ceb66dce095e873743b3539a726852c7.ppt
- Количество слайдов: 52
Protein Analysis Course Day 1: Databases, dotplots and pairwise alignment

Todays timetable l Databases ¡ Exercises l Dotplot ¡ and file formats and pairwise alignment Exercises Coffee breaks during the exercises
Databases and file formats l Sequence file format ¡ FASTA l Uni. Prot ¡ Primary structure l PDB ¡ (Universal protein resource) (Protein Database) Tertiary structure
Sequence file format l FASTA (a. k. a Pearson format) ¡ ¡ Most commonly used Can be easily construted by hand if needed Straightforward way to store multiple sequences – just concatenate multiple FASTA –files Content: l l l First line (Header line) always starts with symbol ”>” followed by identifiers and descriptions Header line is ALWAYS just one line before sequence After header line (from the second line) starts the sequence (presented usingle-letter codes) Sequence normally divided into multiple lines (often required) Recommended line length max 80 chars (also with header line)

FASTA >SEQUENCE_1 MTEITAAMVKELRESTGAGMMDCKNALSETNGDFDKAVQLLREKGLGKAAKKADRLAAEG LVSVKVSDDFTIAAMRPSYLSYEDLDMTFVENEYKALVAELEKENEERRRLKDPNKPEHK IPQFASRKQLSDAILKEAEEKIKEELKAQGKPEKIWDNIIPGKMNSFIADNSQLDSKLTL MGQFYVMDDKKTVEQVIAEKEKEFGGKIKIVEFICFEVGEGLEKKTEDFAAEVAAQL >SEQUENCE_2 SATVSEINSETDFVAKNDQFIALTKDTTAHIQSNSLQSVEELHSSTINGVKFEEYLKSQI ATIGENLVVRRFATLKAGANGVVNGYIHTNGRVGVVIAAACDSAEVASKSRDLLRQICMH …
Databases: Uni. Prot l Uni. Prot is the universal protein resource, a central repository of protein data created by combining Swiss-Prot, Tr. EMBL and PIR. This makes it the world's most comprehensive resource on protein information [wikipedia] l Uni. Prot provides three core database: ¡ ¡ ¡ The Uni. Prot Archive (Uni. Parc) provides a stable, comprehensive sequence collection without redundant sequences by storing the complete body of publicly available protein sequence data The Uni. Prot Reference Clusters (Uni. Ref) databases provide nonredundant reference data collections based on the Uni. Prot knowledgebase in order to obtain complete coverage of sequence space at several resolutions The Uni. Prot Knowledgebase (Uni. Prot. KB) is the central database of protein sequences with accurate, consistent, and rich sequence and functional annotation
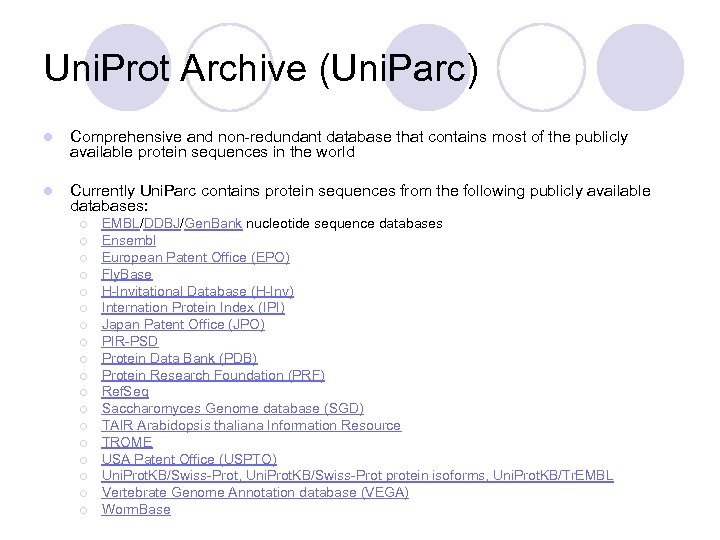
Uni. Prot Archive (Uni. Parc) l Comprehensive and non-redundant database that contains most of the publicly available protein sequences in the world l Currently Uni. Parc contains protein sequences from the following publicly available databases: ¡ ¡ ¡ ¡ ¡ EMBL/DDBJ/Gen. Bank nucleotide sequence databases Ensembl European Patent Office (EPO) Fly. Base H-Invitational Database (H-Inv) Internation Protein Index (IPI) Japan Patent Office (JPO) PIR-PSD Protein Data Bank (PDB) Protein Research Foundation (PRF) Ref. Seq Saccharomyces Genome database (SGD) TAIR Arabidopsis thaliana Information Resource TROME USA Patent Office (USPTO) Uni. Prot. KB/Swiss-Prot, Uni. Prot. KB/Swiss-Prot protein isoforms, Uni. Prot. KB/Tr. EMBL Vertebrate Genome Annotation database (VEGA) Worm. Base

Uni. Prot Reference Clusters (Uni. Ref) Sequence clusters, used to speed up similarity searches l Uni. Ref 100 l ¡ l Uni. Ref 90 ¡ l Cluster is composed of sequences that are identical Cluster is composed of sequences that have at least 90% sequence identity Uni. Ref 50 ¡ Cluster is composed of sequences that have at least 50% sequence identity
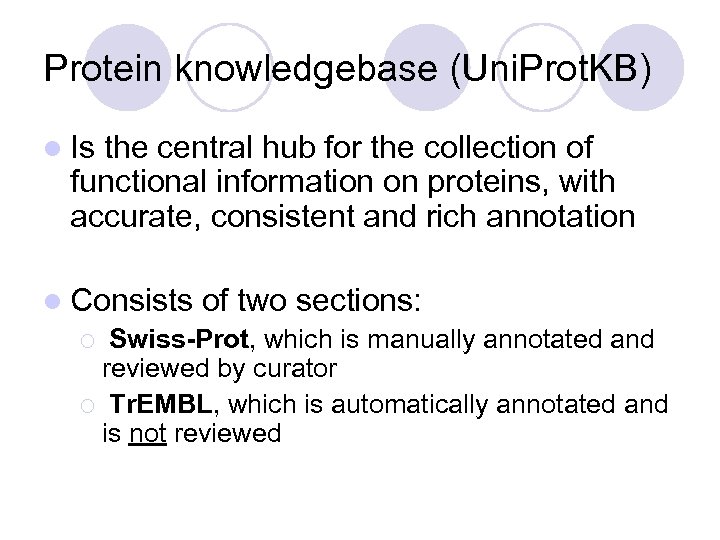
Protein knowledgebase (Uni. Prot. KB) l Is the central hub for the collection of functional information on proteins, with accurate, consistent and rich annotation l Consists of two sections: ¡ Swiss-Prot, which is manually annotated and reviewed by curator ¡ Tr. EMBL, which is automatically annotated and is not reviewed
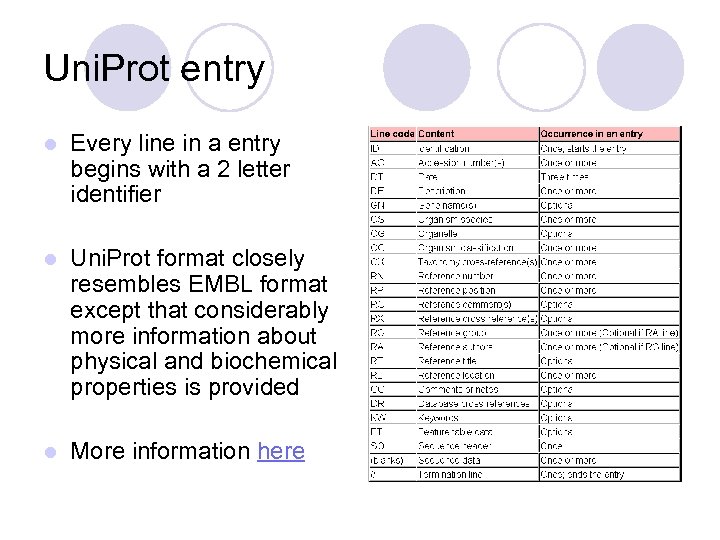
Uni. Prot entry l Every line in a entry begins with a 2 letter identifier l Uni. Prot format closely resembles EMBL format except that considerably more information about physical and biochemical properties is provided l More information here

