6dc57202f5b3c686569c18d5ade6706b.ppt
- Количество слайдов: 34
 Predicting Protein Function Annotation using Protein Interaction Networks By Tamar Eldad Advisor: Dr. Yanay Ofran 89 -385 Computational Biology - Projects Workshop Bar-Ilan University, the Mina and Everard Goodman Faculty of Life Sciences 1
Predicting Protein Function Annotation using Protein Interaction Networks By Tamar Eldad Advisor: Dr. Yanay Ofran 89 -385 Computational Biology - Projects Workshop Bar-Ilan University, the Mina and Everard Goodman Faculty of Life Sciences 1
 Protein Function Prediction § Exponential increase in the number of proteins being identified by sequence genomics projects § Impossible to perform functional assay for every uncharacterized gene § Turn to sophisticated computational methods for assistance in annotating the huge volume of sequence and structure data being produced § § § homology-based annotation transfer sequence patterns structure similarity structure patterns genomic context microarray data 2
Protein Function Prediction § Exponential increase in the number of proteins being identified by sequence genomics projects § Impossible to perform functional assay for every uncharacterized gene § Turn to sophisticated computational methods for assistance in annotating the huge volume of sequence and structure data being produced § § § homology-based annotation transfer sequence patterns structure similarity structure patterns genomic context microarray data 2
 What is Function? § Biological function has more than one aspect § Sub-cellular to whole-organism context § Physiological aspect § Phenotype The need of a well-defined vocabulary 3
What is Function? § Biological function has more than one aspect § Sub-cellular to whole-organism context § Physiological aspect § Phenotype The need of a well-defined vocabulary 3
 Protein Sequence: Protein Structure: 4
