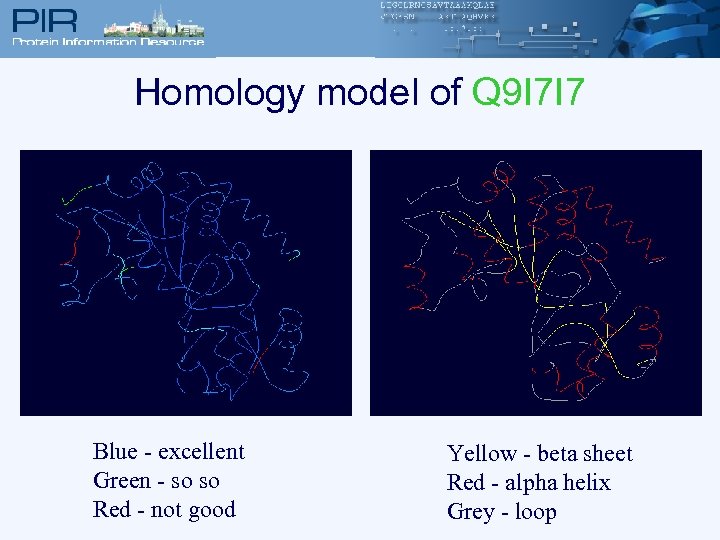 Homology model of Q 9 I 7 I 7 Blue - excellent Green - so so Red - not good Yellow - beta sheet Red - alpha helix Grey - loop
Homology model of Q 9 I 7 I 7 Blue - excellent Green - so so Red - not good Yellow - beta sheet Red - alpha helix Grey - loop
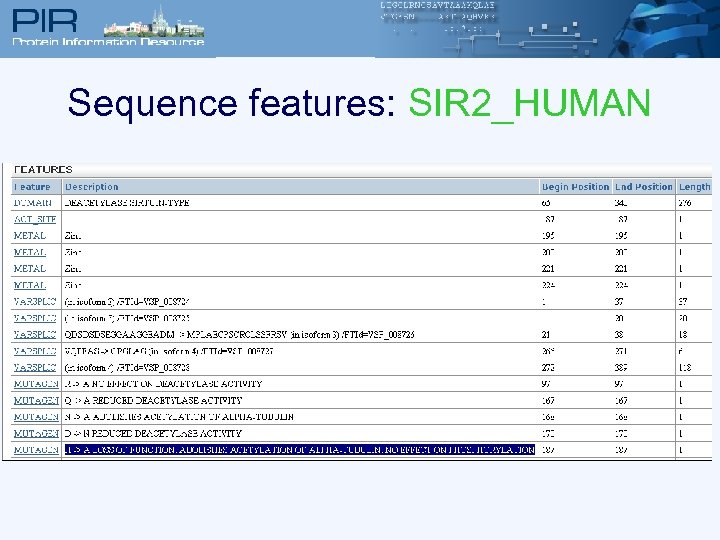 Sequence features: SIR 2_HUMAN
Sequence features: SIR 2_HUMAN
 Multiple sequence alignment
Multiple sequence alignment
