acd1350cffccce78a51d727c7b946846.ppt
- Количество слайдов: 56

Protein NMR Spectroscopy Yuan-Chao Lou 羅元超 Institute of Biomedical Sciences Academia Sinica

Outline A. Classical Protein NMR Spectroscopy (1). 2 D HSQC spectra (2). 3 D Triple-Resonance Spectra (3). Resonance Assignment (4). Nuclear Overhauser Effect (NOE) (5). Dihedral Angle and J Coupling Constant (6). H-bond and Amide Proton Exchange Rate B. Software for Structure Determination (1). X-PLOR, CNS (2). CYANA, Aria
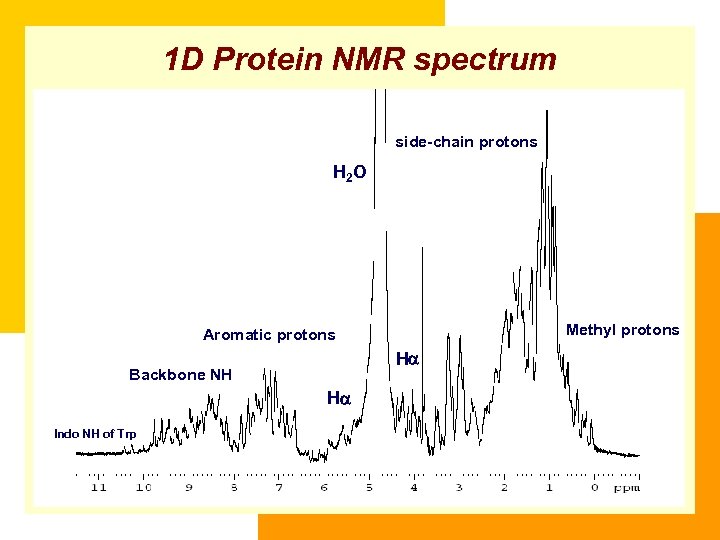
1 D Protein NMR spectrum side-chain protons H 2 O Methyl protons Aromatic protons H Backbone NH H Indo NH of Trp
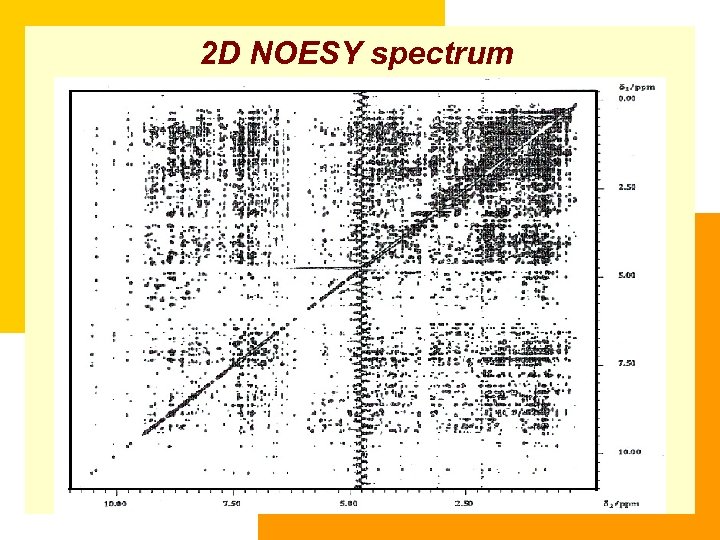
2 D NOESY spectrum

Isotope Labeling 1. 2 D homonuclear NMR spectra are too crowded to make assignment when your protein size is more than 100 amino acids. 2. 3. Stable isotope labeling (normally 15 N and 13 C) can provide additional dimensions to separate the signals. 4. Besides, the large coupling constants between these labeled nuclei facilitate the acquisition of triple resonance NMR spectra.
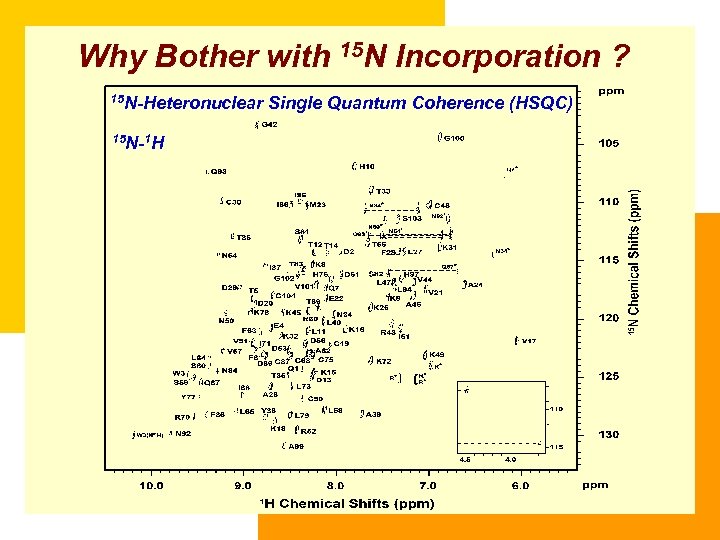
Why Bother with 15 N Incorporation ? 15 N-Heteronuclear 15 N-1 H Single Quantum Coherence (HSQC)
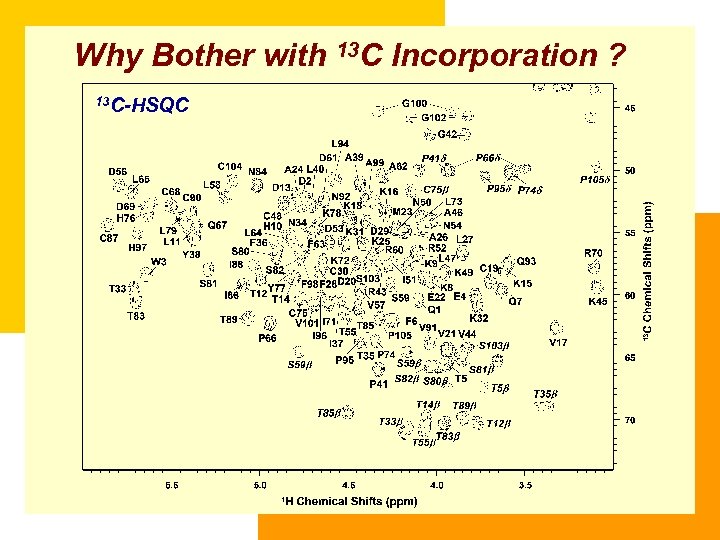
Why Bother with 13 C Incorporation ? 13 C-HSQC
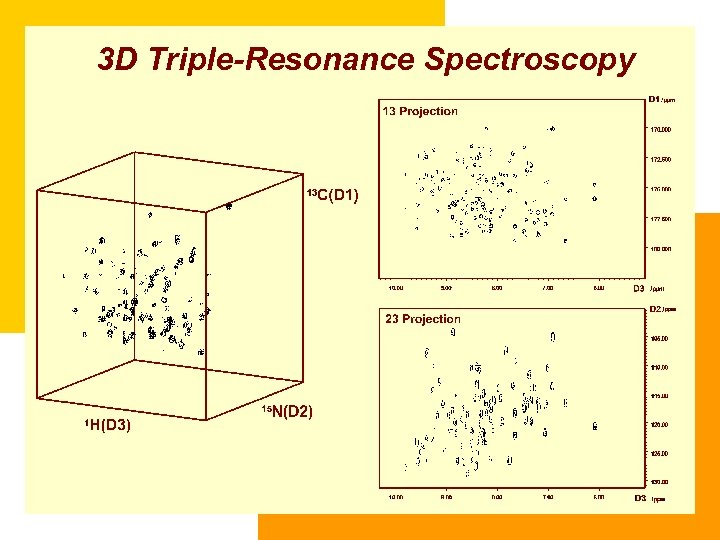
3 D Triple-Resonance Spectroscopy
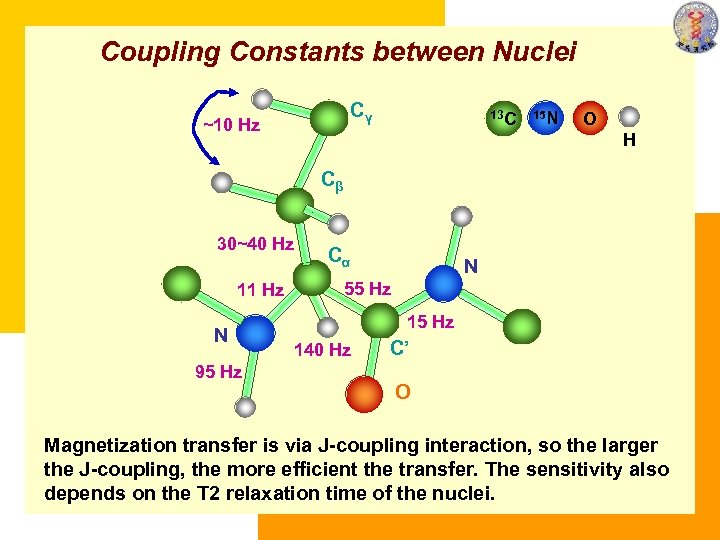