Protein Sequence: Protein Structure: 4
 The Gene Ontology project is a major bioinformatics initiative with the aim of standardizing the representation of gene and gene product attributes across species and databases. The project provides a controlled vocabulary of terms for describing gene product characteristics and gene product annotation data. 6
The Gene Ontology project is a major bioinformatics initiative with the aim of standardizing the representation of gene and gene product attributes across species and databases. The project provides a controlled vocabulary of terms for describing gene product characteristics and gene product annotation data. 6
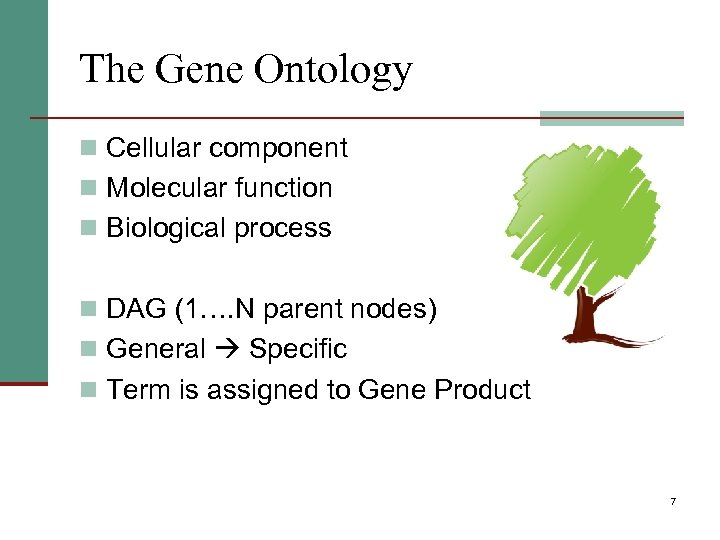 The Gene Ontology n Cellular component n Molecular function n Biological process n DAG (1…. N parent nodes) n General Specific n Term is assigned to Gene Product 7
The Gene Ontology n Cellular component n Molecular function n Biological process n DAG (1…. N parent nodes) n General Specific n Term is assigned to Gene Product 7
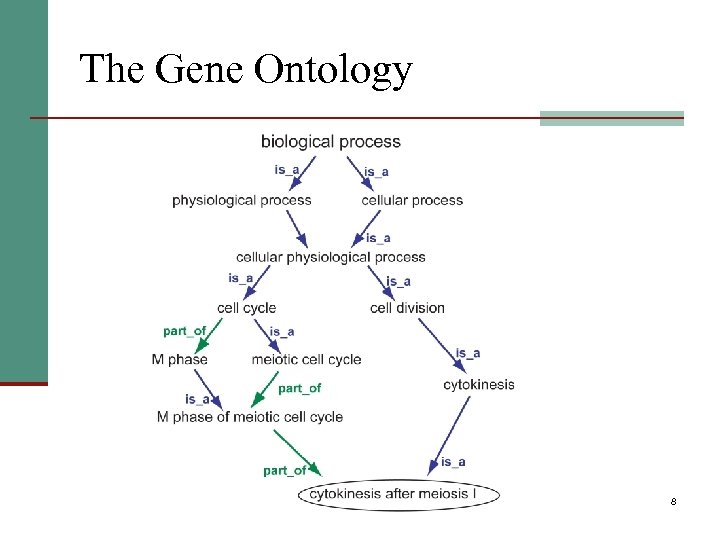 The Gene Ontology 8
The Gene Ontology 8
 A New Approach n Classical Biology – collect a set of features for each protein n Systems Biology – study protein function in the context of a network Assemblies represent more than the sum of their parts 9
A New Approach n Classical Biology – collect a set of features for each protein n Systems Biology – study protein function in the context of a network Assemblies represent more than the sum of their parts 9
 Protein Interactions n Data on thousands of interactions in humans and most model species have become available n mass spectrometry n genome-wide chromatin immunoprecipitation n yeast two-hybrid assays n combinatorial reverse genetic screens n rapid literature mining techniques 10
