b2690ddc9babeb9ce42d10c8a3b77b83.ppt
- Количество слайдов: 56
 Practical tips for cloning, expressing and purifying proteins for structural biology Aled Edwards Banting and Best Department of Medical Research University of Toronto, Canada aled. edwards@utoronto. ca Affinium Pharmaceuticals Toronto, Canada aedwards@afnm. com
Practical tips for cloning, expressing and purifying proteins for structural biology Aled Edwards Banting and Best Department of Medical Research University of Toronto, Canada aled. edwards@utoronto. ca Affinium Pharmaceuticals Toronto, Canada aedwards@afnm. com
 Molecular biological approaches to structural biology An excellent structural sample usually has the following properties • Lack of conformational heterogeneity • Soluble at high concentrations • Pure Molecular biology is probably fastest way to transform “poor” sample into an “excellent” one.
Molecular biological approaches to structural biology An excellent structural sample usually has the following properties • Lack of conformational heterogeneity • Soluble at high concentrations • Pure Molecular biology is probably fastest way to transform “poor” sample into an “excellent” one.
 Outline • Historical perspective on engineering proteins for structural biology • Practical advice for cloning/purification of structural samples • Ancillary benefits of high-throughput studies
Outline • Historical perspective on engineering proteins for structural biology • Practical advice for cloning/purification of structural samples • Ancillary benefits of high-throughput studies
 RNA polymerase II From 15Å to 3Å by eliminating heterogeneity
RNA polymerase II From 15Å to 3Å by eliminating heterogeneity
 Another source of sample heterogeneity Eukaryotic proteins comprise multiple domains • Conformational heterogeneity lowers probability of crystallization • Protein domains • Are resistant to proteolysis • Fold autonomously • Can usually be expressed in bacteria • Are between 15 and 30 k. Da (NMR or X-ray size) • Are fundamental unit of protein function • Domains are often only tractable targets for HTP crystallography
Another source of sample heterogeneity Eukaryotic proteins comprise multiple domains • Conformational heterogeneity lowers probability of crystallization • Protein domains • Are resistant to proteolysis • Fold autonomously • Can usually be expressed in bacteria • Are between 15 and 30 k. Da (NMR or X-ray size) • Are fundamental unit of protein function • Domains are often only tractable targets for HTP crystallography