Coupling Constants between Nuclei Cγ ~10 Hz 13 C 15 N O H Cβ 30~40 Hz 11 Hz N 95 Hz Cα N 55 Hz 140 Hz C’ O Magnetization transfer is via J-coupling interaction, so the larger the J-coupling, the more efficient the transfer. The sensitivity also depends on the T 2 relaxation time of the nuclei. 140 Hz

Triple (1 H, 13 C, and 15 N) Resonance Experiments Through-bond experiments: (a) HNCA and HN(CO)CA (b) HNCACB and CBCA(CO)NH (c) HNCO and HN(CA)CO (d) C(CO)NH and HCC(CO)NH (e) HCCH-TOCSY
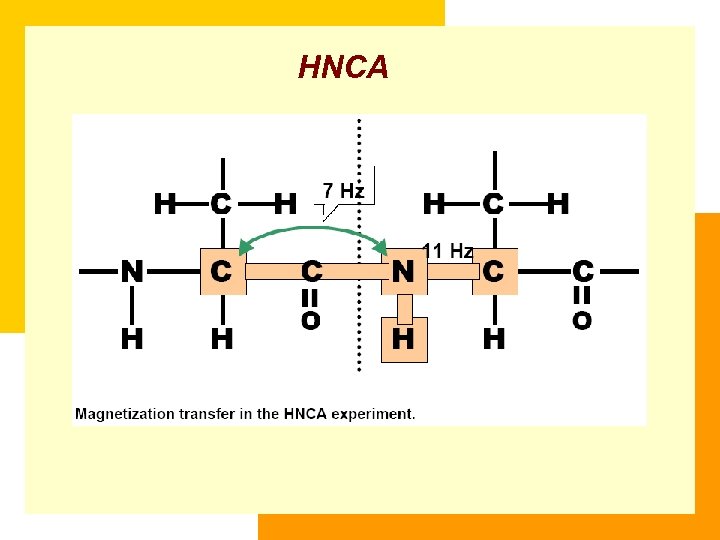
HNCA
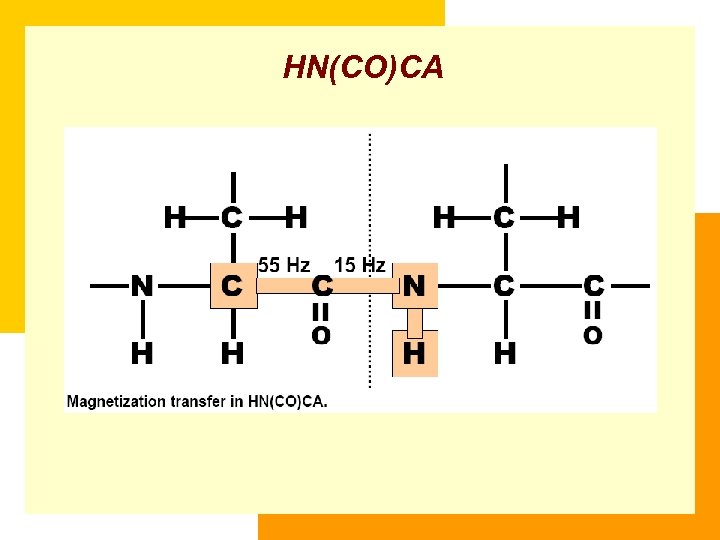
HN(CO)CA
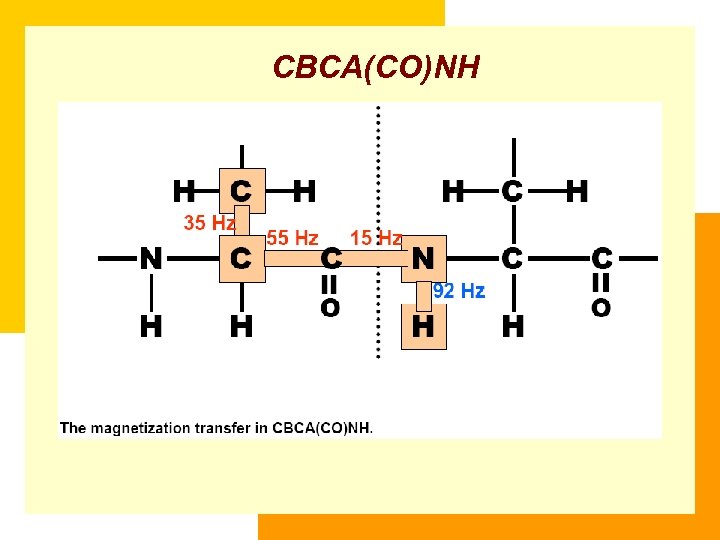
CBCA(CO)NH
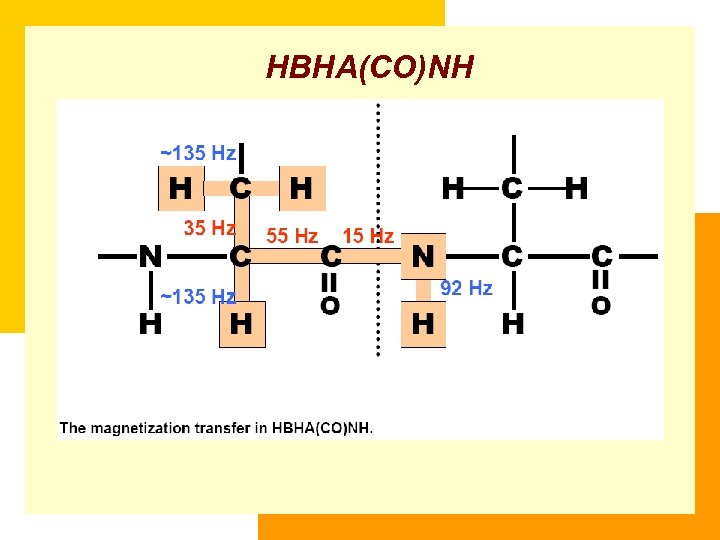
HBHA(CO)NH
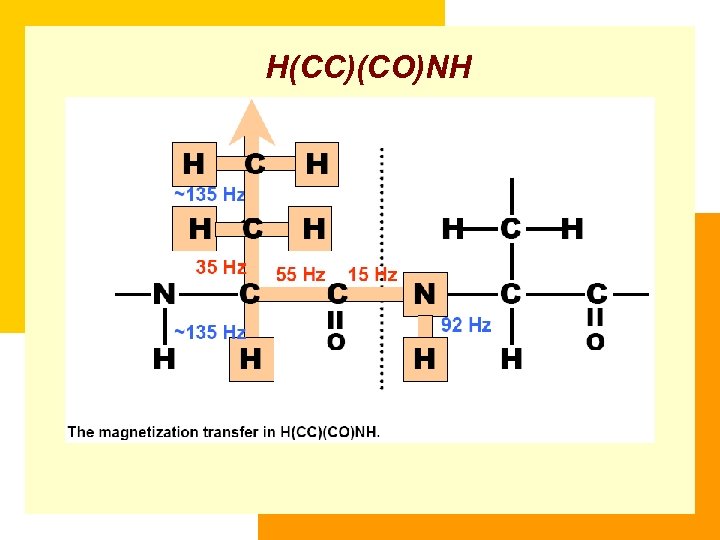
H(CC)(CO)NH
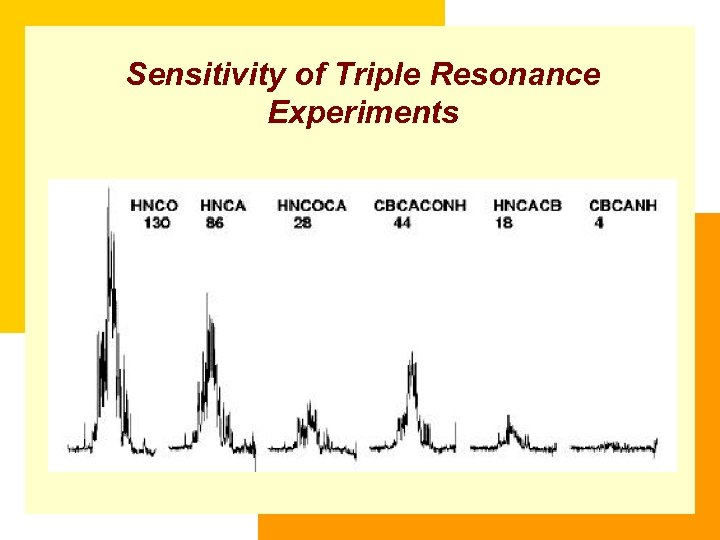
Sensitivity of Triple Resonance Experiments
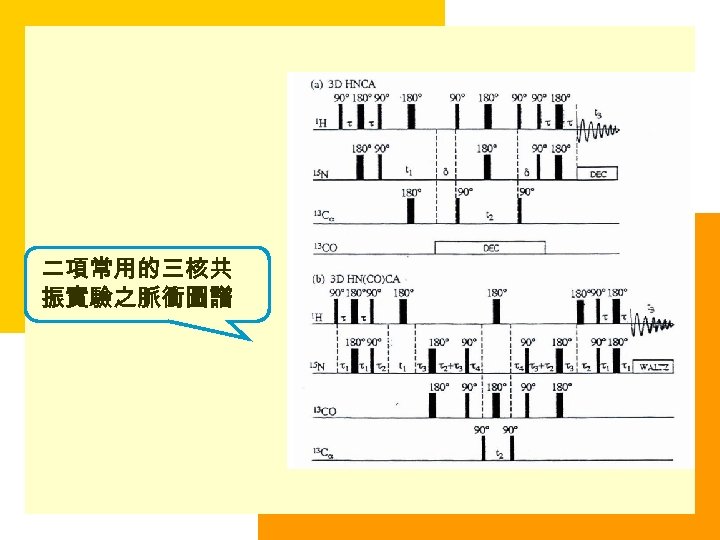
二項常用的三核共 振實驗之脈衝圖譜

NMR Resonance Assignments Using Triple Resonance Experiments (1) Carry out HNCACB, CBCA(CO)NH, HNCO, and HNCACO. HNCA and HN(CO)CA may also be needed. (2) Sequential backbone assignments: a. Identify those residues that have unique chemical shifts. For example, C of Thr, Ile, Val, and Pro (> 60 ppm); C of Thr, and Ser (> 60 ppm) b. C of Ala (~ 50 ppm), C of Gly (~ 45 ppm), and C of Pro (~ 50 ppm) c. C of Leu, Asp, Asn, Ile, Phe, and Tyr (36~ 43 ppm); C of Pro and Val (30 ~ 35 ppm) d. Others (3) Using C(CO)NH or HCC(CO)NH etc to distinguish AMX residues from the long-chain residues.