Protein Interactions n Data on thousands of interactions in humans and most model species have become available n mass spectrometry n genome-wide chromatin immunoprecipitation n yeast two-hybrid assays n combinatorial reverse genetic screens n rapid literature mining techniques 10
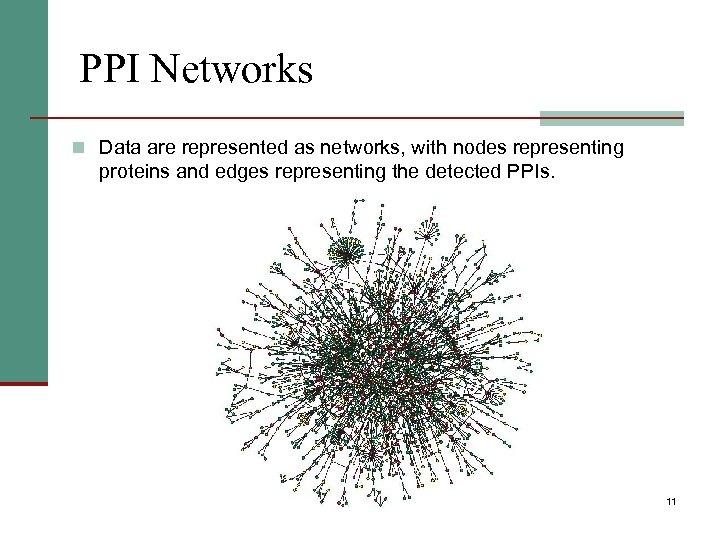 PPI Networks n Data are represented as networks, with nodes representing proteins and edges representing the detected PPIs. 11
PPI Networks n Data are represented as networks, with nodes representing proteins and edges representing the detected PPIs. 11
 Existing Methods § Alignment – aligning sequence-matching proteins between species and checking if they also share network alignment can teach us about conserved pathways between species § Integration - data from different types of networks (i. e. protein, genetic, and transcriptional interaction networks) are integrated in order to get a better picture of the whole biological system § Querying - find sub-networks similar to functional units (by comparing interactions and the proteins themselves) - likely to be functioning units too 12
Existing Methods § Alignment – aligning sequence-matching proteins between species and checking if they also share network alignment can teach us about conserved pathways between species § Integration - data from different types of networks (i. e. protein, genetic, and transcriptional interaction networks) are integrated in order to get a better picture of the whole biological system § Querying - find sub-networks similar to functional units (by comparing interactions and the proteins themselves) - likely to be functioning units too 12
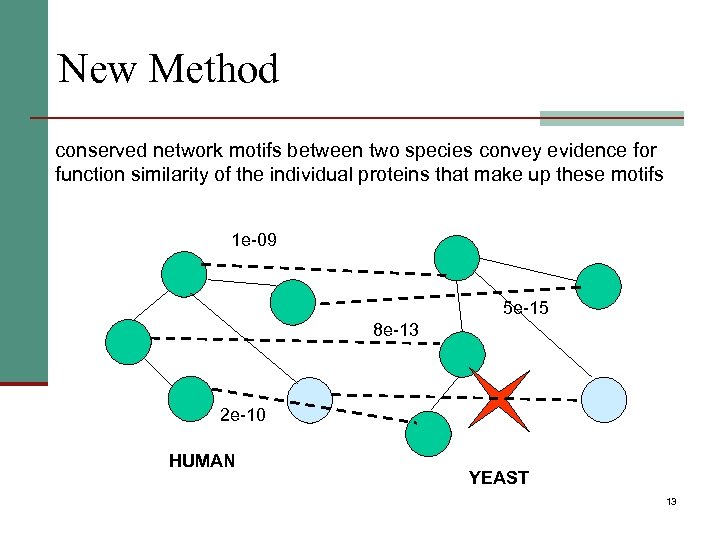 New Method conserved network motifs between two species convey evidence for function similarity of the individual proteins that make up these motifs 1 e-09 5 e-15 8 e-13 2 e-10 HUMAN YEAST 13