Databases: PDB l Founded in 1971 by Brookhaven National Laboratory, New York. l Transferred to the Research Collaboratory for Structural Bioinformatics (RCSB) in 1998. l Currently it holds more than 55, 000 released structures.
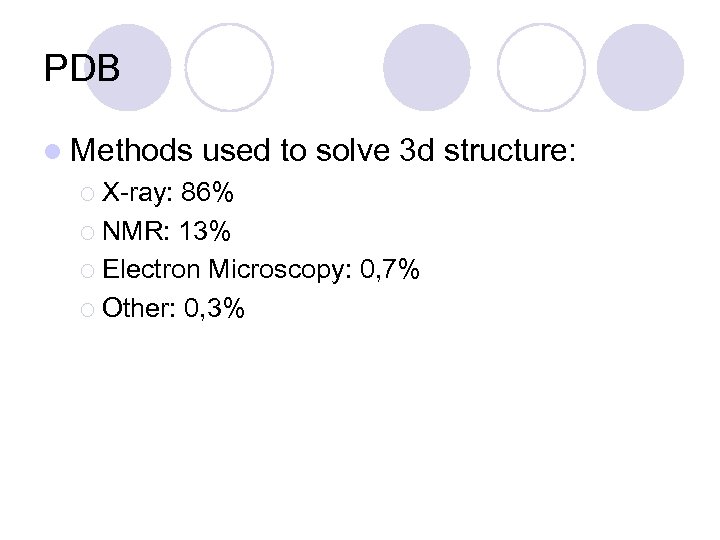
PDB l Methods ¡ X-ray: used to solve 3 d structure: 86% ¡ NMR: 13% ¡ Electron Microscopy: 0, 7% ¡ Other: 0, 3%

PDB file format l Text file – you can edit with a text editor e. g. Word. Pad l Atomic co-ordinates l Rich annotation ¡ Citation ¡ Experimental Method ¡ Biological source e. ¡ Etc.

FYI: Errors in databases l Be aware of errors in the databases: ¡ sequence errors: l l ¡ genome projects’ error rate is 1/10, 000 nts; ESTs’ error rate is 1/100 nts. annotation errors: l Automated computer programs do not always give correct annotations. l Swiss. Prot is a protein database curated annotated manually by biologists. Most reliable database, but is not up-to-date

Exercises l Go to the course web page and start with exercises given in file: database_exercises. doc l http: //ekhidna. biocenter. helsinki. fi/how
Pairwise sequence alignments l l Motivation – Why alignments? Sequence comparison ¡ ¡ l Pairwise alignment algorithms ¡ ¡ ¡ l Dotplot The alignment problem Exact algorithms Heuristic algorithms Database searches Web tools: ¡ ¡ ¡ Build alignments using EBI server, Blast at NCBI, EBI, Pairs. DB, …