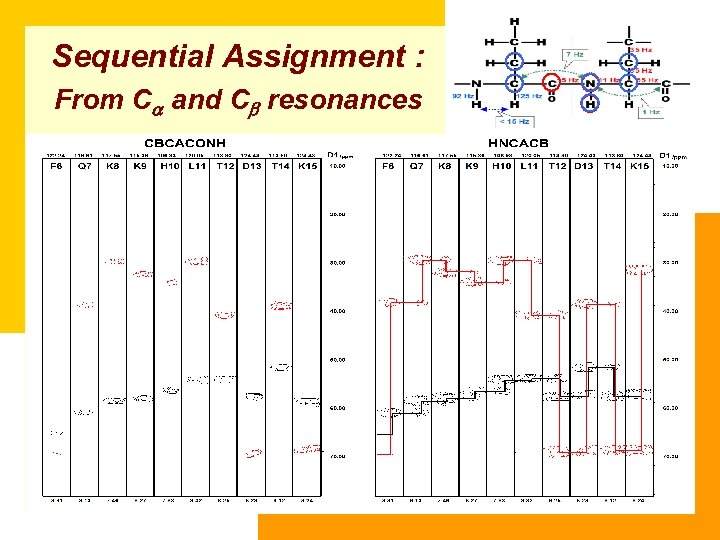
Sequential Assignment : From Ca and Cb resonances
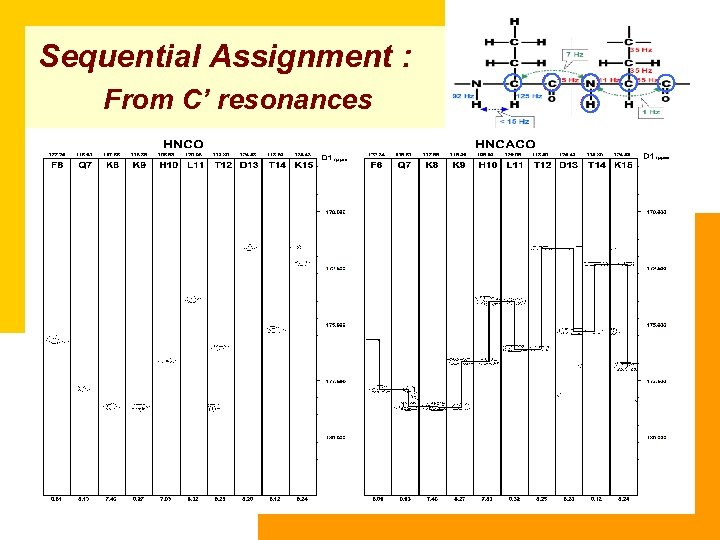
Sequential Assignment : From C’ resonances
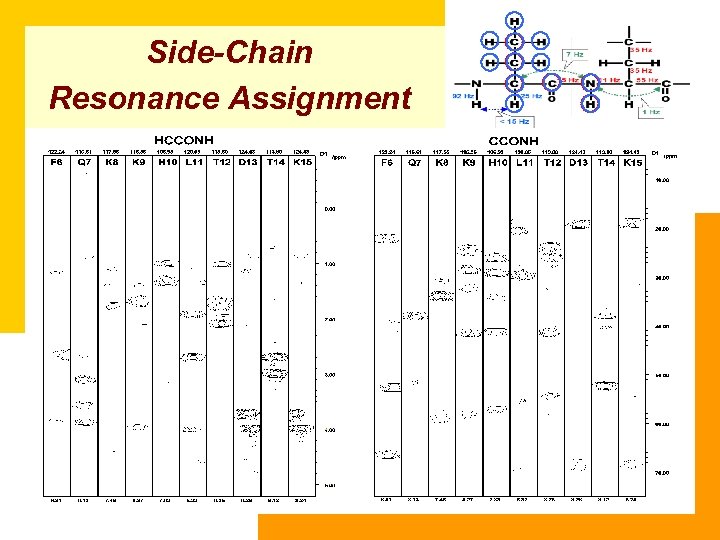
Side-Chain Resonance Assignment
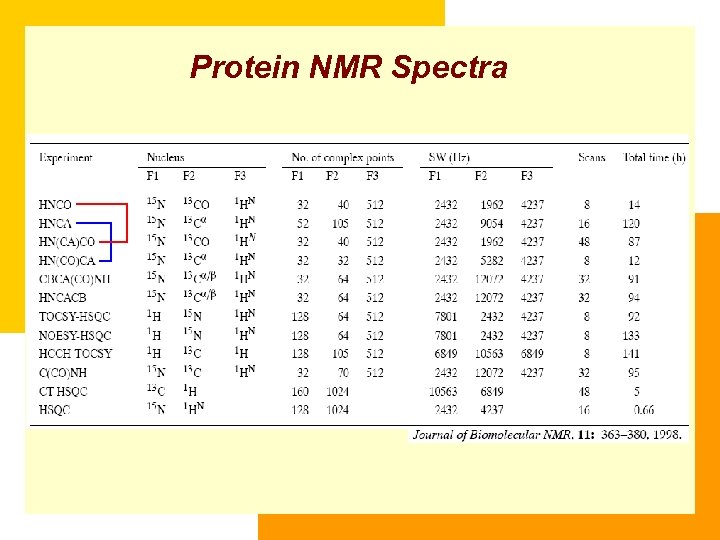
Protein NMR Spectra
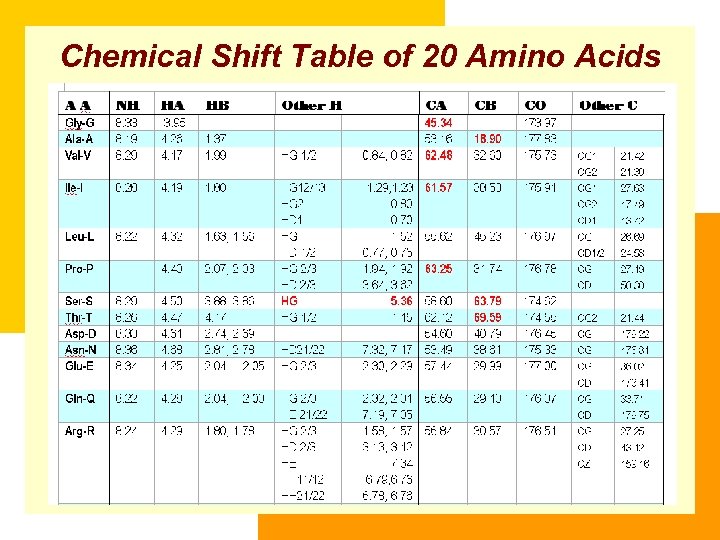
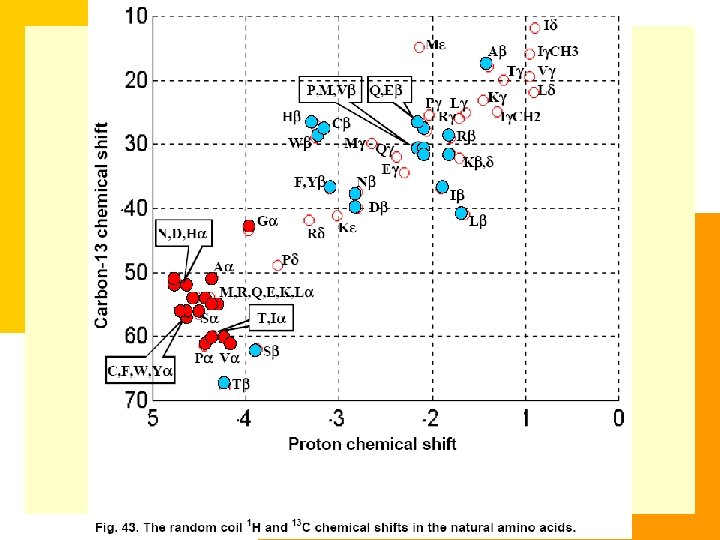
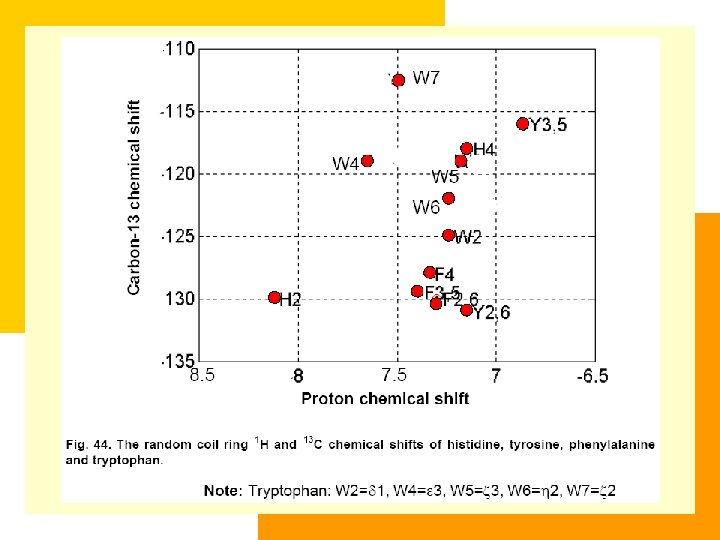
Chemical Shift Table of 20 Amino Acids
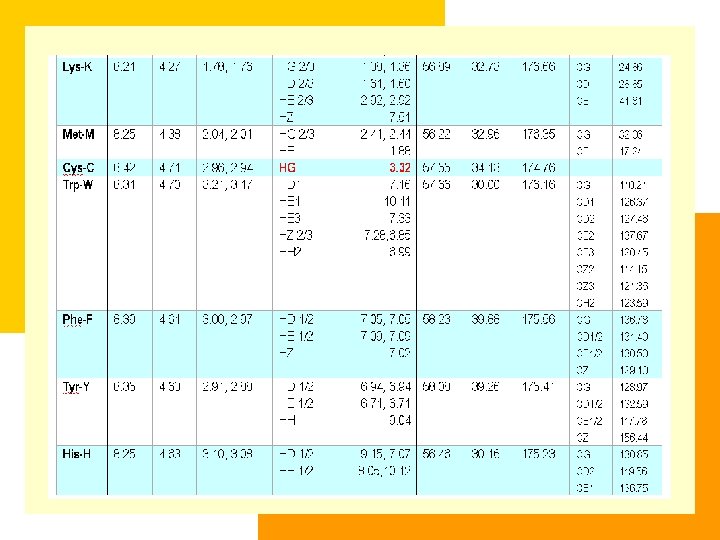
The Completeness of Assignment is an Determinant for NOESY Assignment residue N C C C other Q 1 124. 279 (8. 379) 175. 880 60. 337 (4. 111) 27. 906 (2. 834, 2. 302) C , 31. 404 (2. 644, 2. 644) D 2 114. 136 (7. 959) 174. 728 52. 070 (4. 746) 42. 126 (3. 154, 3. 154) W 3 124. 678 (9. 602) 178. 494 58. 251 (5. 635) 32. 468 (3. 526, 3. 317) C 1, 128. 925 (7. 384); C 3, 124. 926 (8. 290); C 2, 123. 330 (7. 286); C 2, 114. 589 (7. 308); C 3, 120. 036 (6. 811); N 1, 129. 962 (10. 193) E 4 120. 757 (8. 707) 178. 990 59. 771 (3. 782) 27. 665 (2. 021, 2. 021) C , 34. 591 (2. 422, 2. 200) T 5 118. 196 (8. 910) 175. 760 65. 742 (3. 942) 67. 089 (3. 739) C 2, 22. 548 (1. 248) F 6 122. 999 (8. 796) 177. 890 61. 930 (4. 228) 39. 135 (3. 615, 3. 171) C 1, 132. 878 (7. 165); C 2, 132. 878 (7. 165); C 1, 129. 972 (7. 024); C 2, 129. 972 (7. 024); C , 128. 127 (6. 834) Q 7 117. 315 (8. 118) 178. 649 59. 520 (3. 647) 30. 229 (1. 193, 1. 193) C , 34. 492 (1. 851, 1. 851); N 2, 107. 564 (6. 195, 4. 472) K 8 118. 131 (7. 449) 178. 680 58. 946 (4. 036) 32. 895 (1. 807, 1. 770) C , 25. 188 (1. 487, 1. 487); C , 29. 128 (1. 710, 1. 710); C , 42. 023 (2. 944, 2. 944) K 9 115. 307 (8. 261) 176. 634 57. 489 (4. 167) 34. 724 (1. 626, 1. 626) C , 26. 475 (1. 090, 1. 090); C , 29. 549 (1. 398, 1. 398); C , 41. 720 (2. 882, 2. 882) H 10 106. 834 (7. 803) 173. 998 55. 370 (4. 846) 30. 286 (2. 767, 2. 000) C 2, 122. 074 (6. 786); C 1, 137. 835 (8. 755)