New Method conserved network motifs between two species convey evidence for function similarity of the individual proteins that make up these motifs 1 e-09 5 e-15 8 e-13 2 e-10 HUMAN YEAST 13
 New Method What do we need? 1. list of proteins in human cell 2. list of proteins in yeast cell 3. interactions in each cell 4. sequence similarity grades 5. known GO annotations 6. function distance calculation 14
New Method What do we need? 1. list of proteins in human cell 2. list of proteins in yeast cell 3. interactions in each cell 4. sequence similarity grades 5. known GO annotations 6. function distance calculation 14
 Protein Lists - Uni. Prot DB 15
Protein Lists - Uni. Prot DB 15
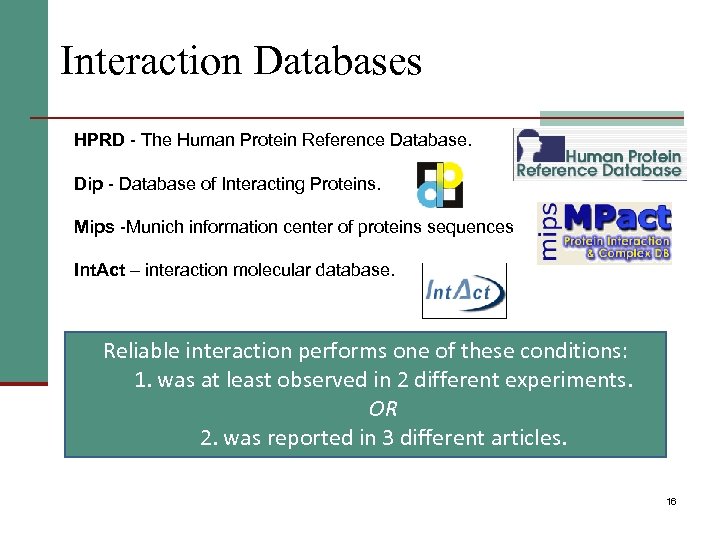 Interaction Databases HPRD - The Human Protein Reference Database. Dip - Database of Interacting Proteins. Mips -Munich information center of proteins sequences Int. Act – interaction molecular database. Reliable interaction performs one of these conditions: 1. was at least observed in 2 different experiments. OR 2. was reported in 3 different articles. 16
Interaction Databases HPRD - The Human Protein Reference Database. Dip - Database of Interacting Proteins. Mips -Munich information center of proteins sequences Int. Act – interaction molecular database. Reliable interaction performs one of these conditions: 1. was at least observed in 2 different experiments. OR 2. was reported in 3 different articles. 16
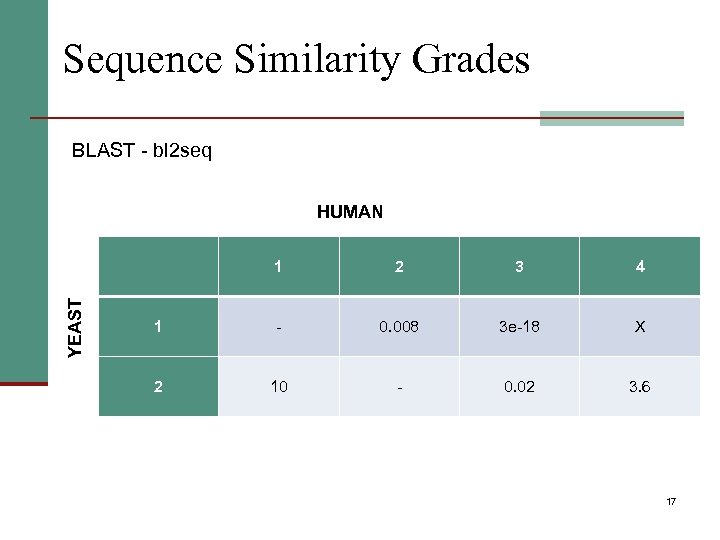 Sequence Similarity Grades BLAST - bl 2 seq HUMAN YEAST 1 2 3 4 1 - 0. 008 3 e-18 X 2 10 - 0. 02 3. 6 17
Sequence Similarity Grades BLAST - bl 2 seq HUMAN YEAST 1 2 3 4 1 - 0. 008 3 e-18 X 2 10 - 0. 02 3. 6 17
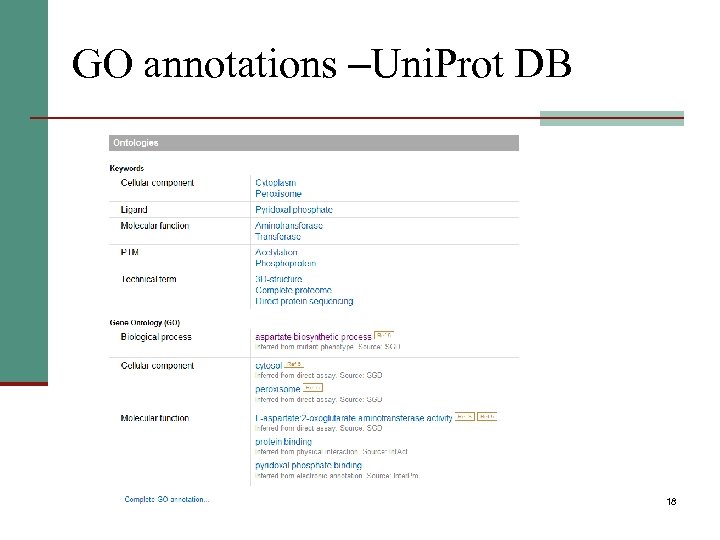 GO annotations –Uni. Prot DB 18
GO annotations –Uni. Prot DB 18
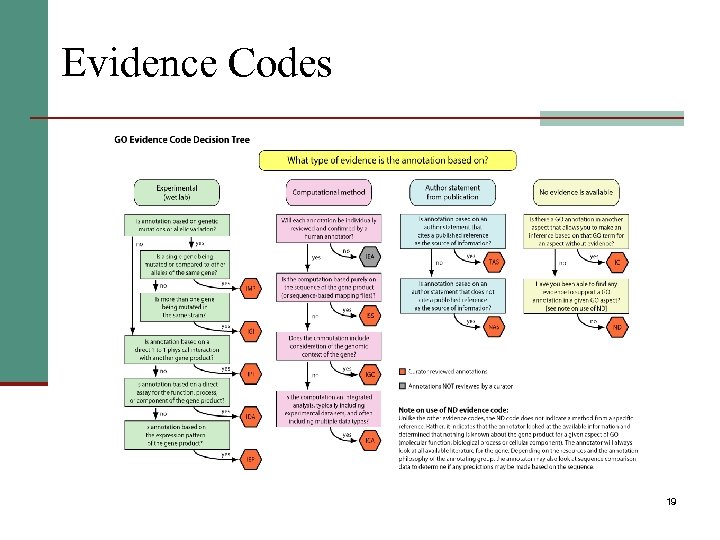 Evidence Codes 19
