 Progress with Sequencing Tomato Chromosome 4 Clare Riddle Tomato Project Group Wellcome Trust Sanger Institute 13 th January 2008 Plant & Animal Genome Conference
Progress with Sequencing Tomato Chromosome 4 Clare Riddle Tomato Project Group Wellcome Trust Sanger Institute 13 th January 2008 Plant & Animal Genome Conference
 A Summary of… Map progress Strategies for additional chromosome coverage Chromosome 4 sequencing and clone selection update BAC Finishing report Copyright Wellcome Trust Medical Photographic Library
A Summary of… Map progress Strategies for additional chromosome coverage Chromosome 4 sequencing and clone selection update BAC Finishing report Copyright Wellcome Trust Medical Photographic Library
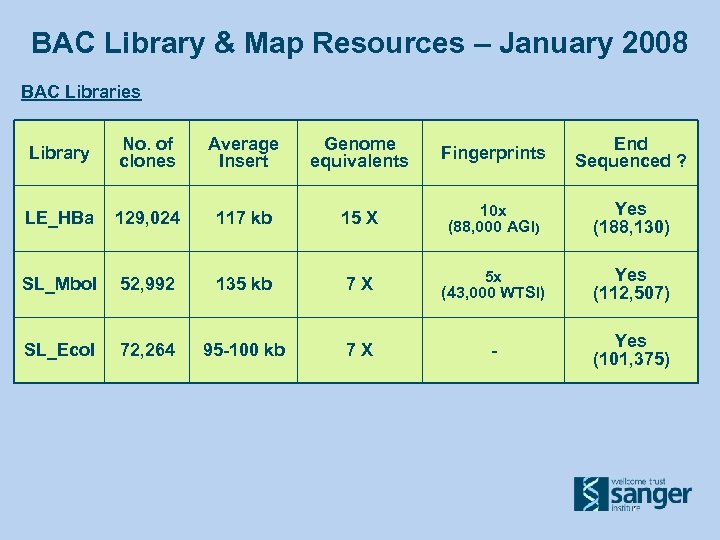 BAC Library & Map Resources – January 2008 BAC Libraries Library No. of clones Average Insert Genome equivalents Fingerprints End Sequenced ? LE_HBa 129, 024 117 kb 15 X 10 x (88, 000 AGI) Yes (188, 130) SL_Mbo. I 52, 992 135 kb 7 X 5 x (43, 000 WTSI) Yes (112, 507) SL_Eco. I 72, 264 95 -100 kb 7 X - Yes (101, 375)
BAC Library & Map Resources – January 2008 BAC Libraries Library No. of clones Average Insert Genome equivalents Fingerprints End Sequenced ? LE_HBa 129, 024 117 kb 15 X 10 x (88, 000 AGI) Yes (188, 130) SL_Mbo. I 52, 992 135 kb 7 X 5 x (43, 000 WTSI) Yes (112, 507) SL_Eco. I 72, 264 95 -100 kb 7 X - Yes (101, 375)
 Additional Resources Fosmid library (end-sequenced Dec 2007) – smaller than BACs – known insert size Ø Contig extension Ø Gap closure Ø Confirm assembly – 107, 681 tomato reads added to trace archive http: //trace. ensembl. org
Additional Resources Fosmid library (end-sequenced Dec 2007) – smaller than BACs – known insert size Ø Contig extension Ø Gap closure Ø Confirm assembly – 107, 681 tomato reads added to trace archive http: //trace. ensembl. org