Motivation l Proteins perform most of the functions required in biological systems: ¡ ¡ ¡ Signaling (kinases, . . . ) Enzymes (proteases, …) Structural (collagen, elastin, …) Immune system (antibodies, . . . ) Storage and transport (hemoglobin, …) … l Large amount of information available in current databanks. l Goal: Want to extrapolate information about the function of a newly discovered sequence by comparing it to annotated sequences.
Does it make sense? All functional information is ultimately contained within the sequence. l Proteins are evolutionary related: l ¡ ¡ l Selective pressure is on function, and thus on residues with functional role (eg: active site or structural key residues are conserved). Modular nature of proteins. Two sequences have the same structure if corresponding residues are similar enough on physico-chemical level.
Application of sequence alignment Determining function of newly discovered genetic or protein sequences. l Identification of functional patterns/domains. l Predicting structure of proteins. l Determining evolutionary relationships among genes, proteins, and entire species. l Aligning and comparing sequences, and searching databases for similar sequences – a cornerstone of bioinformatics!!
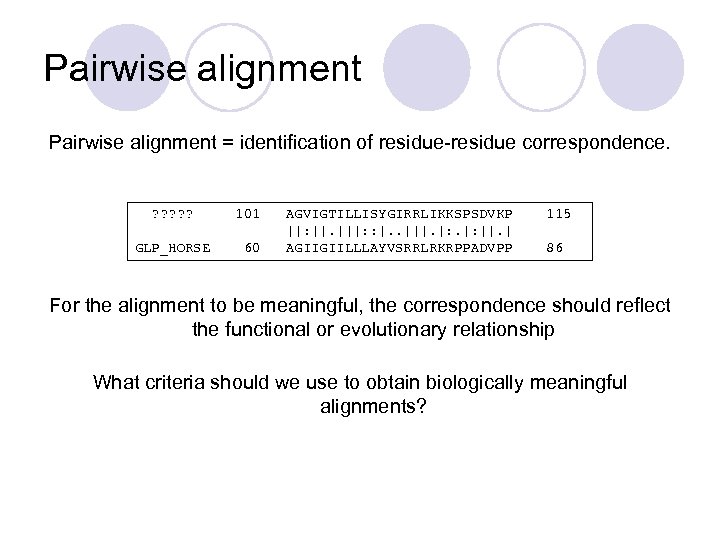
Pairwise alignment = identification of residue-residue correspondence. ? ? ? GLP_HORSE 101 60 AGVIGTILLISYGIRRLIKKSPSDVKP ||: ||. |||: : |. . |||. |: ||. | AGIIGIILLLAYVSRRLRKRPPADVPP 115 86 For the alignment to be meaningful, the correspondence should reflect the functional or evolutionary relationship What criteria should we use to obtain biologically meaningful alignments?
Terminology l Identity: ¡ l percentage of pairs of identical residues between two aligned sequences. Similarity: ¡ ¡ percentage of pairs of similar residues between two aligned sequences. one must define what similar means. Eg: l l l as observed in well studied evolutionary related protein families, physico-chemical amino acid properties: hydropathy, size, … Homology: ¡ ¡ ¡ two sequences are homologous if and only if they have a common ancestor. it´s either yes or no. Two types: orthology and paralogy not to be confused with similarity! don’t mix up with analogy
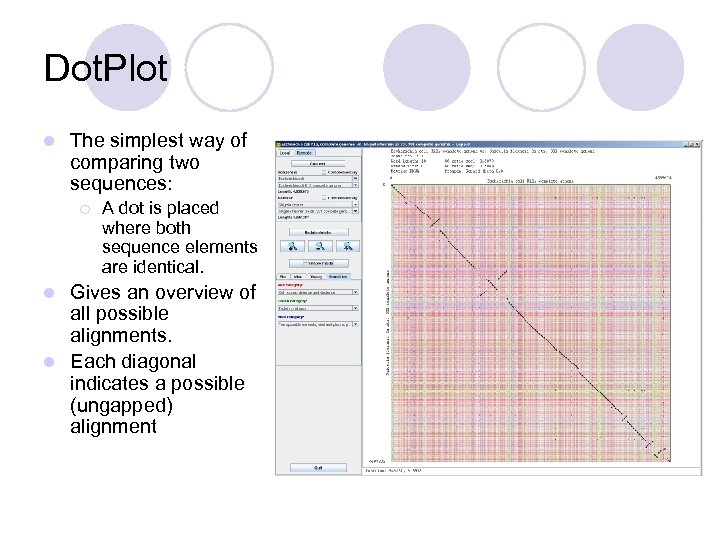
Dot. Plot l The simplest way of comparing two sequences: ¡ A dot is placed where both sequence elements are identical. Gives an overview of all possible alignments. l Each diagonal indicates a possible (ungapped) alignment l
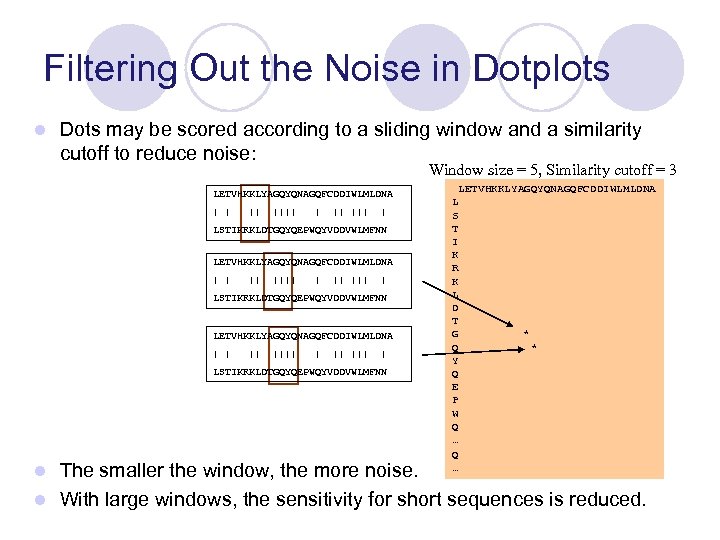
Filtering Out the Noise in Dotplots l Dots may be scored according to a sliding window and a similarity cutoff to reduce noise: Window size = 5, Similarity cutoff = 3 LETVHKKLYAGQYQNAGQFCDDIWLMLDNA | | || |||| ||| | LSTIKRKLDTGQYQEPWQYVDDVWLMFNN LETVHKKLYAGQYQNAGQFCDDIWLMLDNA | | || ||| | LSTIKRKLDTGQYQEPWQYVDDVWLMFNN LETVHKKLYAGQYQNAGQFCDDIWLMLDNA L SLETVHKKLYAGQYQNAGQFCDDIWLMLDNA T L I S K T R I K L R D K T L G * D Q * T Y G Q * E Y P Q W E Q P … W Q … The smaller the window, the more noise. l With large windows, the sensitivity for short sequences is reduced. l
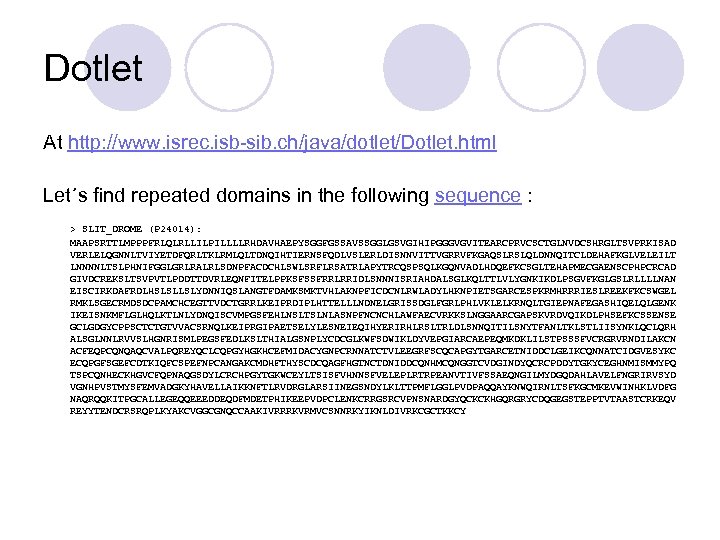