Evidence Codes 19
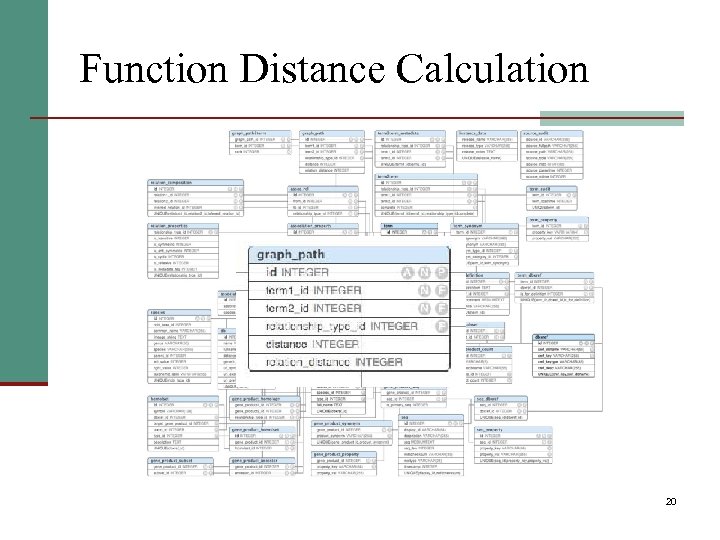 Function Distance Calculation 20
Function Distance Calculation 20
 Implementation 1. Prepare similarity matrix for cutoff e-value 2. Find all components of size N – 1 (DFS search) 3. Compare sub-graphs found using similarity matrix 4. Add N-th non-similar component to each pair of matching graphs 5. Get GO function annotation of N-th components 6. Calculate average distance of N-th component’s function 21
Implementation 1. Prepare similarity matrix for cutoff e-value 2. Find all components of size N – 1 (DFS search) 3. Compare sub-graphs found using similarity matrix 4. Add N-th non-similar component to each pair of matching graphs 5. Get GO function annotation of N-th components 6. Calculate average distance of N-th component’s function 21
 Quality Assurance 1. Compare to random-pair annotation No-sequence similarity 2. Compare to sequence-similar annotation BLAST Only proteins under cut-off value Human genes only 22
Quality Assurance 1. Compare to random-pair annotation No-sequence similarity 2. Compare to sequence-similar annotation BLAST Only proteins under cut-off value Human genes only 22
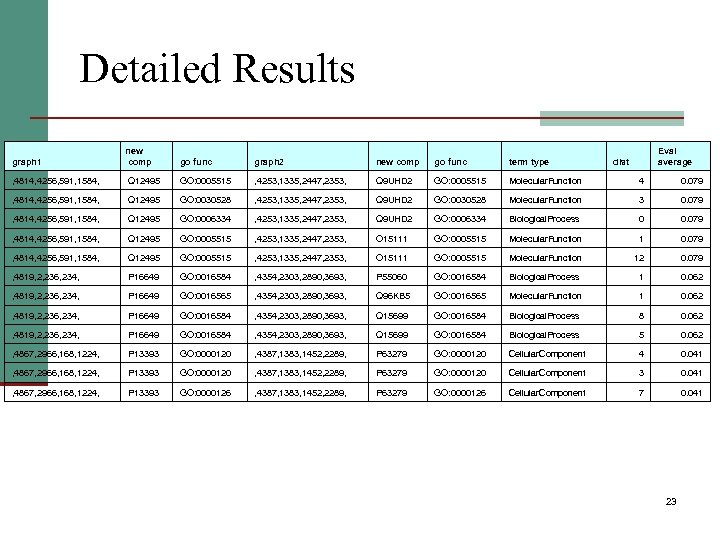 Detailed Results graph 1 new comp go func graph 2 new comp go func term type Eval average , 4814, 4256, 591, 1584, Q 12495 GO: 0005515 , 4253, 1335, 2447, 2353, Q 9 UHD 2 GO: 0005515 Molecular. Function 4 0. 079 , 4814, 4256, 591, 1584, Q 12495 GO: 0030528 , 4253, 1335, 2447, 2353, Q 9 UHD 2 GO: 0030528 Molecular. Function 3 0. 079 , 4814, 4256, 591, 1584, Q 12495 GO: 0006334 , 4253, 1335, 2447, 2353, Q 9 UHD 2 GO: 0006334 Biological. Process 0 0. 079 , 4814, 4256, 591, 1584, Q 12495 GO: 0005515 , 4253, 1335, 2447, 2353, O 15111 GO: 0005515 Molecular. Function 12 0. 079 , 4819, 2, 236, 234, P 16649 GO: 0016584 , 4354, 2303, 2890, 3693, P 55060 GO: 0016584 Biological. Process 1 0. 