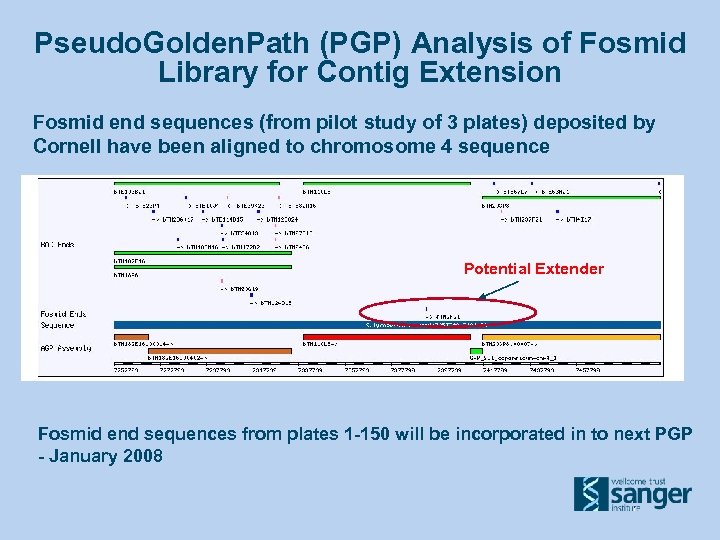 Pseudo. Golden. Path (PGP) Analysis of Fosmid Library for Contig Extension Fosmid end sequences (from pilot study of 3 plates) deposited by Cornell have been aligned to chromosome 4 sequence Potential Extender Fosmid end sequences from plates 1 -150 will be incorporated in to next PGP - January 2008
Pseudo. Golden. Path (PGP) Analysis of Fosmid Library for Contig Extension Fosmid end sequences (from pilot study of 3 plates) deposited by Cornell have been aligned to chromosome 4 sequence Potential Extender Fosmid end sequences from plates 1 -150 will be incorporated in to next PGP - January 2008
 Chromosome 4 Map – January 2008 • 44 chromosome 4 FPC contigs • Tiling paths selected across contigs • Additional contig coverage required Strategies to identify further coverage: • Screen unanchored markers • Walk from contig ends by hybridization Libraries gridded inhouse
Chromosome 4 Map – January 2008 • 44 chromosome 4 FPC contigs • Tiling paths selected across contigs • Additional contig coverage required Strategies to identify further coverage: • Screen unanchored markers • Walk from contig ends by hybridization Libraries gridded inhouse
 Chromosome Position and Localisation • FISH analysis for Chr 4 Placement • Repeat Assessment of clones to determine heterochromatic/euchromatic boundaries
Chromosome Position and Localisation • FISH analysis for Chr 4 Placement • Repeat Assessment of clones to determine heterochromatic/euchromatic boundaries
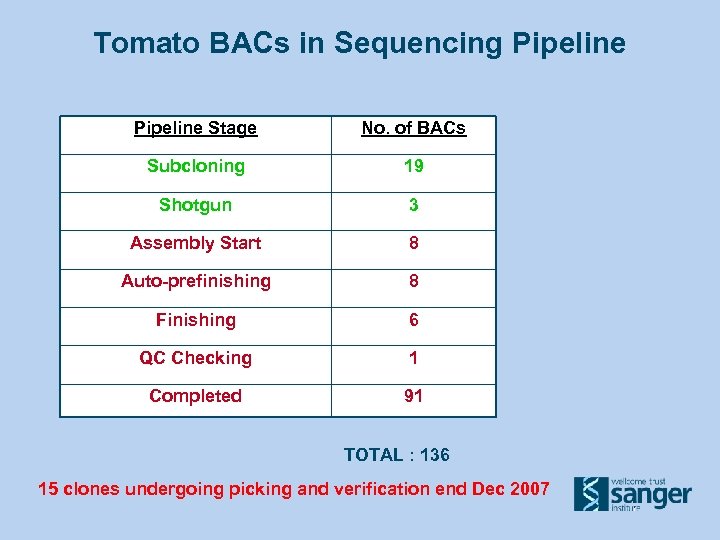 Tomato BACs in Sequencing Pipeline Stage No. of BACs Subcloning 19 Shotgun 3 Assembly Start 8 Auto-prefinishing 8 Finishing 6 QC Checking 1 Completed 91 TOTAL : 136 15 clones undergoing picking and verification end Dec 2007
Tomato BACs in Sequencing Pipeline Stage No. of BACs Subcloning 19 Shotgun 3 Assembly Start 8 Auto-prefinishing 8 Finishing 6 QC Checking 1 Completed 91 TOTAL : 136 15 clones undergoing picking and verification end Dec 2007
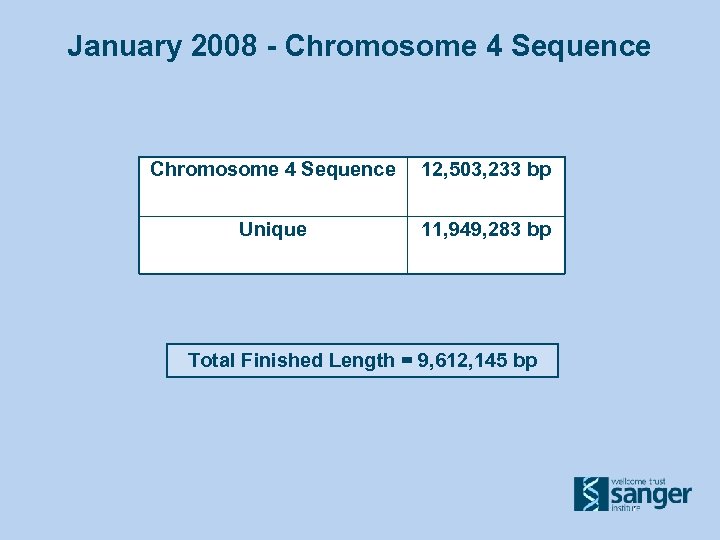 January 2008 - Chromosome 4 Sequence 12, 503, 233 bp Unique 11, 949, 283 bp Total Finished Length = 9, 612, 145 bp