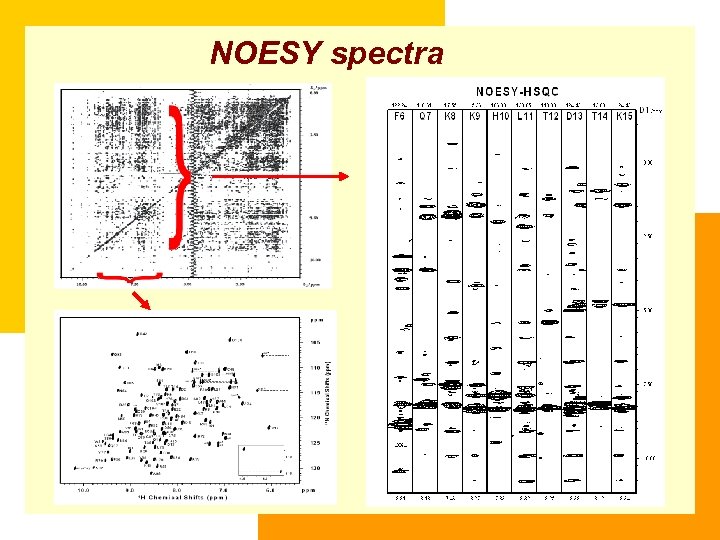
NOESY spectra
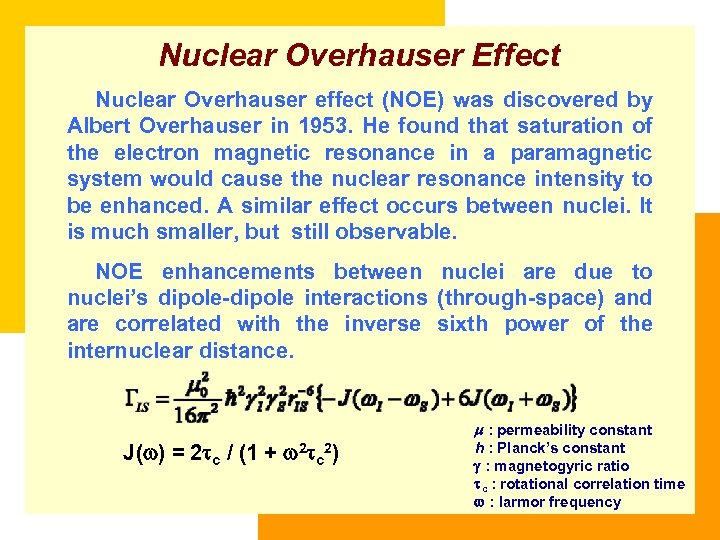
Nuclear Overhauser Effect Nuclear Overhauser effect (NOE) was discovered by Albert Overhauser in 1953. He found that saturation of the electron magnetic resonance in a paramagnetic system would cause the nuclear resonance intensity to be enhanced. A similar effect occurs between nuclei. It is much smaller, but still observable. NOE enhancements between nuclei are due to nuclei’s dipole-dipole interactions (through-space) and are correlated with the inverse sixth power of the internuclear distance. J( ) = 2 c / (1 + 2 c 2) : permeability constant h : Planck’s constant : magnetogyric ratio c : rotational correlation time : larmor frequency
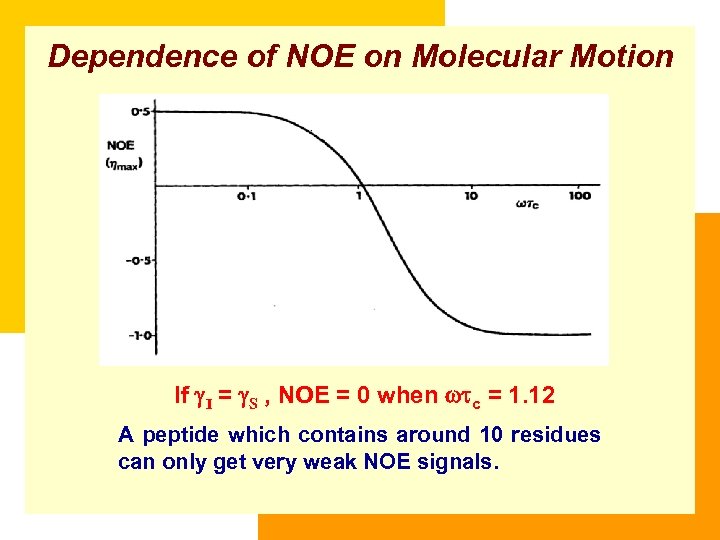
Dependence of NOE on Molecular Motion If I = S , NOE = 0 when c = 1. 12 A peptide which contains around 10 residues can only get very weak NOE signals.
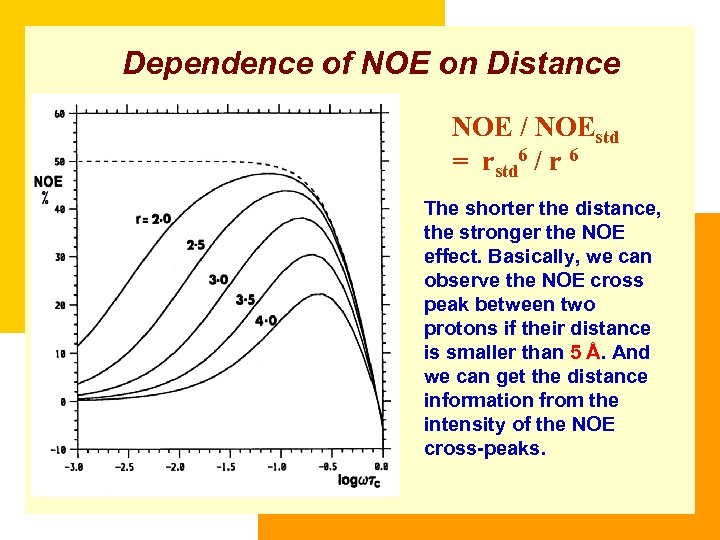
Dependence of NOE on Distance NOE / NOEstd = rstd 6 / r 6 The shorter the distance, the stronger the NOE effect. Basically, we can observe the NOE cross peak between two protons if their distance is smaller than 5 Å. And we can get the distance information from the intensity of the NOE cross-peaks.
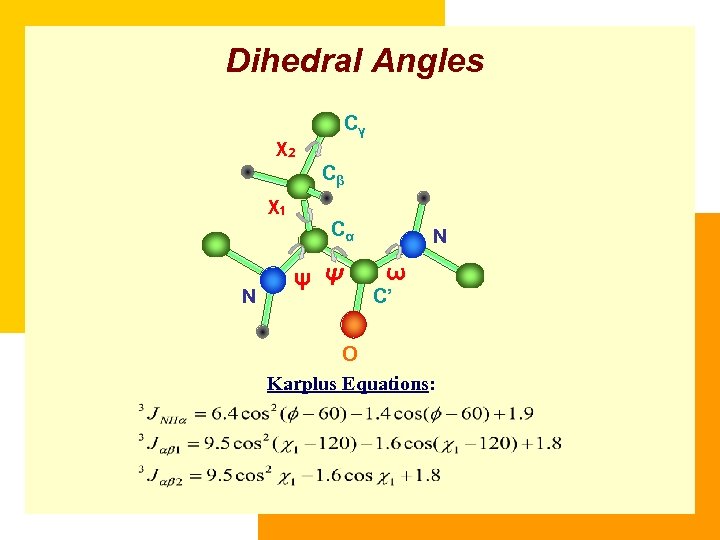
Dihedral Angles Cγ χ2 Cβ χ1 Cα N ψ Ψ N ω C’ O Karplus Equations:
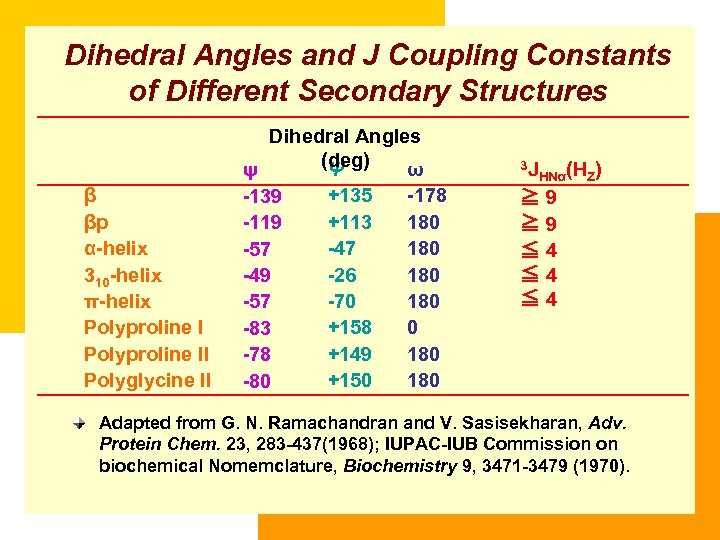
Dihedral Angles and J Coupling Constants of Different Secondary Structures β βp α-helix 310 -helix π-helix Polyproline II Polyglycine II Dihedral Angles (deg) Ψ ω ψ +135 -178 -139 +113 180 -119 -47 180 -57 -26 180 -49 -70 180 -57 +158 0 -83 +149 180 -78 +150 180 -80 3 J HNα(HZ) ≧ 9 ≦ 4 ≦ 4 Adapted from G. N. Ramachandran and V. Sasisekharan, Adv. Protein Chem. 23, 283 -437(1968); IUPAC-IUB Commission on biochemical Nomemclature, Biochemistry 9, 3471 -3479 (1970).