062 , 4819, 2, 236, 234, P 16649 GO: 0016565 , 4354, 2303, 2890, 3693, Q 96 KB 5 GO: 0016565 Molecular. Function 1 0. 062 , 4819, 2, 236, 234, P 16649 GO: 0016584 , 4354, 2303, 2890, 3693, Q 15699 GO: 0016584 Biological. Process 8 0. 062 , 4819, 2, 236, 234, P 16649 GO: 0016584 , 4354, 2303, 2890, 3693, Q 15699 GO: 0016584 Biological. Process 5 0. 062 , 4867, 2966, 168, 1224, P 13393 GO: 0000120 , 4387, 1383, 1452, 2289, P 63279 GO: 0000120 Cellular. Component 4 0. 041 , 4867, 2966, 168, 1224, P 13393 GO: 0000120 , 4387, 1383, 1452, 2289, P 63279 GO: 0000120 Cellular. Component 3 0. 041 , 4867, 2966, 168, 1224, P 13393 GO: 0000126 , 4387, 1383, 1452, 2289, P 63279 GO: 0000126 Cellular. Component 7 0. 041 dist 23
Detailed Results graph 1 new comp go func graph 2 new comp go func term type Eval average , 4814, 4256, 591, 1584, Q 12495 GO: 0005515 , 4253, 1335, 2447, 2353, Q 9 UHD 2 GO: 0005515 Molecular. Function 4 0. 079 , 4814, 4256, 591, 1584, Q 12495 GO: 0030528 , 4253, 1335, 2447, 2353, Q 9 UHD 2 GO: 0030528 Molecular. Function 3 0. 079 , 4814, 4256, 591, 1584, Q 12495 GO: 0006334 , 4253, 1335, 2447, 2353, Q 9 UHD 2 GO: 0006334 Biological. Process 0 0. 079 , 4814, 4256, 591, 1584, Q 12495 GO: 0005515 , 4253, 1335, 2447, 2353, O 15111 GO: 0005515 Molecular. Function 12 0. 079 , 4819, 2, 236, 234, P 16649 GO: 0016584 , 4354, 2303, 2890, 3693, P 55060 GO: 0016584 Biological. Process 1 0. 062 , 4819, 2, 236, 234, P 16649 GO: 0016565 , 4354, 2303, 2890, 3693, Q 96 KB 5 GO: 0016565 Molecular. Function 1 0. 062 , 4819, 2, 236, 234, P 16649 GO: 0016584 , 4354, 2303, 2890, 3693, Q 15699 GO: 0016584 Biological. Process 8 0. 062 , 4819, 2, 236, 234, P 16649 GO: 0016584 , 4354, 2303, 2890, 3693, Q 15699 GO: 0016584 Biological. Process 5 0. 062 , 4867, 2966, 168, 1224, P 13393 GO: 0000120 , 4387, 1383, 1452, 2289, P 63279 GO: 0000120 Cellular. Component 4 0. 