a96d27a3c6f8871e0b470692983a24a7.ppt
- Количество слайдов: 24

Protein Databank in Europe (PDBe) An Introduction EBI is an Outstation of the European Molecular Biology Laboratory.
Introduction • • • 2 18. 02. 09 Based at the European Bioinformatics Institute (EBI), an outstation of the European Molecular Biology Laboratory (EMBL) at Hinxton, UK Started in 1996 with the goal of providing an autonomous structural database capability in Europe The aims of the group are to provide: • a deposition site via which macromolecular structures can be added to the PDB (Auto. Dep) or EM (EMDep). • a stable and clean repository of macromolecular structure data • services that allow users to access, search and retrieve structural data Protein Databank in Europe
Protein Databank in Europe (PDBe) group • Is one of the four sites around the world that where 3 D structures may be deposited. • Provides stable and clean repository of macromolecular structure data. • Has services that allow users to access, search and retrieve structural data from a single web access point. 3 18. 02. 09 Protein Databank in Europe
worldwide Protein Data Bank (ww. PDB) • • • 4 Consists of four sites • RCSB (USA), PDB-j (Japan) BMRB (USA) and PDBe. PDB is the single repository of all publicly available macromolecular structures. The PDB started in 1971 and now has around 54, 000 entries and new entries are added weekly. Structures are deposited by scientists and contents are freely available. The format of the archive is flat-files with fixed line format, although an improved flat-file format (mm. CIF) is available. 18. 02. 09 Protein Databank in Europe

PDBe Tasks Deposition site Data clean-up Database design and implementation Retrieve data EBI is an Outstation of the European Molecular Biology Laboratory.