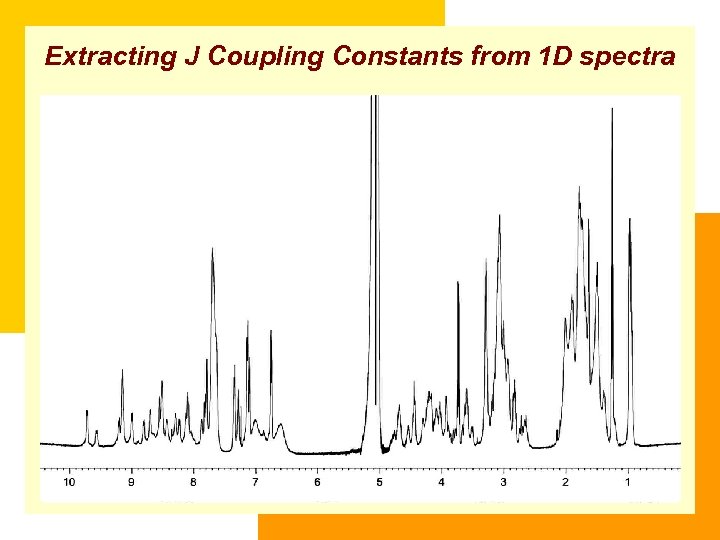
Extracting J Coupling Constants from 1 D spectra 3 J HNα
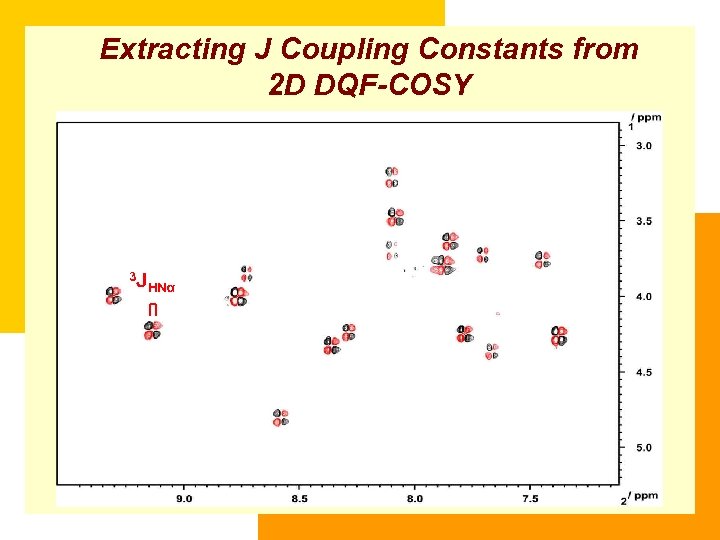
Extracting J Coupling Constants from 2 D DQF-COSY 3 J HNα
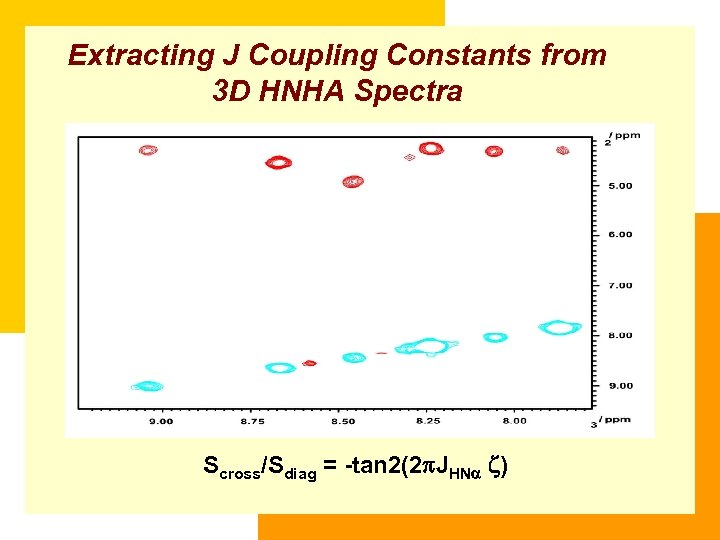
Extracting J Coupling Constants from 3 D HNHA Spectra Scross/Sdiag = -tan 2(2 JHN )
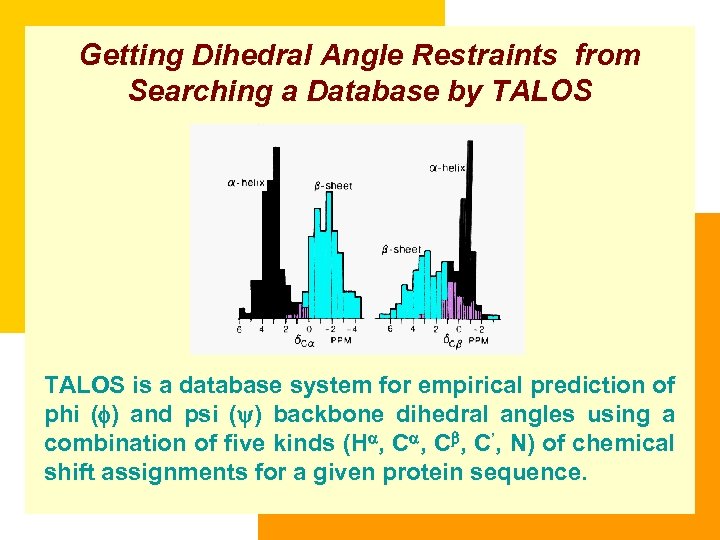
Getting Dihedral Angle Restraints from Searching a Database by TALOS is a database system for empirical prediction of phi (f) and psi (y) backbone dihedral angles using a combination of five kinds (H , C , C’, N) of chemical shift assignments for a given protein sequence.
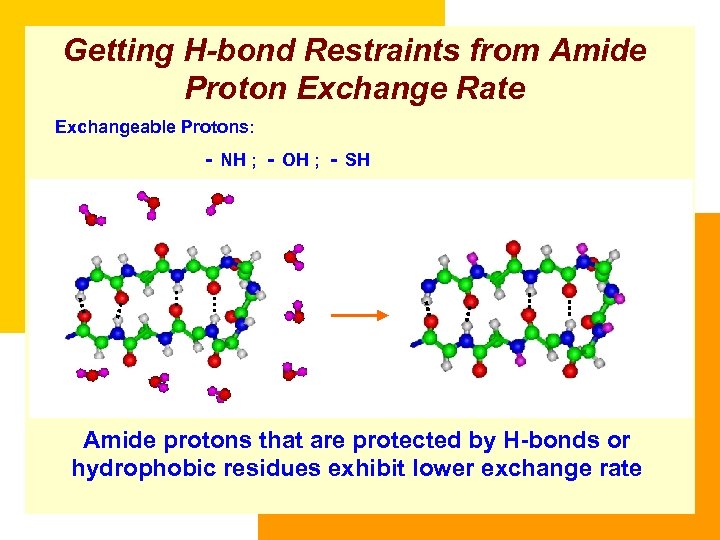
Getting H-bond Restraints from Amide Proton Exchange Rate Exchangeable Protons: - NH ; - OH ; - SH Amide protons that are protected by H-bonds or hydrophobic residues exhibit lower exchange rate
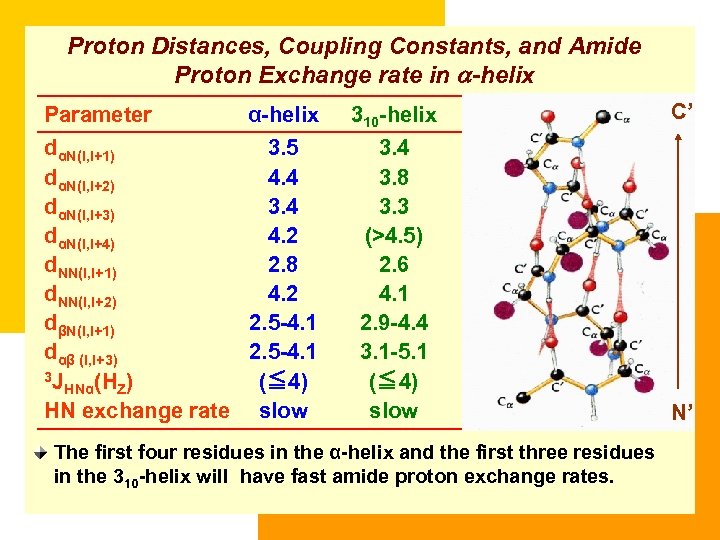
Proton Distances, Coupling Constants, and Amide Proton Exchange rate in a-helix Parameter α-helix dαN(i, i+1) 3. 5 dαN(i, i+2) 4. 4 dαN(i, i+3) 3. 4 dαN(i, i+4) 4. 2 d. NN(i, i+1) 2. 8 d. NN(i, i+2) 4. 2 dβN(i, i+1) 2. 5 -4. 1 dαβ (i, i+3) 2. 5 -4. 1 3 J (≦ 4) HNα(HZ) HN exchange rate slow 310 -helix C’ 3. 4 3. 8 3. 3 (>4. 5) 2. 6 4. 1 2. 9 -4. 4 3. 1 -5. 1 (≦ 4) slow N’ The first four residues in the α-helix and the first three residues in the 310 -helix will have fast amide proton exchange rates.