041 , 4867, 2966, 168, 1224, P 13393 GO: 0000120 , 4387, 1383, 1452, 2289, P 63279 GO: 0000120 Cellular. Component 3 0. 041 , 4867, 2966, 168, 1224, P 13393 GO: 0000126 , 4387, 1383, 1452, 2289, P 63279 GO: 0000126 Cellular. Component 7 0. 041 dist 23
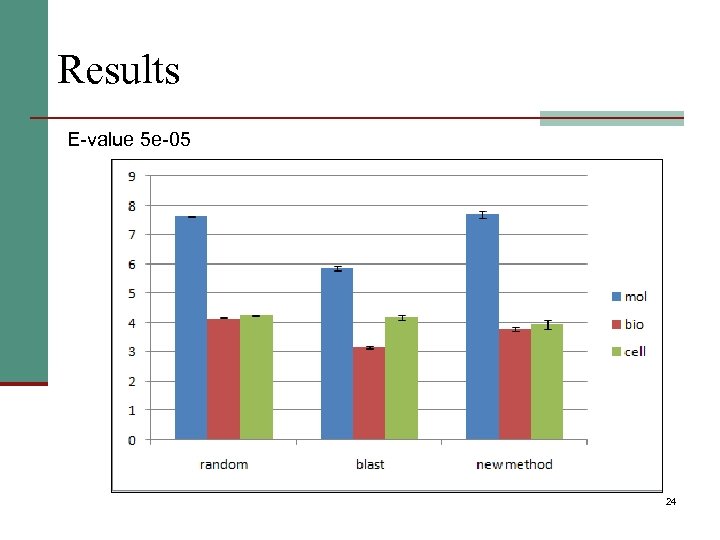 Results E-value 5 e-05 24
Results E-value 5 e-05 24
 Play with Parameters • Change graph size • Lower e-value • Start with larger amount of connected components • Use only graphs with higher connectivity • Non-similar proteins can be any protein in the graph • Different network topology • Limit number of paired proteins 25
Play with Parameters • Change graph size • Lower e-value • Start with larger amount of connected components • Use only graphs with higher connectivity • Non-similar proteins can be any protein in the graph • Different network topology • Limit number of paired proteins 25
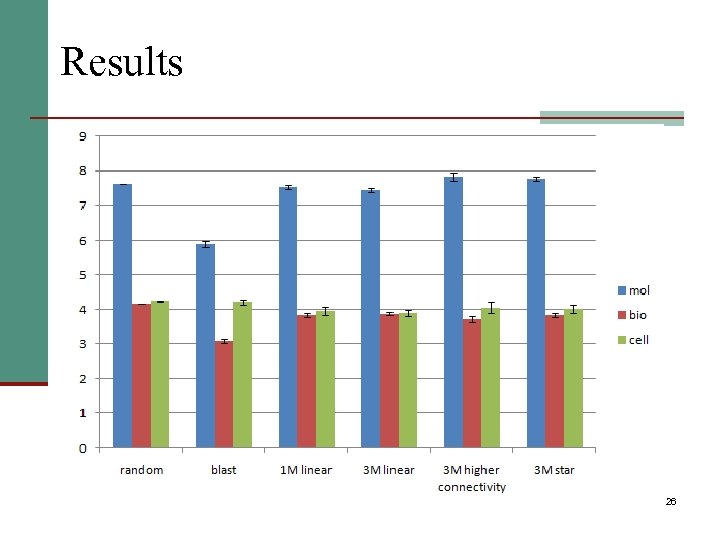 Results 26
Results 26
 Conclusions n Most results are random n Significant improvement only for Biological Process prediction n Still far behind Homology Based Transfer 27
Conclusions n Most results are random n Significant improvement only for Biological Process prediction n Still far behind Homology Based Transfer 27
 Summary n Functional annotation is one of the greatest challenges in the post-genomic era n PPI data for functional annotation as a new approach for promoting this field n Method tried out is unsuccessful n Other Ideas: n n Find a more specific search pattern Start from best results – what specializes them? 28
Summary n Functional annotation is one of the greatest challenges in the post-genomic era n PPI data for functional annotation as a new approach for promoting this field n Method tried out is unsuccessful n Other Ideas: n n Find a more specific search pattern Start from best results – what specializes them? 28