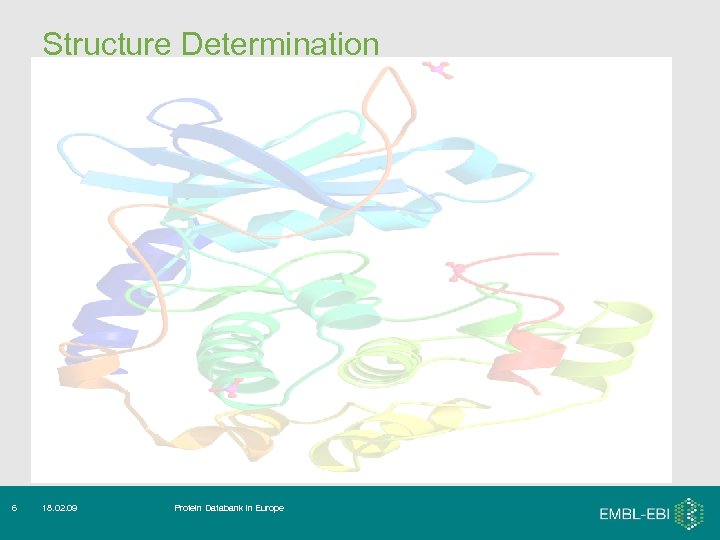
Structure Determination 6 18. 02. 09 Protein Databank in Europe

Depositions and Curation Full deposition site from June 1999 18% of all submissions via the EBI. Closely collaborate with the other ww. PDB members for a single unified archive. . Depositions started June 2002 7 18. 02. 09 Protein Databank in Europe

Auto. Dep 4. 0 • A structure deposition and archiving system. • Based on Java/XML technology. • Available free under license for academic and industry use. • Easy to install and use for in-house archiving before deposition to the PDB via the PDBe interface. http: //www. ebi. ac. uk/msd-srv/autodep 4

Disadvantages of Flat files… • Macromolecular structures are very complex. • Existing PDB format is incapable of fully describing even existing structures. • Format is not readily extensible, to cope, for example, with structural genomics data. • Historical archive is non-uniform and poorly populated. • Search and retrieval of flat files is difficult and/or inaccurate. 9 18. 02. 09 Protein Databank in Europe
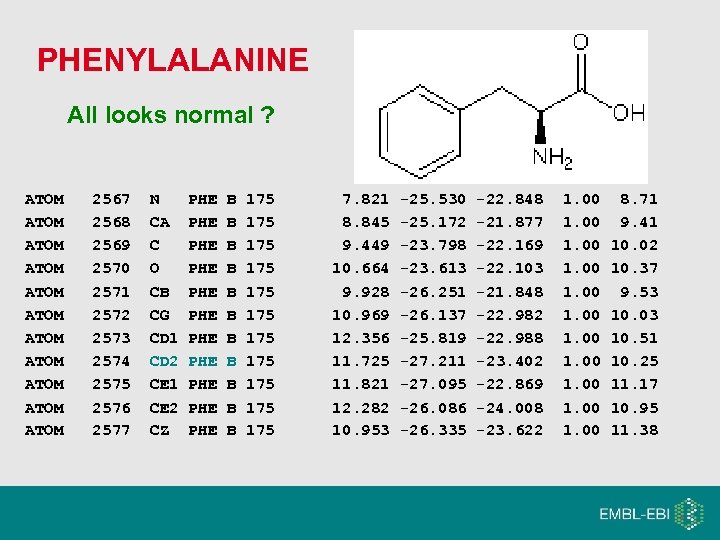
PHENYLALANINE All looks normal ? ATOM ATOM ATOM 2567 2568 2569 2570 2571 2572 2573 2574 2575 2576 2577 N CA C O CB CG CD 1 CD 2 CE 1 CE 2 CZ PHE PHE PHE B B B 175 175 175 7. 821 8. 845 9. 449 10. 664 9. 928 10. 969 12. 356 11. 725 11. 821 12. 282 10. 953 -25. 530 -25. 172 -23. 798 -23. 613 -26. 251 -26. 137 -25. 819 -27. 211 -27. 095 -26. 086 -26. 335 -22. 848 -21. 877 -22. 169 -22. 103 -21. 848 -22. 982 -22. 988 -23. 402 -22. 869 -24. 008 -23. 622 1. 00 1. 00 8. 71 9. 41 10. 02 10. 37 9. 53 10. 03 10. 51 10. 25 11. 17 10. 95 11. 38
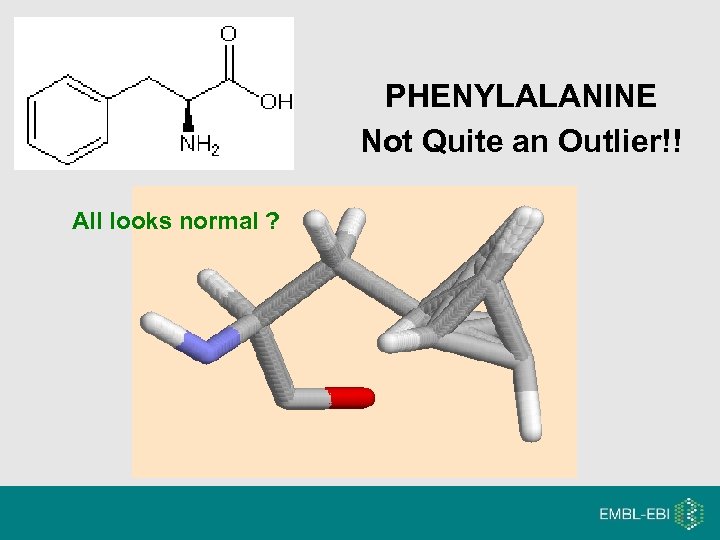
PHENYLALANINE Not Quite an Outlier!! All looks normal ?