Dotlet At http: //www. isrec. isb-sib. ch/java/dotlet/Dotlet. html Let´s find repeated domains in the following sequence : > SLIT_DROME (P 24014): MAAPSRTTLMPPPFRLQLRLLILPILLLLRHDAVHAEPYSGGFGSSAVSSGGLGSVGIHIPGGGVGVITEARCPRVCSCTGLNVDCSHRGLTSVPRKISAD VERLELQGNNLTVIYETDFQRLTKLRMLQLTDNQIHTIERNSFQDLVSLERLDISNNVITTVGRRVFKGAQSLRSLQLDNNQITCLDEHAFKGLVELEILT LNNNNLTSLPHNIFGGLGRLRALRLSDNPFACDCHLSWLSRFLRSATRLAPYTRCQSPSQLKGQNVADLHDQEFKCSGLTEHAPMECGAENSCPHPCRCAD GIVDCREKSLTSVPVTLPDDTTDVRLEQNFITELPPKSFSSFRRLRRIDLSNNNISRIAHDALSGLKQLTTLVLYGNKIKDLPSGVFKGLGSLRLLLLNAN EISCIRKDAFRDLHSLSLLSLYDNNIQSLANGTFDAMKSMKTVHLAKNPFICDCNLRWLADYLHKNPIETSGARCESPKRMHRRRIESLREEKFKCSWGEL RMKLSGECRMDSDCPAMCHCEGTTVDCTGRRLKEIPRDIPLHTTELLLNDNELGRISSDGLFGRLPHLVKLELKRNQLTGIEPNAFEGASHIQELQLGENK IKEISNKMFLGLHQLKTLNLYDNQISCVMPGSFEHLNSLTSLNLASNPFNCNCHLAWFAECVRKKSLNGGAARCGAPSKVRDVQIKDLPHSEFKCSSENSE GCLGDGYCPPSCTCTGTVVACSRNQLKEIPRGIPAETSELYLESNEIEQIHYERIRHLRSLTRLDLSNNQITILSNYTFANLTKLSTLIISYNKLQCLQRH ALSGLNNLRVVSLHGNRISMLPEGSFEDLKSLTHIALGSNPLYCDCGLKWFSDWIKLDYVEPGIARCAEPEQMKDKLILSTPSSSFVCRGRVRNDILAKCN ACFEQPCQNQAQCVALPQREYQCLCQPGYHGKHCEFMIDACYGNPCRNNATCTVLEEGRFSCQCAPGYTGARCETNIDDCLGEIKCQNNATCIDGVESYKC ECQPGFSGEFCDTKIQFCSPEFNPCANGAKCMDHFTHYSCDCQAGFHGTNCTDNIDDCQNHMCQNGGTCVDGINDYQCRCPDDYTGKYCEGHNMISMMYPQ TSPCQNHECKHGVCFQPNAQGSDYLCRCHPGYTGKWCEYLTSISFVHNNSFVELEPLRTRPEANVTIVFSSAEQNGILMYDGQDAHLAVELFNGRIRVSYD VGNHPVSTMYSFEMVADGKYHAVELLAIKKNFTLRVDRGLARSIINEGSNDYLKLTTPMFLGGLPVDPAQQAYKNWQIRNLTSFKGCMKEVWINHKLVDFG NAQRQQKITPGCALLEGEQQEEEDDEQDFMDETPHIKEEPVDPCLENKCRRGSRCVPNSNARDGYQCKCKHGQRGRYCDQGEGSTEPPTVTAASTCRKEQV REYYTENDCRSRQPLKYAKCVGGCGNQCCAAKIVRRRKVRMVCSNNRKYIKNLDIVRKCGCTKKCY
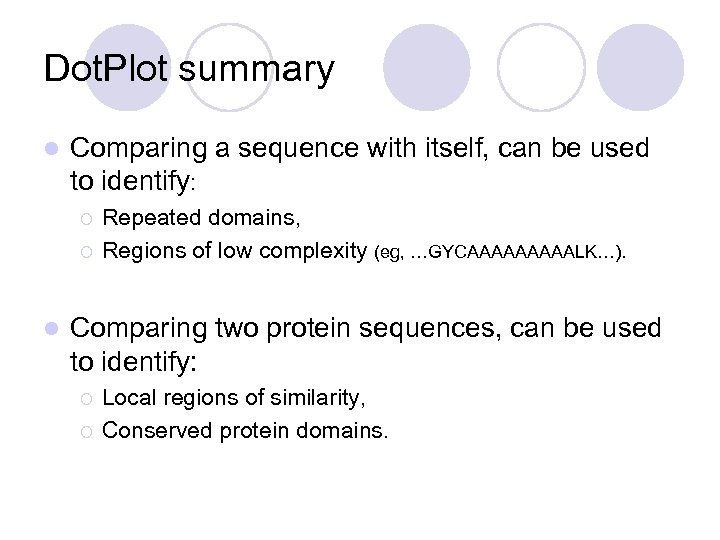
Dot. Plot summary l Comparing a sequence with itself, can be used to identify: ¡ ¡ l Repeated domains, Regions of low complexity (eg, …GYCAAAAALK…). Comparing two protein sequences, can be used to identify: ¡ ¡ Local regions of similarity, Conserved protein domains.
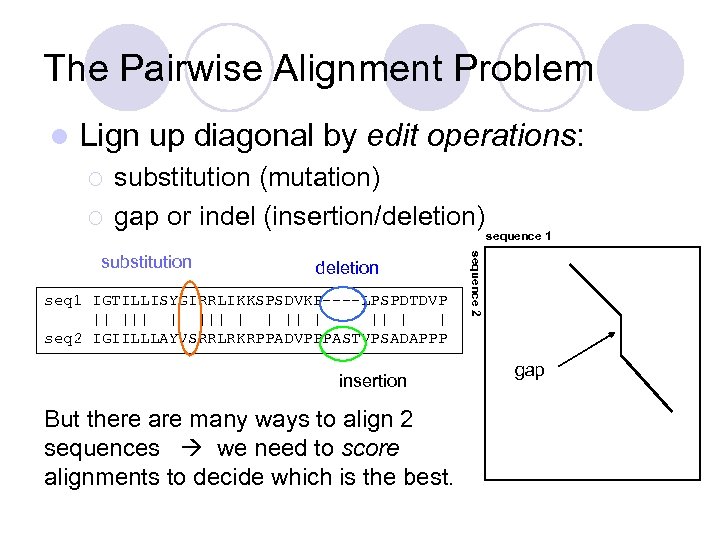
The Pairwise Alignment Problem l Lign up diagonal by edit operations: ¡ ¡ substitution (mutation) gap or indel (insertion/deletion) deletion seq 1 IGTILLISYGIRRLIKKSPSDVKP----LPSPDTDVP || | ||| | | seq 2 IGIILLLAYVSRRLRKRPPADVPPPASTVPSADAPPP insertion But there are many ways to align 2 sequences we need to score alignments to decide which is the best. sequence 2 substitution sequence 1 gap
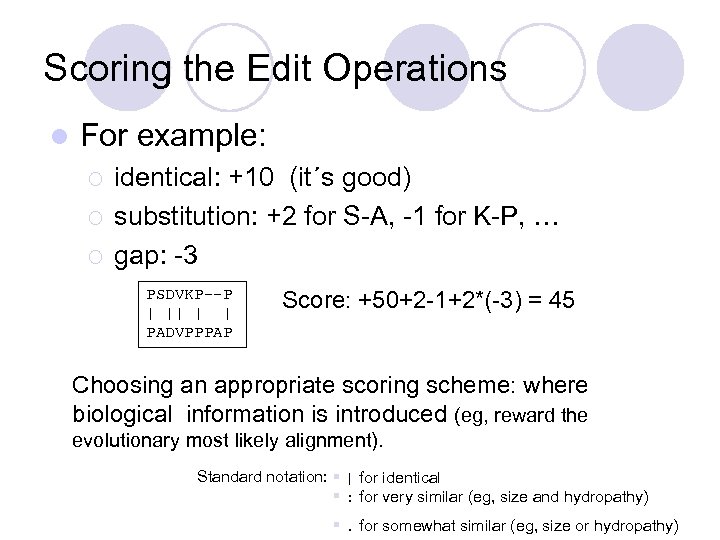
Scoring the Edit Operations l For example: ¡ ¡ ¡ identical: +10 (it´s good) substitution: +2 for S-A, -1 for K-P, … gap: -3 PSDVKP--P | || | | PADVPPPAP Score: +50+2 -1+2*(-3) = 45 Choosing an appropriate scoring scheme: where biological information is introduced (eg, reward the evolutionary most likely alignment). Standard notation: § | for identical § : for very similar (eg, size and hydropathy) §. for somewhat similar (eg, size or hydropathy)
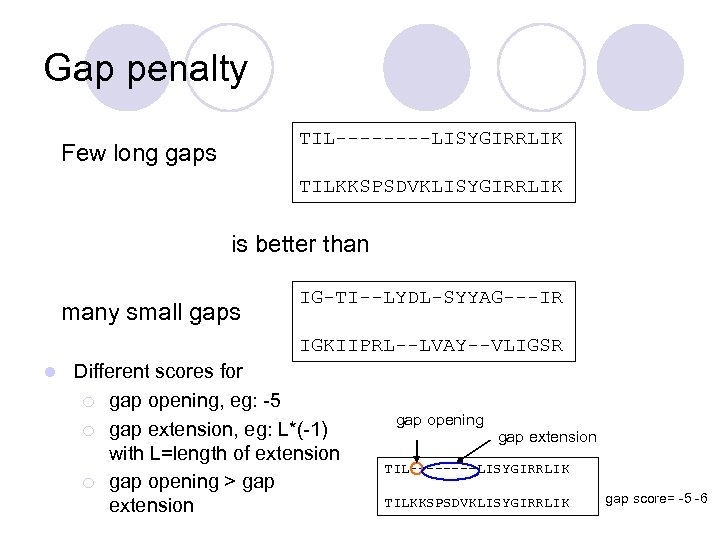
Gap penalty TIL----LISYGIRRLIK Few long gaps TILKKSPSDVKLISYGIRRLIK is better than many small gaps IG-TI--LYDL-SYYAG---IR IGKIIPRL--LVAY--VLIGSR l Different scores for ¡ gap opening, eg: -5 ¡ gap extension, eg: L*(-1) with L=length of extension ¡ gap opening > gap extension gap opening gap extension TIL----LISYGIRRLIK TILKKSPSDVKLISYGIRRLIK gap score= -5 -6