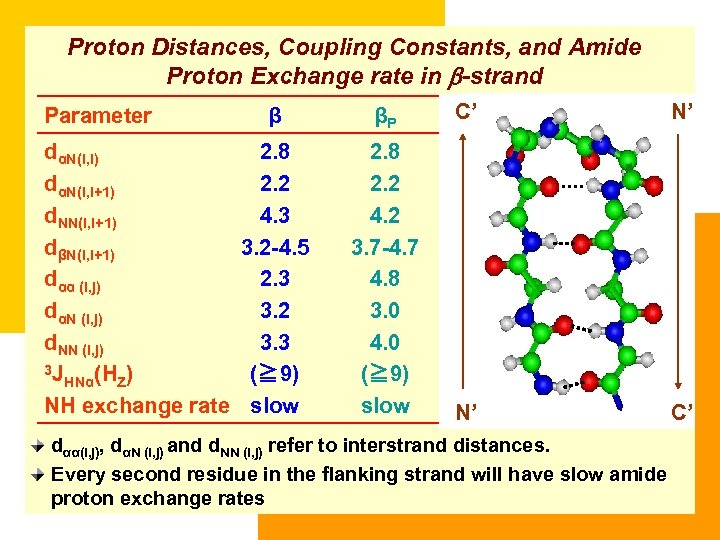
Proton Distances, Coupling Constants, and Amide Proton Exchange rate in b-strand Parameter β dαN(i, i) 2. 8 dαN(i, i+1) 2. 2 d. NN(i, i+1) 4. 3 dβN(i, i+1) 3. 2 -4. 5 dαα (i, j) 2. 3 dαN (i, j) 3. 2 d. NN (i, j) 3. 3 3 J (≧ 9) HNα(HZ) NH exchange rate slow βP C’ N’ 2. 8 2. 2 4. 2 3. 7 -4. 7 4. 8 3. 0 4. 0 (≧ 9) slow N’ C’ dαα(i, j), dαN (i, j) and d. NN (i, j) refer to interstrand distances. Every second residue in the flanking strand will have slow amide proton exchange rates
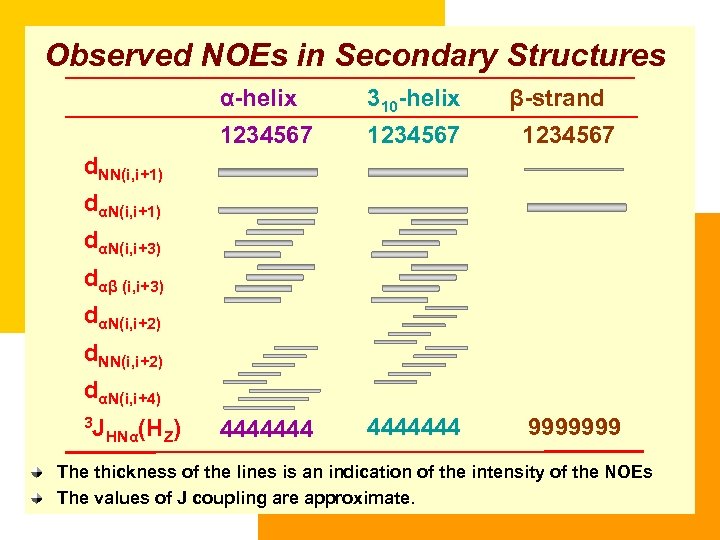
Observed NOEs in Secondary Structures α-helix 310 -helix β-strand 1234567 4444444 9999999 d. NN(i, i+1) dαN(i, i+3) dαβ (i, i+3) dαN(i, i+2) d. NN(i, i+2) dαN(i, i+4) 3 J HNα(HZ) The thickness of the lines is an indication of the intensity of the NOEs The values of J coupling are approximate.
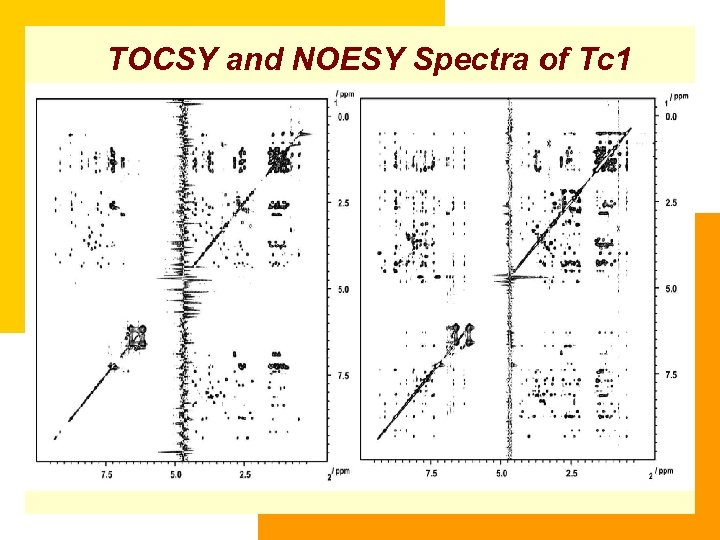
TOCSY and NOESY Spectra of Tc 1
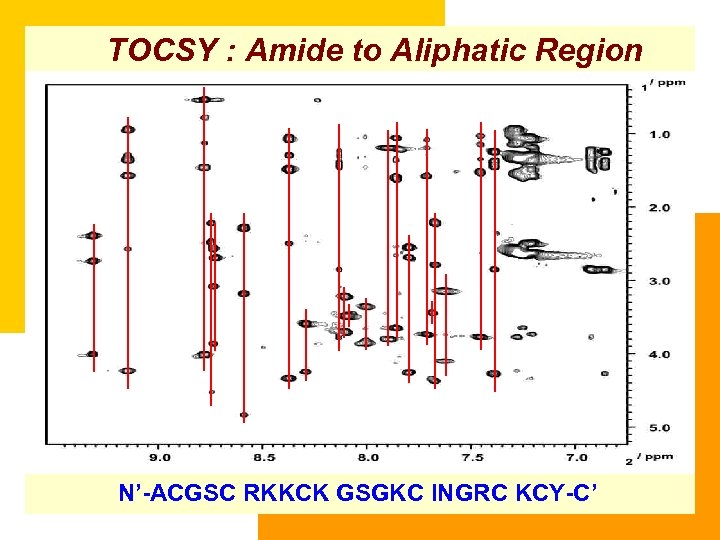
TOCSY : Amide to Aliphatic Region N’-ACGSC RKKCK GSGKC INGRC KCY-C’
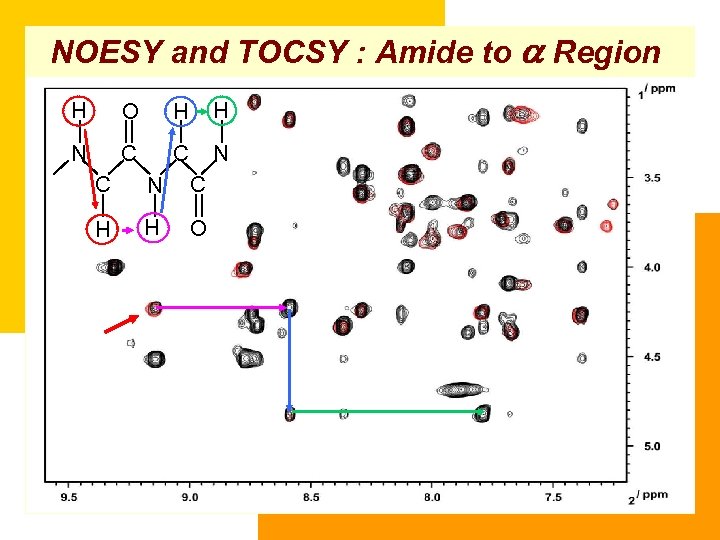
NOESY and TOCSY : Amide to a Region H O H H N C C N C H H O
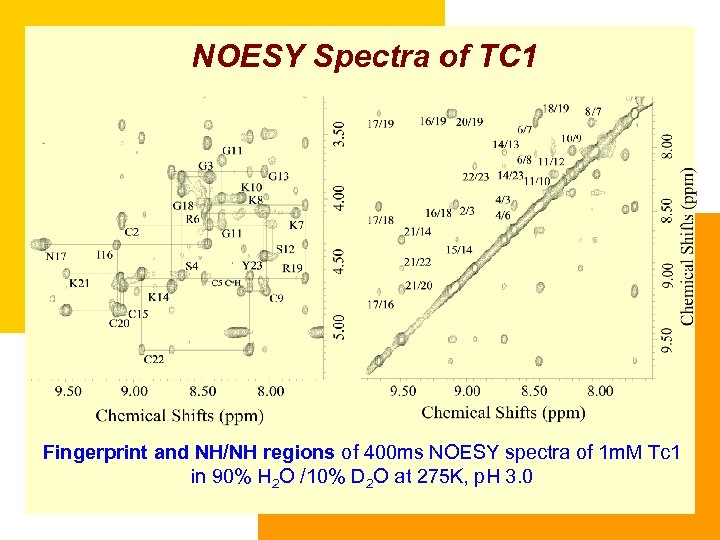
NOESY Spectra of TC 1 Fingerprint and NH/NH regions of 400 ms NOESY spectra of 1 m. M Tc 1 in 90% H 2 O /10% D 2 O at 275 K, p. H 3. 0
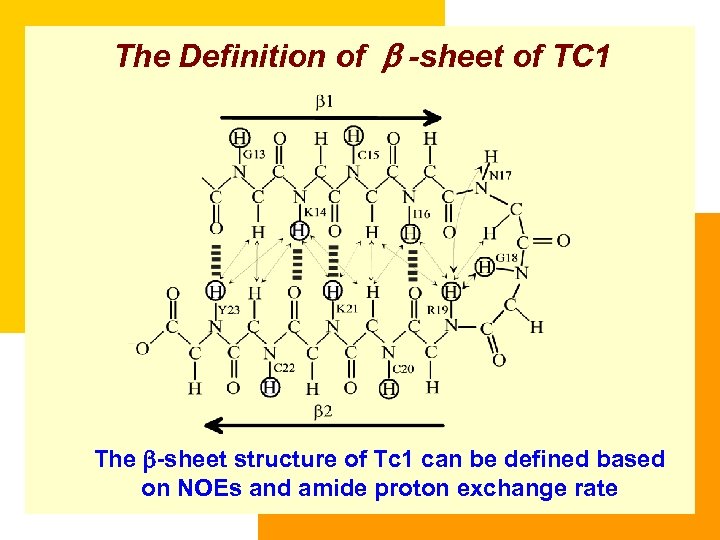
The Definition of b -sheet of TC 1 The -sheet structure of Tc 1 can be defined based on NOEs and amide proton exchange rate
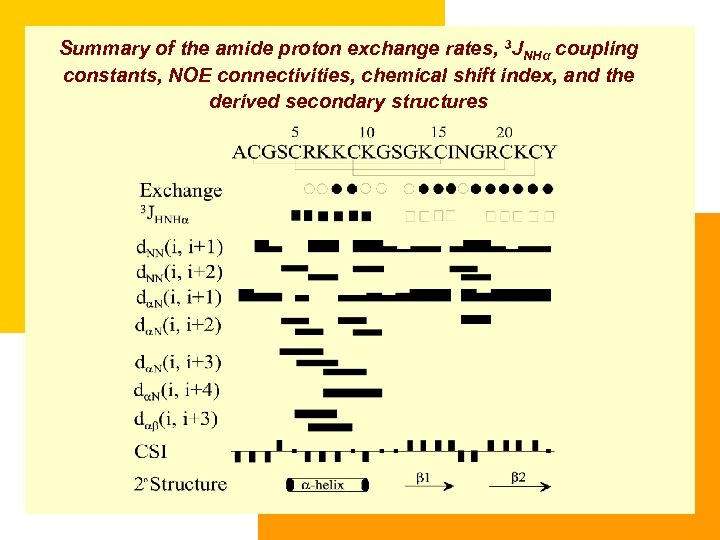
Summary of the amide proton exchange rates, 3 JNHα coupling constants, NOE connectivities, chemical shift index, and the derived secondary structures
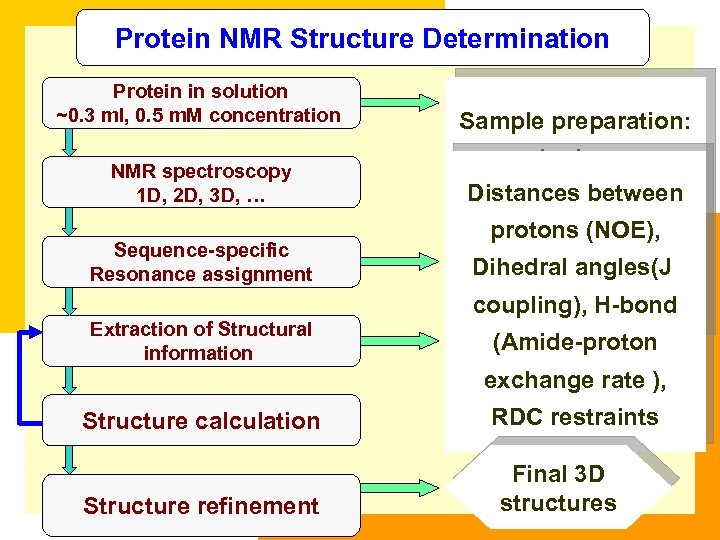
Protein NMR Structure Determination Protein in solution ~0. 3 ml, 0. 5 m. M concentration NMR spectroscopy 1 D, 2 D, 3 D, … Sequence-specific Resonance assignment Extraction of Structural information Sample preparation: cloning, Distances between protein expression Secondary protons (NOE), purification, structure of Dihedral angles(J characterization, protein coupling), H-bond isotopic labeling. (Amide-proton exchange rate ), Structure calculation Structure refinement RDC restraints Final 3 D structures
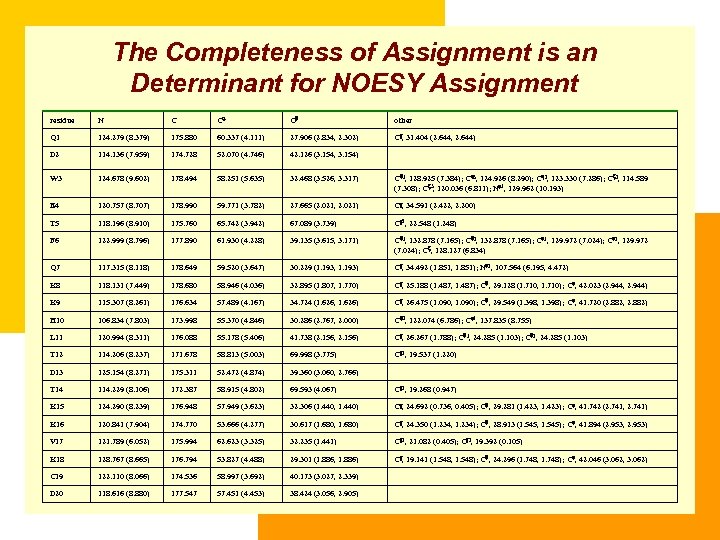
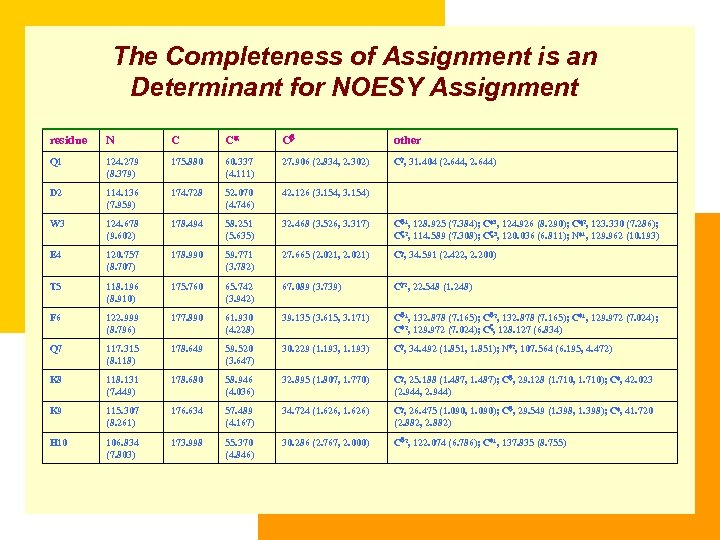