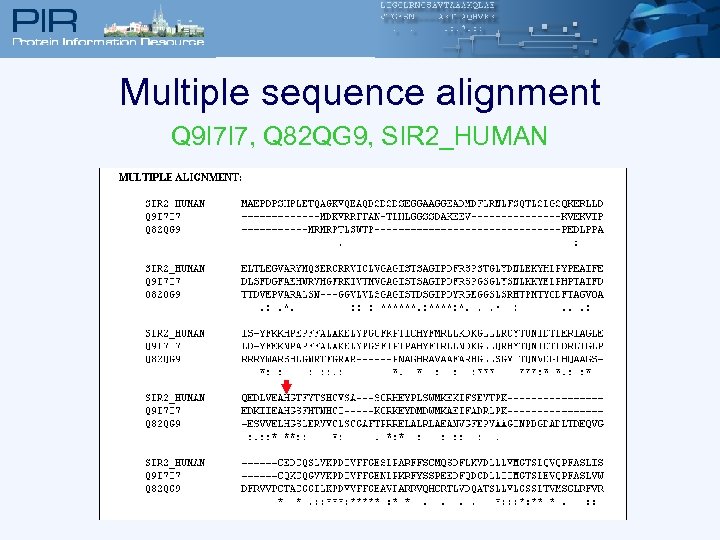 Multiple sequence alignment Q 9 I 7 I 7, Q 82 QG 9, SIR 2_HUMAN
Multiple sequence alignment Q 9 I 7 I 7, Q 82 QG 9, SIR 2_HUMAN
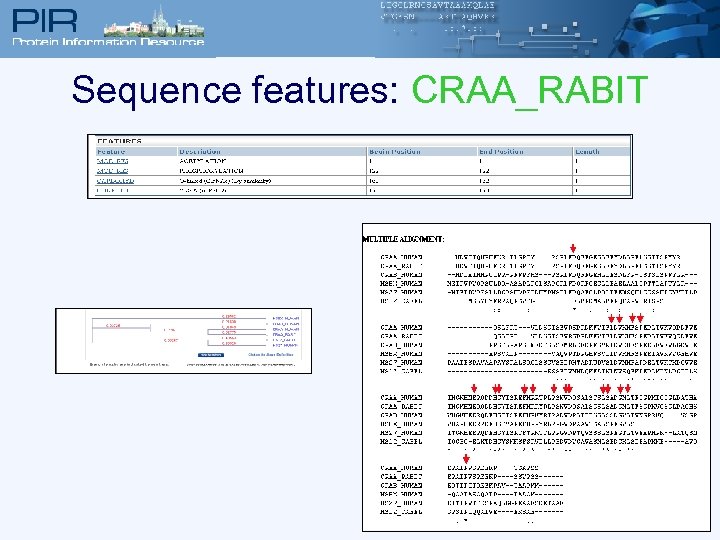 Sequence features: CRAA_RABIT
Sequence features: CRAA_RABIT
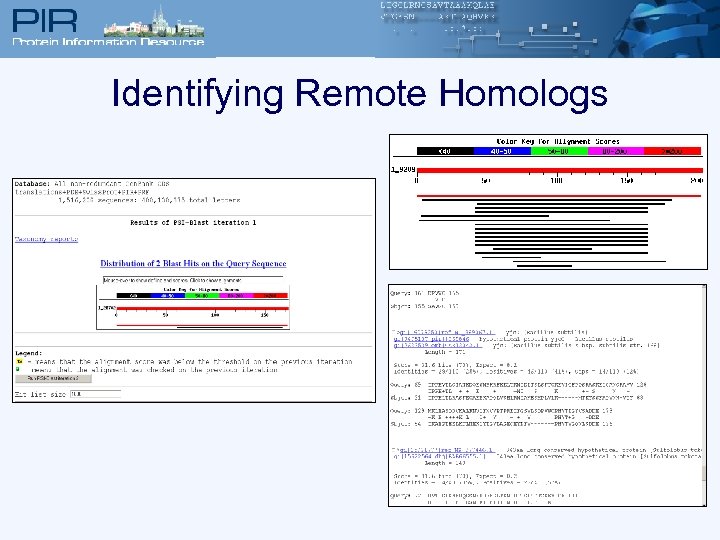 Identifying Remote Homologs
Identifying Remote Homologs
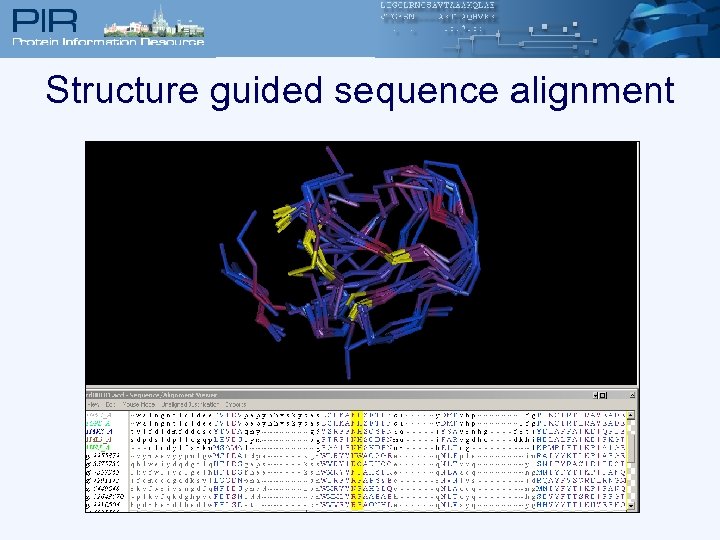 Structure guided sequence alignment
Structure guided sequence alignment