Gap penalty l Can also consider special penalty for gaps at end/beginning of alignment (eg, zero penalty). l Need to be careful in adjusting the gap score to the substitution score: ¡ too strong penalty no gaps, ¡ too weak penalty too many gaps. l Insertions and deletions have been found to occur in nature at significantly lower frequency than mutations.
Residue Substitution l A substitution score for each aa pair a substitution matrix. l Most used: based on evolutionary relationship. l Two types: ¡ PAM series, ¡ BLOSUM series.
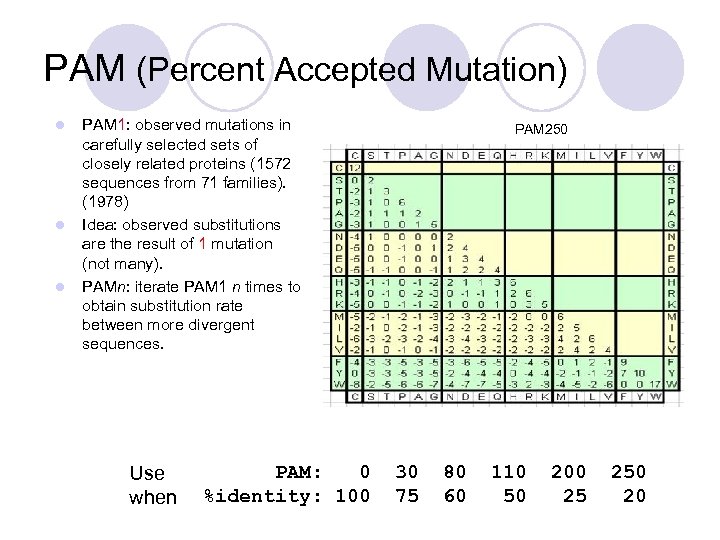
PAM (Percent Accepted Mutation) l l l PAM 1: observed mutations in carefully selected sets of closely related proteins (1572 sequences from 71 families). (1978) Idea: observed substitutions are the result of 1 mutation (not many). PAMn: iterate PAM 1 n times to obtain substitution rate between more divergent sequences. Use when PAM: 0 %identity: 100 PAM 250 30 75 80 60 110 50 200 25 250 20
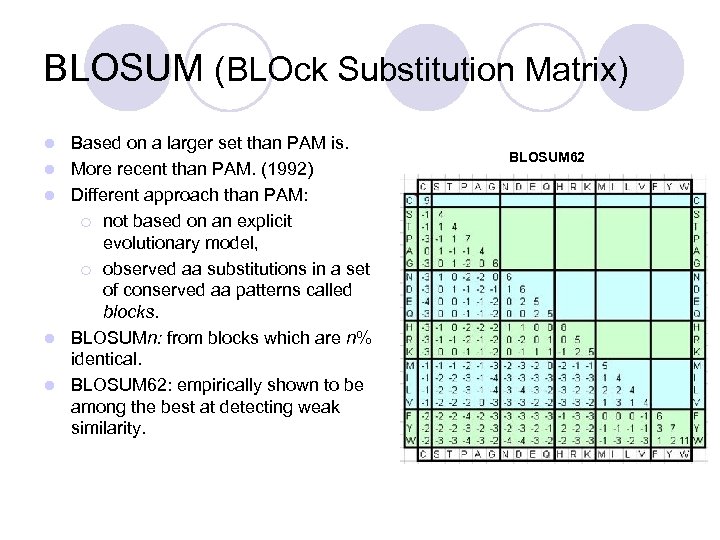
BLOSUM (BLOck Substitution Matrix) l l l Based on a larger set than PAM is. More recent than PAM. (1992) Different approach than PAM: ¡ not based on an explicit evolutionary model, ¡ observed aa substitutions in a set of conserved aa patterns called blocks. BLOSUMn: from blocks which are n% identical. BLOSUM 62: empirically shown to be among the best at detecting weak similarity. BLOSUM 62
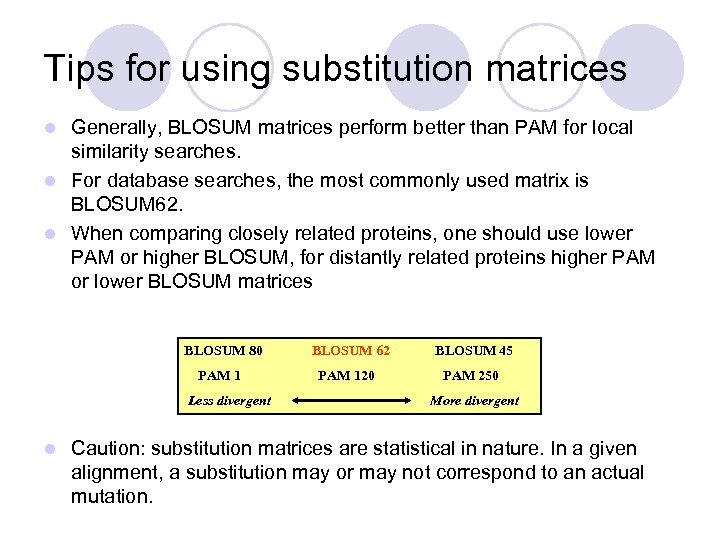
Tips for using substitution matrices Generally, BLOSUM matrices perform better than PAM for local similarity searches. l For database searches, the most commonly used matrix is BLOSUM 62. l When comparing closely related proteins, one should use lower PAM or higher BLOSUM, for distantly related proteins higher PAM or lower BLOSUM matrices l BLOSUM 80 BLOSUM 62 BLOSUM 45 PAM 120 PAM 250 Less divergent l More divergent Caution: substitution matrices are statistical in nature. In a given alignment, a substitution may or may not correspond to an actual mutation.
Pairwise Alignment Algorithms l Given a scoring scheme, an alignment algorithm tries to find the best alignment between 2 sequences according to that scheme. Exact algorithms: ¡ guaranteed to return an alignment with the best possible score. l Heuristic alignments: ¡ not guaranteed to return best alignments. ¡ but they are quicker (and hopefully still return good alignments). l l Two types of alignment: ¡ Global: forced over the entire length of 2 sequences. ¡ Local: between substrings of 2 sequences. .
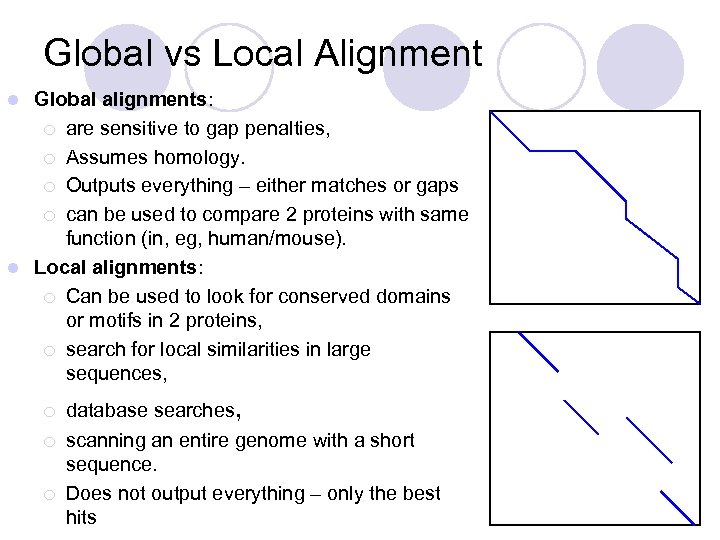
Global vs Local Alignment Global alignments: ¡ are sensitive to gap penalties, ¡ Assumes homology. ¡ Outputs everything – either matches or gaps ¡ can be used to compare 2 proteins with same function (in, eg, human/mouse). l Local alignments: ¡ Can be used to look for conserved domains or motifs in 2 proteins, ¡ search for local similarities in large sequences, l ¡ ¡ ¡ database searches, scanning an entire genome with a short sequence. Does not output everything – only the best hits
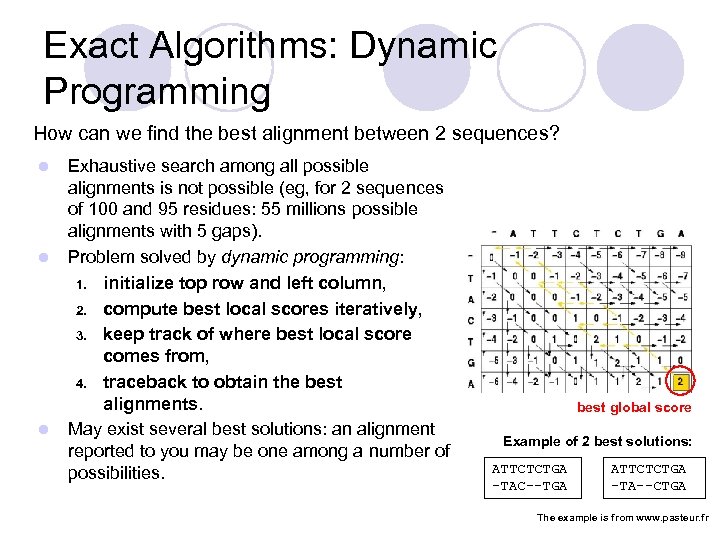
Exact Algorithms: Dynamic Programming How can we find the best alignment between 2 sequences? l l l Exhaustive search among all possible alignments is not possible (eg, for 2 sequences of 100 and 95 residues: 55 millions possible alignments with 5 gaps). Problem solved by dynamic programming: 1. initialize top row and left column, 2. compute best local scores iteratively, 3. keep track of where best local score comes from, 4. traceback to obtain the best alignments. May exist several best solutions: an alignment reported to you may be one among a number of possibilities. best global score Example of 2 best solutions: ATTCTCTGA -TAC--TGA ATTCTCTGA -TA--CTGA The example is from www. pasteur. fr