The Completeness of Assignment is an Determinant for NOESY Assignment residue N C C C other Q 1 124. 279 (8. 379) 175. 880 60. 337 (4. 111) 27. 906 (2. 834, 2. 302) C , 31. 404 (2. 644, 2. 644) D 2 114. 136 (7. 959) 174. 728 52. 070 (4. 746) 42. 126 (3. 154, 3. 154) W 3 124. 678 (9. 602) 178. 494 58. 251 (5. 635) 32. 468 (3. 526, 3. 317) C 1, 128. 925 (7. 384); C 3, 124. 926 (8. 290); C 2, 123. 330 (7. 286); C 2, 114. 589 (7. 308); C 3, 120. 036 (6. 811); N 1, 129. 962 (10. 193) E 4 120. 757 (8. 707) 178. 990 59. 771 (3. 782) 27. 665 (2. 021, 2. 021) C , 34. 591 (2. 422, 2. 200) T 5 118. 196 (8. 910) 175. 760 65. 742 (3. 942) 67. 089 (3. 739) C 2, 22. 548 (1. 248) F 6 122. 999 (8. 796) 177. 890 61. 930 (4. 228) 39. 135 (3. 615, 3. 171) C 1, 132. 878 (7. 165); C 2, 132. 878 (7. 165); C 1, 129. 972 (7. 024); C 2, 129. 972 (7. 024); C , 128. 127 (6. 834) Q 7 117. 315 (8. 118) 178. 649 59. 520 (3. 647) 30. 229 (1. 193, 1. 193) C , 34. 492 (1. 851, 1. 851); N 2, 107. 564 (6. 195, 4. 472) K 8 118. 131 (7. 449) 178. 680 58. 946 (4. 036) 32. 895 (1. 807, 1. 770) C , 25. 188 (1. 487, 1. 487); C , 29. 128 (1. 710, 1. 710); C , 42. 023 (2. 944, 2. 944) K 9 115. 307 (8. 261) 176. 634 57. 489 (4. 167) 34. 724 (1. 626, 1. 626) C , 26. 475 (1. 090, 1. 090); C , 29. 549 (1. 398, 1. 398); C , 41. 720 (2. 882, 2. 882) H 10 106. 834 (7. 803) 173. 998 55. 370 (4. 846) 30. 286 (2. 767, 2. 000) C 2, 122. 074 (6. 786); C 1, 137. 835 (8. 755) L 11 120. 994 (8. 311) 176. 088 55. 178 (5. 406) 41. 738 (2. 156, 2. 156) C , 26. 267 (1. 788); C 1, 24. 285 (1. 103); C 2, 24. 285 (1. 103) T 12 114. 206 (8. 237) 171. 678 58. 813 (5. 003) 69. 998 (3. 775) C 2, 19. 537 (1. 220) D 13 125. 154 (8. 271) 175. 311 52. 472 (4. 874) 39. 360 (3. 060, 2. 766) T 14 114. 229 (8. 106) 172. 387 58. 915 (4. 802) 69. 593 (4. 067) C 2, 19. 268 (0. 947) K 15 124. 290 (8. 239) 176. 948 57. 949 (3. 623) 32. 306 (1. 440, 1. 440) C , 24. 692 (0. 736, 0. 405); C , 29. 281 (1. 423, 1. 423); C , 41. 742 (2. 741, 2. 741) K 16 120. 841 (7. 904) 174. 770 53. 666 (4. 277) 30. 617 (1. 680, 1. 680) C , 24. 350 (1. 234, 1. 234); C , 28. 913 (1. 545, 1. 545); C , 41. 894 (2. 953, 2. 953) V 17 121. 789 (6. 052) 175. 994 62. 623 (3. 325) 32. 235 (1. 441) C 1, 21. 082 (0. 405); C 2, 19. 392 (0. 105) K 18 128. 767 (8. 665) 176. 794 53. 827 (4. 488) 29. 301 (1. 886, 1. 886) C , 19. 141 (1. 548, 1. 548); C , 24. 296 (1. 748, 1. 748); C , 42. 046 (3. 062, 3. 062) C 19 122. 110 (8. 066) 174. 536 58. 997 (3. 692) 40. 173 (3. 027, 2. 339) D 20 118. 616 (8. 880) 177. 547 57. 451 (4. 453) 38. 424 (3. 056, 2. 905)