PDBe Curation • Authentication of source That the protein is from human and not rabbit, for example ! • Authentication of structure Comparison of structure against raw data. Geometry and Stereochemistry. Provide results back to depositor. • Validation of correct methodology used Whether X-Ray, NMR or EM. • Conformity to standards Follows PDB format specifications • Error checks • Consistency checks - to identify simple typos Homo sapiens and not Homo sapien (single human? ). • Outlier detection - to identify suspect records
Adopt standards • Use NCBI taxonomy database to ensure correct organism names • Use Uniprot database to ensure correct protein description • Enzyme database • Annotated ligand information

• What happens when these checks fail? Raise issue with the depositor • But the depositor might: be unavailable not interested not know the answer anyway not be sure about which data have the problem • The older the entry, the less likely the depositor can/will help 14 18. 02. 09 Protein Databank in Europe
What is the solution? • Don’t rush and define another format • Represent the structure data in a meaningful way (use data model)
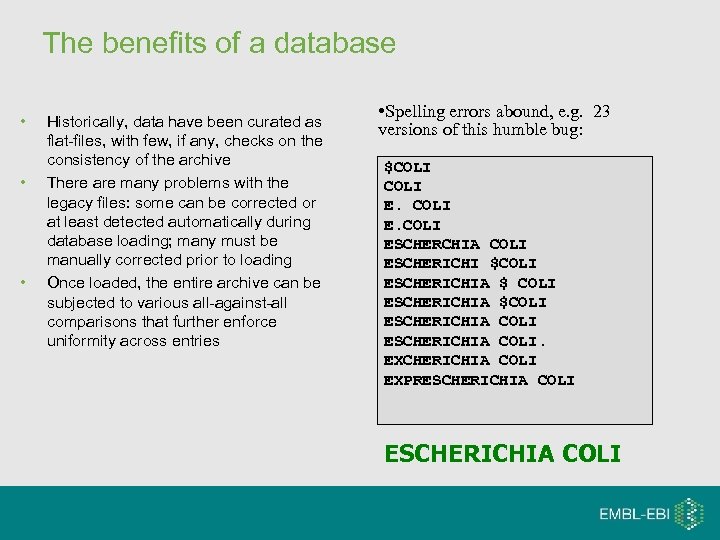
The benefits of a database • • • Historically, data have been curated as flat-files, with few, if any, checks on the consistency of the archive There are many problems with the legacy files: some can be corrected or at least detected automatically