Local and global Alignment Servers (Exact Algorithm) Use the Needleman-Wunsch algorithm (1970) and the Smith-Waterman algorithm (1981). l Server at EBI: EMBOSS-Align ¡ Let´s submit to http: //www. ebi. ac. uk/emboss/align/index. html the sequence : >uniprot|P 35858|ALS_HUMAN Insulin-like growth factor-binding protein complex MALRKGGLALALLLLSWVALGPRSLEGADPGTPGEAEGPACPAACVCSYDDDADELSVFC SSRNLTRLPDGVPGGTQALWLDGNNLSSVPPAAFQNLSSLGFLNLQGGQLGSLEPQALLG LENLCHLHLERNQLRSLALGTFAHTPALASLGLSNNRLSRLEDGLFEGLGSLWDLNLGWN SLAVLPDAAFRGLGSLRELVLAGNRLAYLQPALFSGLAELRELDLSRNALRAIKANVFVQ LPRLQKLYLDRNLIAAVAPGAFLGLKALRWLDLSHNRVAGLLEDTFPGLLGLRVLRLSHN AIASLRPRTFKDLHFLEELQLGHNRIRQLAERSFEGLGQLEVLTLDHNQLQEVKAGAFLG LTNVAVMNLSGNCLRNLPEQVFRGLGKLHSLHLEGSCLGRIRPHTFTGLSGLRRLFLKDN GLVGIEEQSLWGLAELLELDLTSNQLTHLPHRLFQGLGKLEYLLLSRNRLAELPADALGP LQRAFWLDVSHNRLEALPNSLLAPLGRLRYLSLRNNSLRTFTPQPPGLERLWLEGNPWDC GCPLKALRDFALQNPSAVPRFVQAICEGDDCQPPAYTYNNITCASPPEVVGLDLRDLSEA HFAPC >uniprot|O 08770|GPV_RAT Platelet glycoprotein V precursor (GPV) (CD 42 D). MLRSVLLSAVLSLVGAQPFPCPKTCKCVVRDAVQCSGGSVAHIAELGLPTNLTHILLFRM DRGVLQSHSFSGMTVLQRLMLSDSHISAIDPGTFNDLVKLKTLRLTRNKISHLPRAILDK MVLLEQLFLDHNALRDLDQNLFQKLLNLRDLCLNQNQLSFLPANLFSSLGKLKVLDLSRN NLTHLPQGLLGAQIKLEKLLLYSNRLMSLDSGLLANLGALTELRLERNHLRSIAPGAFDS LGNLSTLTLSGNLLESLPPALFLHVSWLTRLTLFENPLEELPEVLFGEMAGLRELWLNGT HLRTLPAAAFRNLSGLQTLGLTRNPLLSALPPGMFHGLTELRVLAVHTNALEELPEDALR GLGRLRQVSLRHNRLRALPRTLFRNLSSLVTVQLEHNQLKTLPGDVFAALPQLTRVLLGH NPWLCDCGLWPFLQWLRHHLELLGRDEPPQCNGPESRASLTFWELLQGDQWCPSSRGLPP DPPTENALKAPDPTQRPNSSQSWAWVQLVARGESPDNRFYWNLYILLLIAQATIAGFIVF AMIKIGQLFRTLIREELLFEAMGKSSN
Heuristic Algorithms l Motivations: ¡ Exact algorithms are exhaustive but computationally expensive. ¡ Exact algorithms are impractical for comparing a query sequence to millions of other sequences in a database (database scanning), ¡ and so, database scanning requires faster alignment algorithm (at the cost of optimality).
Heuristic Algorithms l Probing a database with a query is similar to aligning a query with a very long sequence. need fast local alignment methods. Main idea: ¡ Use dynamic programming, but limited to (sub-)sequences which are likely to produce interesting alignments with the query. ¡ Heuristic part of the algorithm: eliminate from search uninteresting sequences (need to make a guess). l Algorithms: ¡ FASTA : Lipman-Pearson (1985). ¡ BLAST (Basic Local Alignment Search Tool) : Altshul et al. (1990). l