 References n n n Friedberg, I. (2006) Automated function prediction: the genomic challenge. Brief. Bioinform. Accepted for publication Sharan R, Ulitsky I, Shamir R: Network-based prediction of protein function. Mol Syst Biol 2007, 3: 88. Sharan R, Ideker T: Modeling cellular machinery through biological network comparison. Nature Biotechnology 24, 4: 427 - 433. http: //www. geneontology. org/ http: //www. chem. qmul. ac. uk/iubmb/enzyme/ 29
References n n n Friedberg, I. (2006) Automated function prediction: the genomic challenge. Brief. Bioinform. Accepted for publication Sharan R, Ulitsky I, Shamir R: Network-based prediction of protein function. Mol Syst Biol 2007, 3: 88. Sharan R, Ideker T: Modeling cellular machinery through biological network comparison. Nature Biotechnology 24, 4: 427 - 433. http: //www. geneontology. org/ http: //www. chem. qmul. ac. uk/iubmb/enzyme/ 29
 Thanks n Advisor – Dr. Yanay Ofran n Guys at the lab – Rotem, Vered, Sivan n Roi Adadi & Omer Erel 30
Thanks n Advisor – Dr. Yanay Ofran n Guys at the lab – Rotem, Vered, Sivan n Roi Adadi & Omer Erel 30
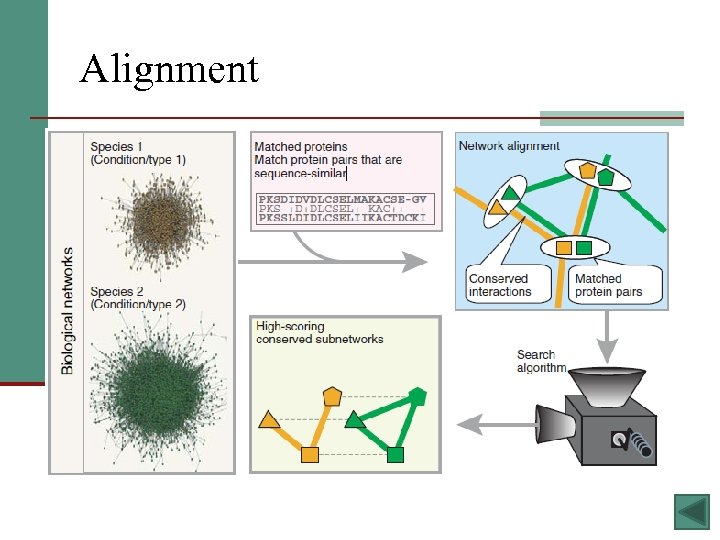 Alignment
Alignment
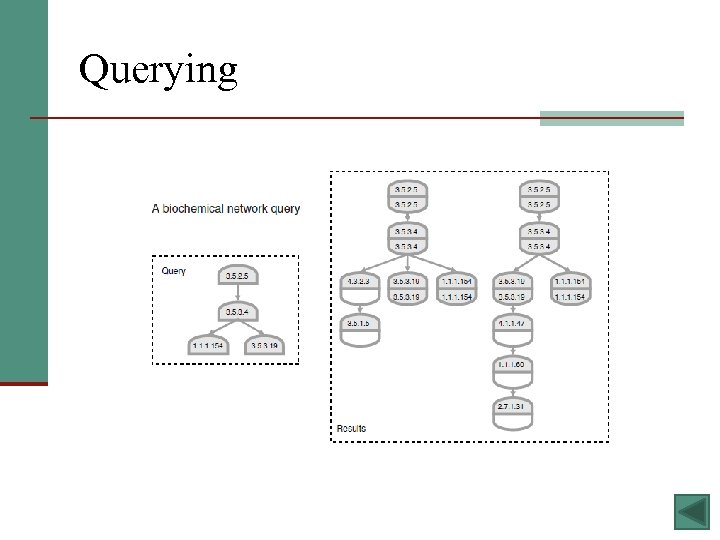 Querying
Querying
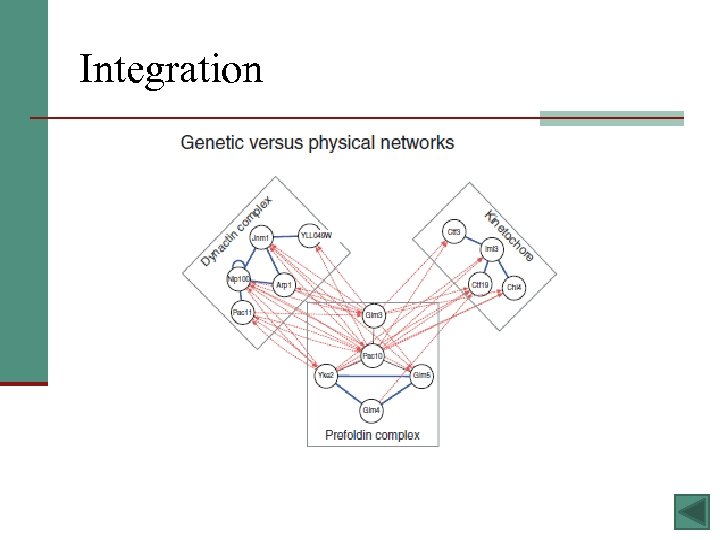 Integration
Integration
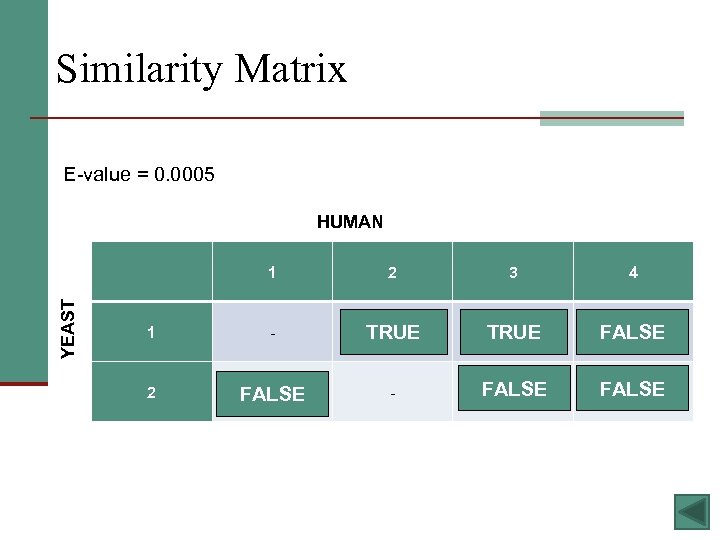 Similarity Matrix E-value = 0. 0005 HUMAN YEAST 1 2 3 4 1 - 0. 008 TRUE 3 e-18 TRUE X FALSE 2 10 FALSE - FALSE 0. 02 FALSE 3. 6
Similarity Matrix E-value = 0. 0005 HUMAN YEAST 1 2 3 4 1 - 0. 008 TRUE 3 e-18 TRUE X FALSE 2 10 FALSE - FALSE 0. 02 FALSE 3. 6
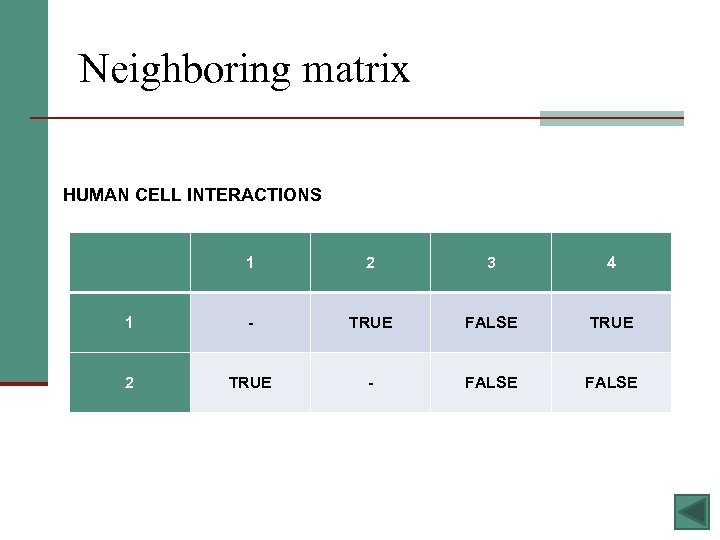 Neighboring matrix HUMAN CELL INTERACTIONS 1 2 3 4 1 - TRUE FALSE TRUE 2 TRUE - FALSE
Neighboring matrix HUMAN CELL INTERACTIONS 1 2 3 4 1 - TRUE FALSE TRUE 2 TRUE - FALSE