during database loading; many must be manually corrected prior to loading Once loaded, the entire archive can be subjected to various all-against-all comparisons that further enforce uniformity across entries • Spelling errors abound, e. g. 23 versions of this humble bug: $COLI ESCHERCHIA COLI ESCHERICHI $COLI ESCHERICHIA $COLI ESCHERICHIA COLI. EXCHERICHIA COLI EXPRESCHERICHIA COLI
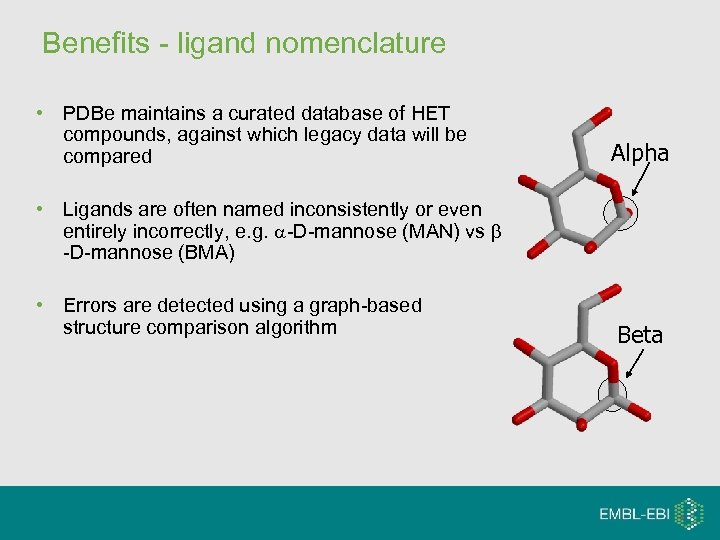
Benefits - ligand nomenclature • PDBe maintains a curated database of HET compounds, against which legacy data will be compared Alpha • Ligands are often named inconsistently or even entirely incorrectly, e. g. -D-mannose (MAN) vs -D-mannose (BMA) • Errors are detected using a graph-based structure comparison algorithm Beta
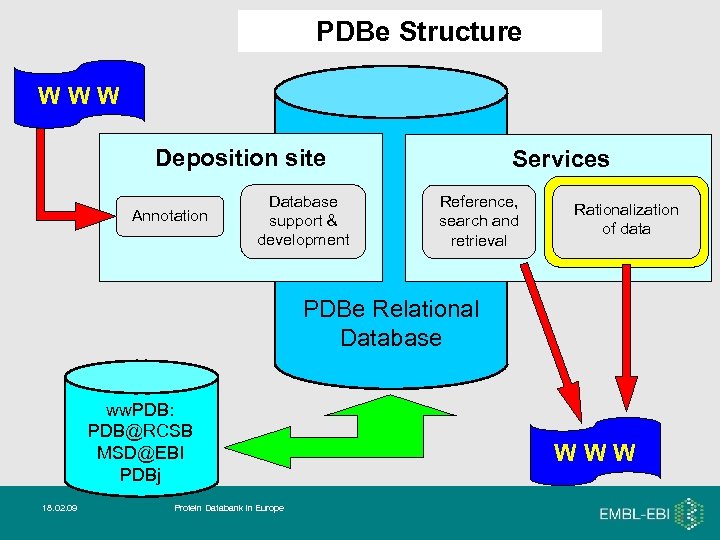
PDBe Structure WWW Deposition site Annotation Database support & development Services Reference, search and retrieval Rationalization of data PDBe Relational Database ww. PDB: PDB@RCSB MSD@EBI PDBj 18. 02. 09 Protein Databank in Europe WWW
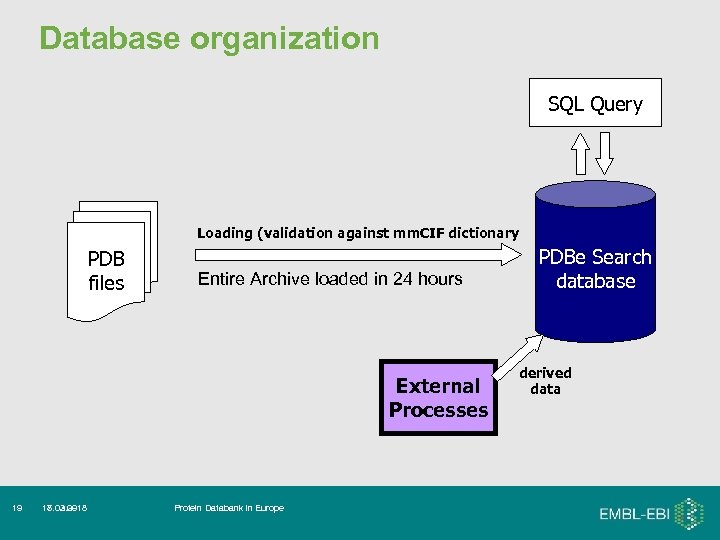
Database organization SQL Query Loading (validation against mm. CIF dictionary PDB files Entire Archive loaded in 24 hours External Processes 19 18. 02. 09 17. 03. 2018 Protein Databank in Europe PDBe Search database derived data
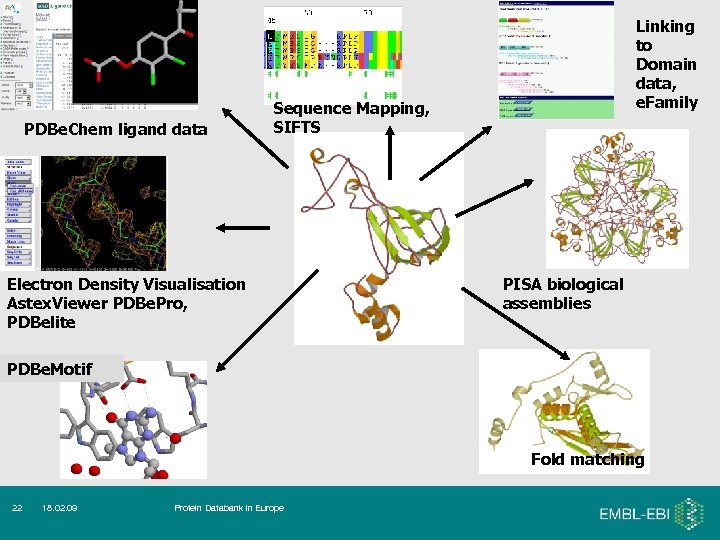