BLAST Overview Many versions for different query-database cases: ¡ blastp: protein - protein ¡ blastn: nucleotide - nucleotide ¡ blastx: nucleotide protein - protein ¡ tblastn: protein - protein nucleotide ¡ tblastx: nucleotide protein - protein nucleotide l Comes in many flavours. l Fast and reliable. l Easy to use. l

BLAST Overview BLAST computes “an alignment”, not necessarily the exact optimal alignment. l Given the query and the database (long sequence): ¡ Find all words of length k (default: k=3 for AA and k=11 for DNA) that match the query with a score high enough. ¡ Look for subsequences in the database that contain these words. ¡ Extend subsequences to see if match score can be increased. ¡ Compute total score when no more extensions are possible. l Rank the alignments. l

BLAST at NCBI Let´s submit the query sequence >1 IGR: A INSULIN-LIKE GROWTH FACTOR RECEPTOR EICGPGIDIRNDYQQLKRLENCTVIEGYLHILLISKAEDYRSYR FPKLTVITEYSLGDLFPNLTVIRGWKLFYNYALVIFEMTNLKDI GLYNLRNITRGAIRIEKNADLCYLSTVDWSLILDAVSNNYIVGN KPPKECGDLCPGTMEEKPMCEKTTINNEYNYRCWTTNRCQKMCP STCGKRACTENNECCHPECLGSCSAPDNDTACVACRHYYYAGVC VPACPPNTYRFEGWRCVDRDFCANILSAESSDSEGFVIHDGECM QECPSGFIRNGSQSMYCIPCEGPCPKVCEEEKKTKTIDSVTSAQ MLQGCTIFKGNLLINIRRGNNIASELENFMGLIEVVTGYVKIRH SHALVSLSFLKNLRLILGEEQLEGNYSFYVLDNQNLQQLWDWDH RNLTIKAGKMYFAFNPKLCVSEIYRMEEVTGTKGRQSKGDINTR NNGERASCESDVDDDDKEQKLISEEDLN At http: //www. ncbi. nlm. nih. gov/BLAST/
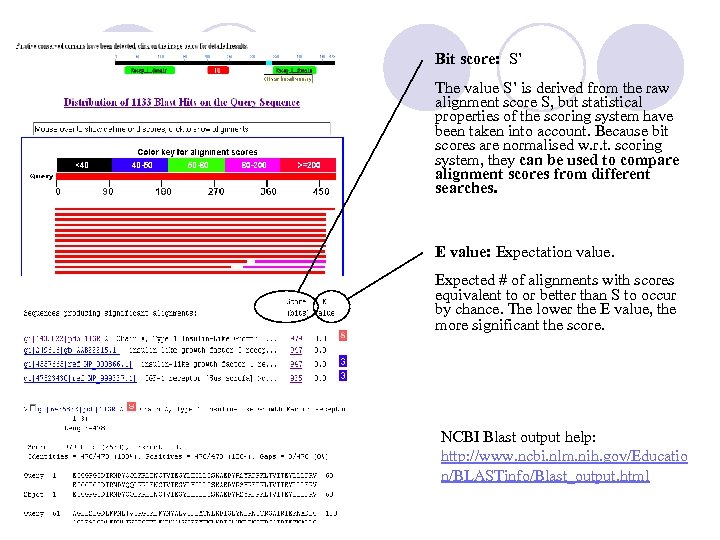
Bit score: S’ The value S’ is derived from the raw alignment score S, but statistical properties of the scoring system have been taken into account. Because bit scores are normalised w. r. t. scoring system, they can be used to compare alignment scores from different searches. E value: Expectation value. Expected # of alignments with scores equivalent to or better than S to occur by chance. The lower the E value, the more significant the score. NCBI Blast output help: http: //www. ncbi. nlm. nih. gov/Educatio n/BLASTinfo/Blast_output. html
BLAST servers Pairwise alignment: ¡ BLAST: http: //www. ncbi. nlm. nih. gov/blast/bl 2 seq/wblast 2. cgi l Database screening: ¡ BLAST: l http: //www. ncbi. nlm. nih. gov/BLAST/ l http: //www. ebi. ac. uk/blast/index. html l http: //www. ch. embnet. org/software/b. BLAST. html l http: //www. ch. embnet. org/software/a. BLAST. html l Remark: there is a server with a powerful implementation of Smith-Waterman for database screening: http: //www. ebi. ac. uk/MPsrch/. Runs about 50 times slower, but is more sensitive and returns less false positives than Blast.