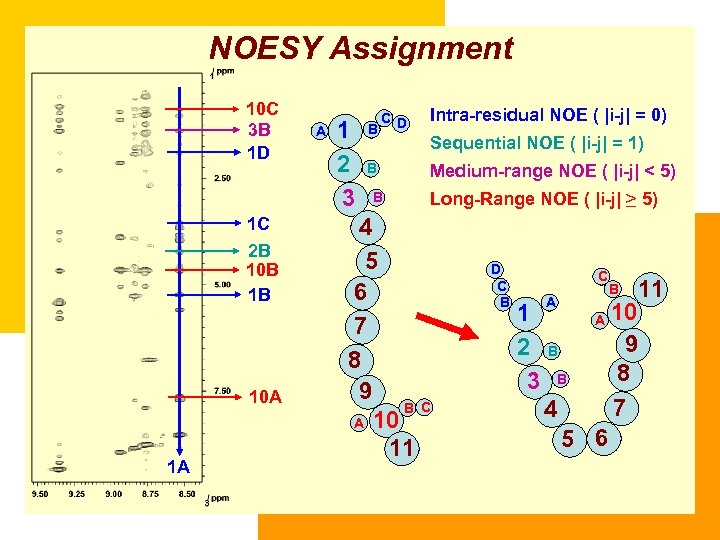
NOESY Assignment 10 C 3 B 1 D 1 C 2 B 10 B 1 B 10 A 1 A A 1 2 3 B C D B B Intra-residual NOE ( |i-j| = 0) Sequential NOE ( |i-j| = 1) Medium-range NOE ( |i-j| < 5) Long-Range NOE ( |i-j| ≥ 5) 4 5 6 7 8 9 B C A 10 11 D C B C 1 2 3 A A B B 4 5 6 B 11 10 9 8 7
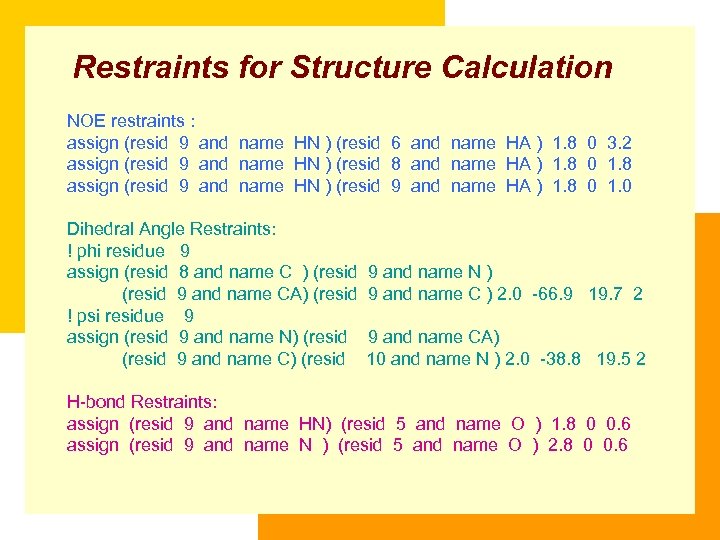
Restraints for Structure Calculation NOE restraints : assign (resid 9 and name HN ) (resid 6 and name HA ) 1. 8 0 3. 2 assign (resid 9 and name HN ) (resid 8 and name HA ) 1. 8 0 1. 8 assign (resid 9 and name HN ) (resid 9 and name HA ) 1. 8 0 1. 0 Dihedral Angle Restraints: ! phi residue 9 assign (resid 8 and name C ) (resid 9 and name CA) (resid ! psi residue 9 assign (resid 9 and name N) (resid 9 and name C) (resid 9 and name N ) 9 and name C ) 2. 0 -66. 9 19. 7 2 9 and name CA) 10 and name N ) 2. 0 -38. 8 19. 5 2 H-bond Restraints: assign (resid 9 and name HN) (resid 5 and name O ) 1. 8 0 0. 6 assign (resid 9 and name N ) (resid 5 and name O ) 2. 8 0 0. 6
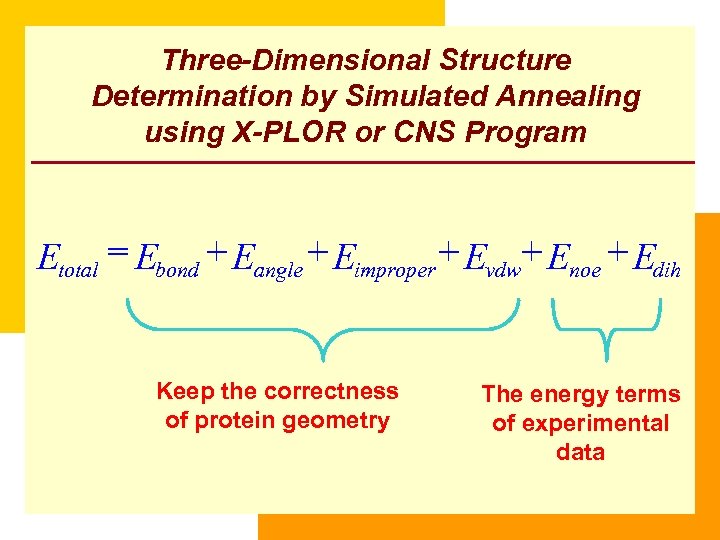
Three-Dimensional Structure Determination by Simulated Annealing using X-PLOR or CNS Program Etotal = Ebond + Eangle + Eimproper + Evdw+ Enoe + Edih Keep the correctness of protein geometry The energy terms of experimental data
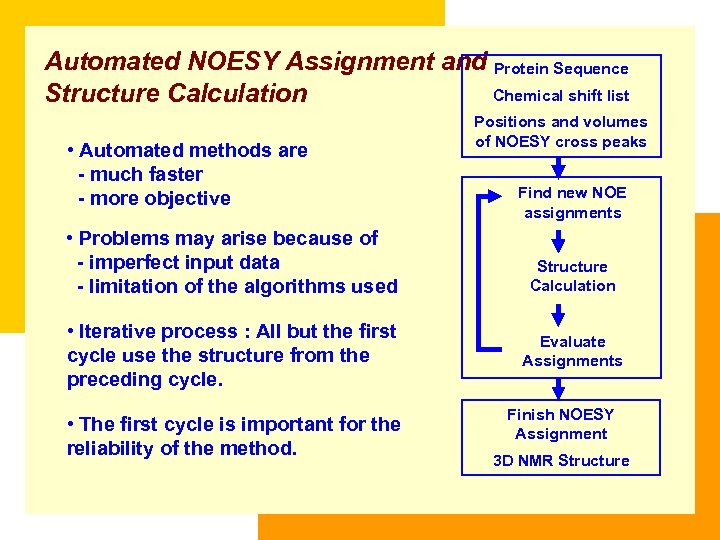
Automated NOESY Assignment and Protein Sequence Chemical shift list Structure Calculation • Automated methods are - much faster - more objective • Problems may arise because of - imperfect input data - limitation of the algorithms used • Iterative process : All but the first cycle use the structure from the preceding cycle. • The first cycle is important for the reliability of the method. Positions and volumes of NOESY cross peaks Find new NOE assignments Structure Calculation Evaluate Assignments Finish NOESY Assignment 3 D NMR Structure
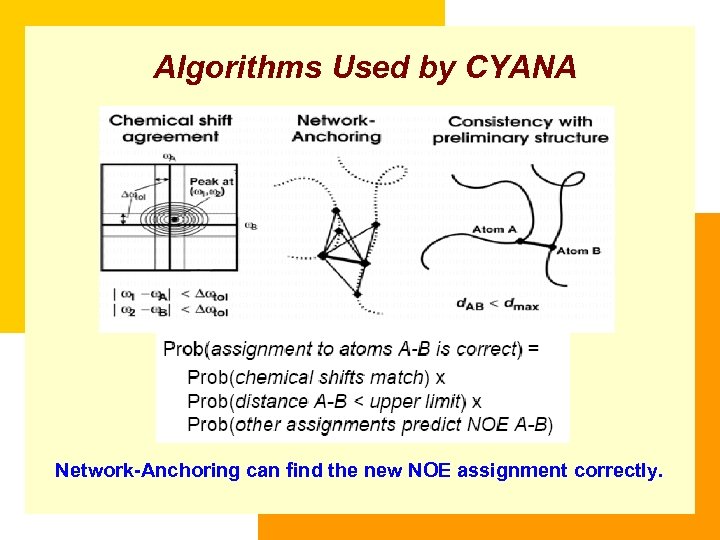
Algorithms Used by CYANA Network-Anchoring can find the new NOE assignment correctly.
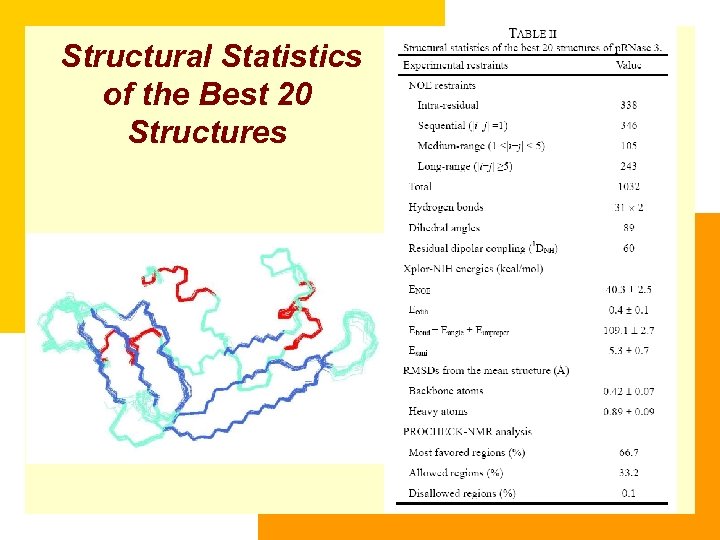
Structural Statistics of the Best 20 Structures
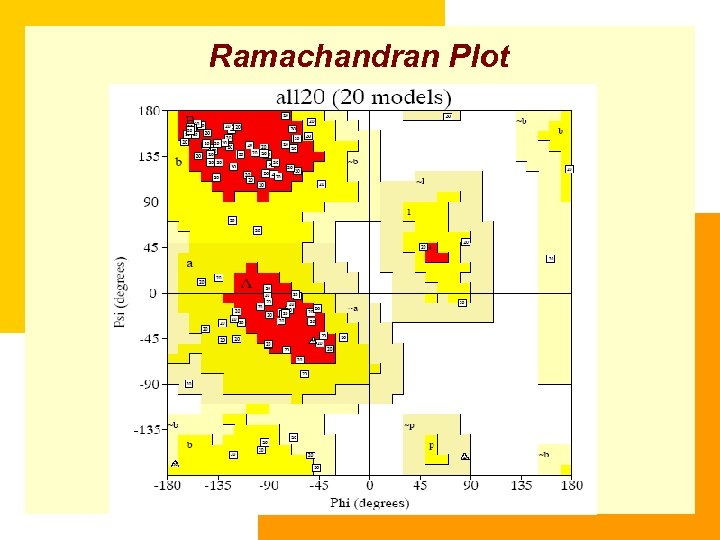
Ramachandran Plot
acd1350cffccce78a51d727c7b946846.ppt