January 2008 - Chromosome 4 Sequence 12, 503, 233 bp Unique 11, 949, 283 bp Total Finished Length = 9, 612, 145 bp
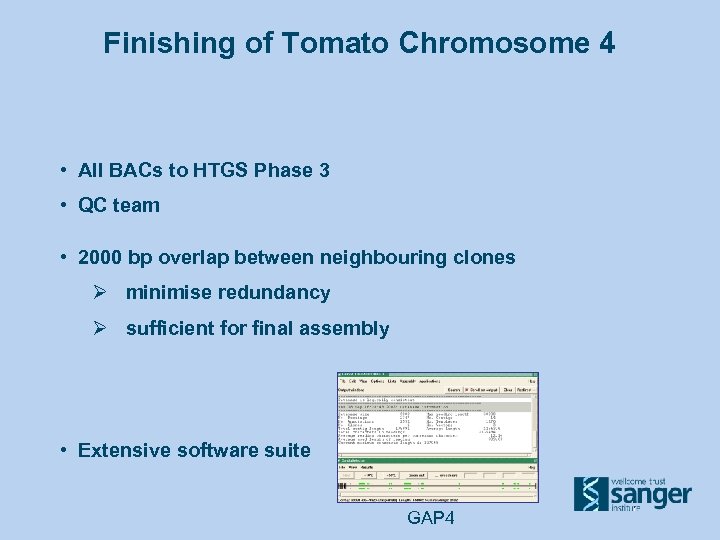 Finishing of Tomato Chromosome 4 • All BACs to HTGS Phase 3 • QC team • 2000 bp overlap between neighbouring clones Ø minimise redundancy Ø sufficient for final assembly • Extensive software suite GAP 4
Finishing of Tomato Chromosome 4 • All BACs to HTGS Phase 3 • QC team • 2000 bp overlap between neighbouring clones Ø minimise redundancy Ø sufficient for final assembly • Extensive software suite GAP 4
 Acknowledgements Wellcome Trust Sanger Institute: Karen Mc. Laren Clare Riddle Sean Humphray Christine Nicholson Carol Scott Stuart Mc. Laren Matt Jones Christine Lloyd Sarah Sims Karen Oliver Cornell University: Lukas Mueller Jim Giovannoni Steve Tanksley Jane Rogers Arizona Genomics Institute: Rod Wing Seunghee Lee Imperial College London: Gerard Bishop Daniel Buchan James Abbott Sarah Butcher University of Nottingham: Graham Seymour Scottish Crop Research Institute: Glenn Bryan FUNDING Colorado State University: Stephen Stack Suzanne Royer Song-Bin Chang MIPS/IBI Institute for Bioinformatics: Klaus Mayer Remy Bruggmann Wageningen University : Hans de Jong Dora Szinay
Acknowledgements Wellcome Trust Sanger Institute: Karen Mc. Laren Clare Riddle Sean Humphray Christine Nicholson Carol Scott Stuart Mc. Laren Matt Jones Christine Lloyd Sarah Sims Karen Oliver Cornell University: Lukas Mueller Jim Giovannoni Steve Tanksley Jane Rogers Arizona Genomics Institute: Rod Wing Seunghee Lee Imperial College London: Gerard Bishop Daniel Buchan James Abbott Sarah Butcher University of Nottingham: Graham Seymour Scottish Crop Research Institute: Glenn Bryan FUNDING Colorado State University: Stephen Stack Suzanne Royer Song-Bin Chang MIPS/IBI Institute for Bioinformatics: Klaus Mayer Remy Bruggmann Wageningen University : Hans de Jong Dora Szinay