PDBe. Chem ligand data Linking to Domain data, e. Family Sequence Mapping, SIFTS Electron Density Visualisation Astex. Viewer PDBe. Pro, PDBelite PISA biological assemblies PDBe. Motif Fold matching 22 18. 02. 09 Protein Databank in Europe
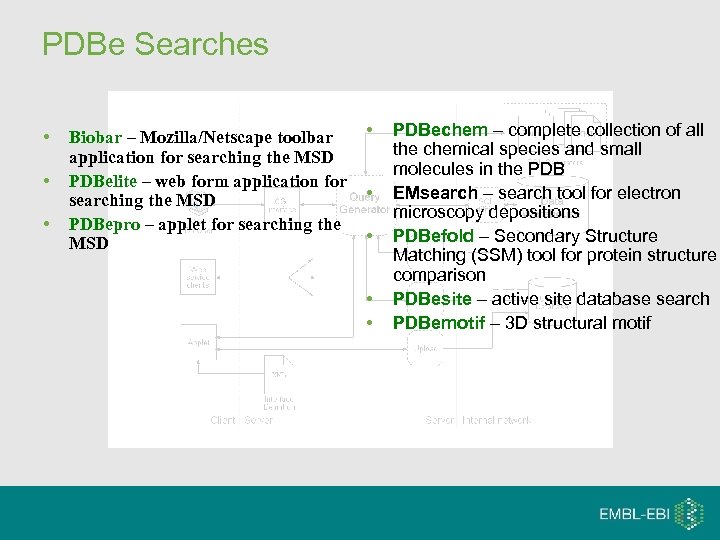
PDBe Searches • • • Biobar – Mozilla/Netscape toolbar application for searching the MSD PDBelite – web form application for searching the MSD PDBepro – applet for searching the MSD • • • PDBechem – complete collection of all the chemical species and small molecules in the PDB EMsearch – search tool for electron microscopy depositions PDBefold – Secondary Structure Matching (SSM) tool for protein structure comparison PDBesite – active site database search PDBemotif – 3 D structural motif
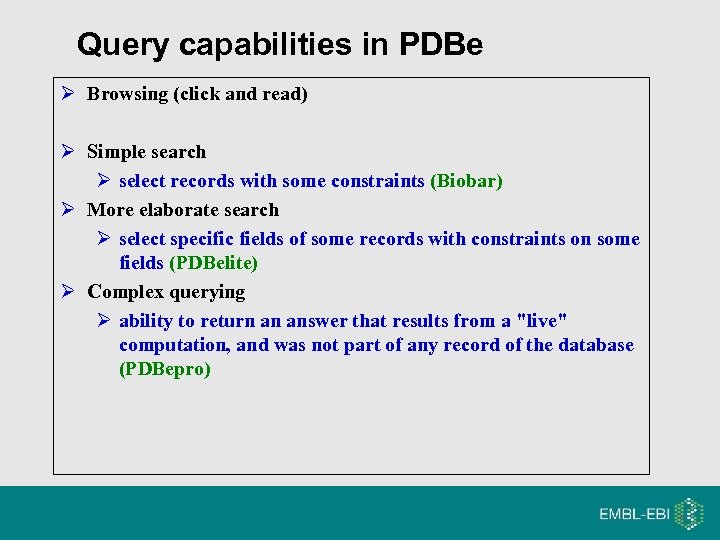
Query capabilities in PDBe Browsing (click and read) Simple search select records with some constraints (Biobar) More elaborate search select specific fields of some records with constraints on some fields (PDBelite) Complex querying ability to return an answer that results from a "live" computation, and was not part of any record of the database (PDBepro)

PDBe provides… • Clean biological data • Integrated data • A single web access point • Query interfaces for different users (Beginner, Occasional or expert). • Interconnected views of the data relating structure, sequence, text & experimental details. 28 18. 02. 09 Protein Databank in Europe
A database for all Search database
a96d27a3c6f8871e0b470692983a24a7.ppt