PSI-BLAST l Position-Specific Iterated Blast: ¡ More sensitive, ie better at detecting distant relationships, than BLAST. ¡ Computes position-specific substitution matrices (PSSMs) to score matches between query and database sequences. (Blast uses precomputed substitution matrices, eg BLOSUM 62. )

PSI-BLAST l Repeatedly searches the target databases. l At each round: ¡ compute a multiple alignment of high scoring sequences to generate a new PSSM for next round of searching. l Iterates until no new sequences found (or until a maximal number of iteration is reached).
Significance of Alignments Scores cannot be used to rank alignments: ¡ a bad but long alignment may have a higher score than a good but short alignment. l We need a normalized scoring scheme that would allow to compare alignments, and evaluate their biological significance. l Idea: ¡ Probe the database with random sequences. ¡ This gives a distribution of scores (it follows the extremevalue distribution). ¡ Establish a threshold for significance. l
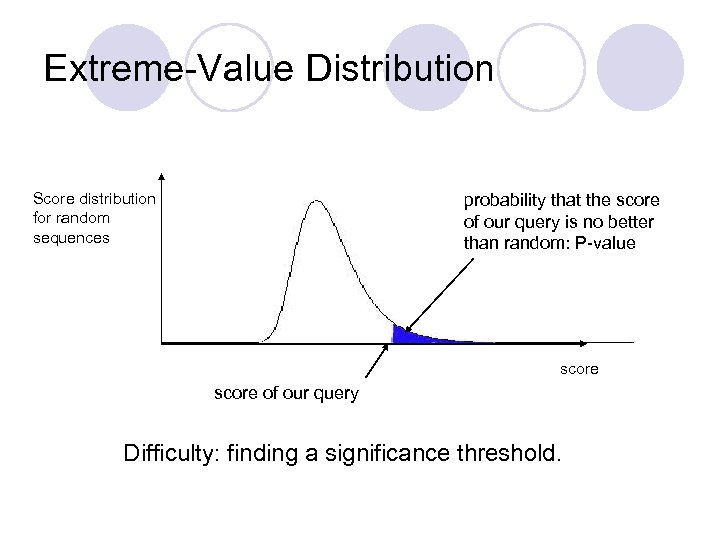
Extreme-Value Distribution Score distribution for random sequences probability that the score of our query is no better than random: P-value score of our query Difficulty: finding a significance threshold.
Quantifying the Significance of Alignments For an alignment with raw score S: l P-value: ¡ The probability of an alignment occurring with score S or better if the aligned-against sequence is random. ¡ The lower the P-value, the more significant the alignment. l E-value: ¡ Expected number of alignments with scores equivalent to or better than S to occur by chance only. ¡ The lower the E-value, the more significant the alignment. ¡ E-value = P-value * size of database.
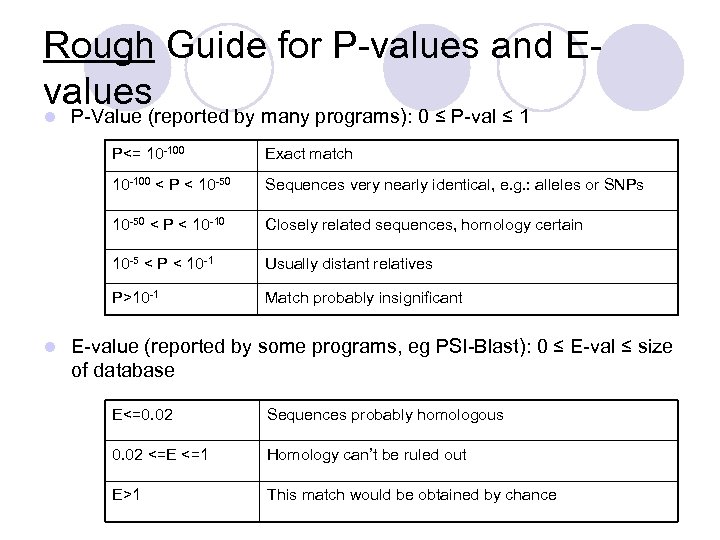
Rough Guide for P-values and Evalues l P-Value (reported by many programs): 0 ≤ P-val ≤ 1 P<= 10 -100 < P < 10 -50 Sequences very nearly identical, e. g. : alleles or SNPs 10 -50 < P < 10 -10 Closely related sequences, homology certain 10 -5 < P < 10 -1 Usually distant relatives P>10 -1 l Exact match Match probably insignificant E-value (reported by some programs, eg PSI-Blast): 0 ≤ E-val ≤ size of database E<=0. 02 Sequences probably homologous 0. 02 <=E <=1 Homology can’t be ruled out E>1 This match would be obtained by chance

Rules of thumb for pairwise alignment Use server defaults in the absence of any other information. l Adjust the substitution matrix to the expected divergence of the 2 sequences. Use BLOSUM 62 if no a priori information. l For distantly related sequences, use PSI-Blast rather than BLAST. If PSI-BLAST doesn’t give you anything use GTG. l Many ways of aligning 2 sequences. ¡ A returned alignment is not the absolute truth. ¡ Inspect the alignment from the biologist´s perspective. l

Exercises l Go to the course web page and start with exercises given in file: p_alignment_exercises. doc l http: //ekhidna. biocenter. helsinki. fi/how
ceb66dce095e873743b3539a726852c7.ppt