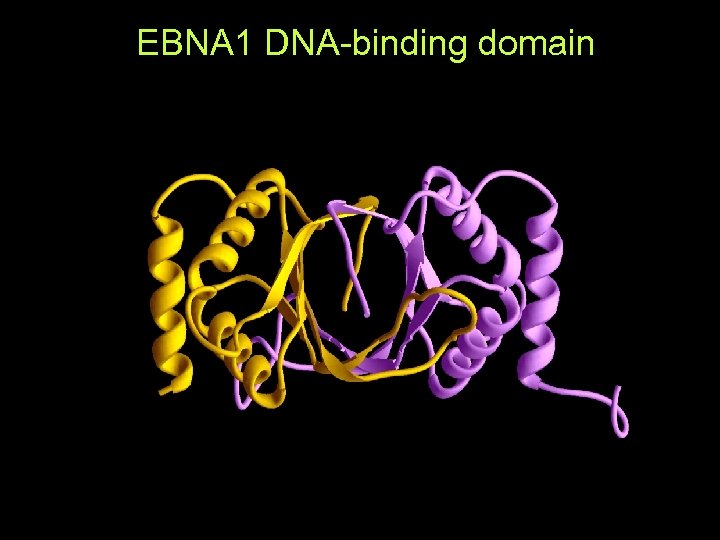 EBNA 1 DNA-binding domain (No sequence homologue in database)
EBNA 1 DNA-binding domain (No sequence homologue in database)
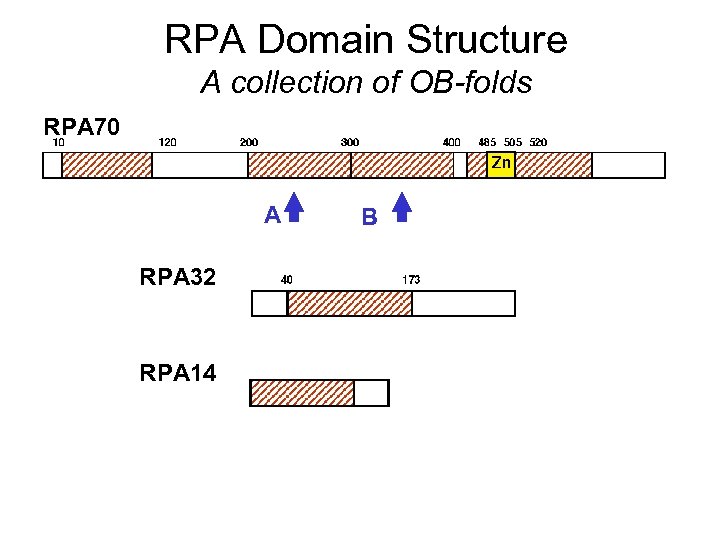 RPA Domain Structure A collection of OB-folds RPA 70 A RPA 32 RPA 14 B
RPA Domain Structure A collection of OB-folds RPA 70 A RPA 32 RPA 14 B
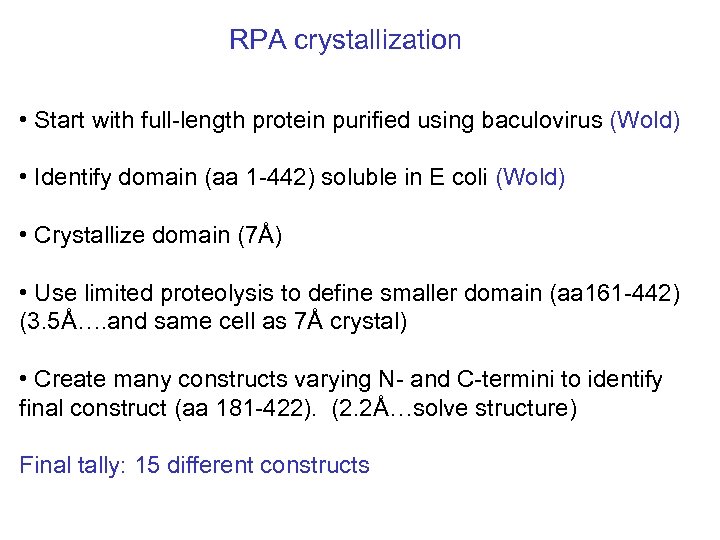 RPA crystallization • Start with full-length protein purified using baculovirus (Wold) • Identify domain (aa 1 -442) soluble in E coli (Wold) • Crystallize domain (7Å) • Use limited proteolysis to define smaller domain (aa 161 -442) (3. 5Å…. and same cell as 7Å crystal) • Create many constructs varying N- and C-termini to identify final construct (aa 181 -422). (2. 2Å…solve structure) Final tally: 15 different constructs
RPA crystallization • Start with full-length protein purified using baculovirus (Wold) • Identify domain (aa 1 -442) soluble in E coli (Wold) • Crystallize domain (7Å) • Use limited proteolysis to define smaller domain (aa 161 -442) (3. 5Å…. and same cell as 7Å crystal) • Create many constructs varying N- and C-termini to identify final construct (aa 181 -422). (2. 2Å…solve structure) Final tally: 15 different constructs
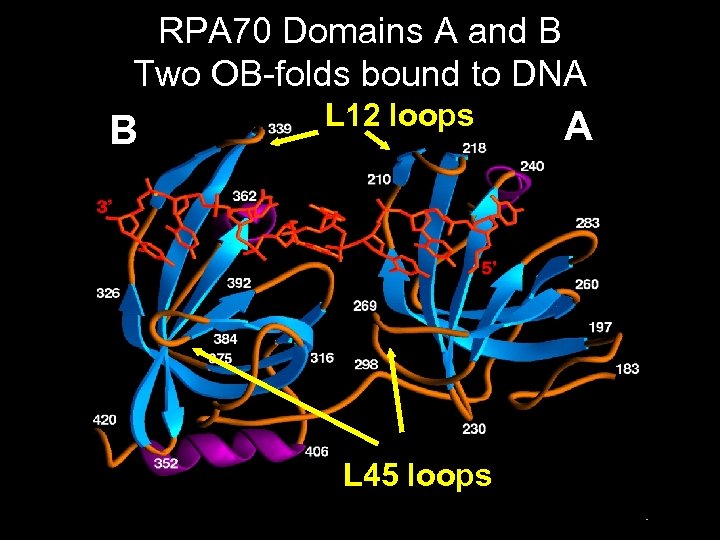 RPA 70 Domains A and B Two OB-folds bound to DNA B L 12 loops L 45 loops A
RPA 70 Domains A and B Two OB-folds bound to DNA B L 12 loops L 45 loops A
 How does one map domains?
How does one map domains?
 Domain mapping using limited proteolysis TFIIS Protease Integrative Proteomics
Domain mapping using limited proteolysis TFIIS Protease Integrative Proteomics
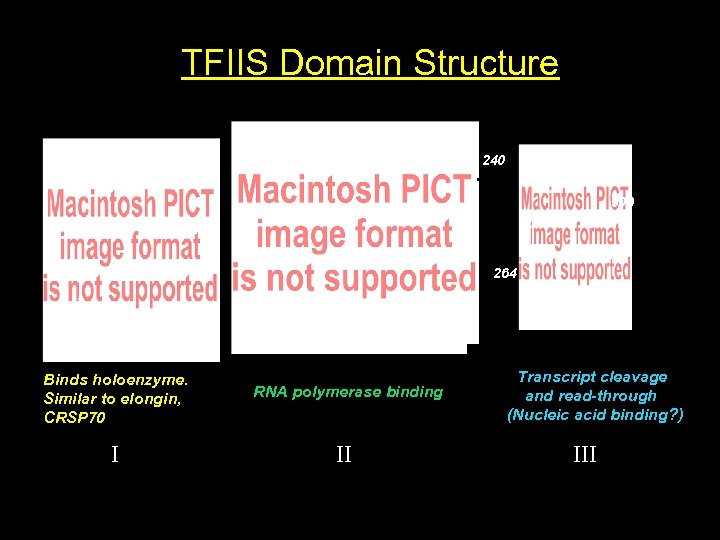 TFIIS Domain Structure 240 309 264 1 124 131 Binds holoenzyme. Similar to elongin, CRSP 70 RNA polymerase binding I II Transcript cleavage and read-through (Nucleic acid binding? ) III
TFIIS Domain Structure 240 309 264 1 124 131 Binds holoenzyme. Similar to elongin, CRSP 70 RNA polymerase binding I II Transcript cleavage and read-through (Nucleic acid binding? ) III
 Domain. Hunter. TM Industrialized Domain Mapping • Partial proteolysis in 96 well plates • Optimized set of proteases • Low protein requirement • No SDS-PAGE • No N-terminal sequencing • Direct identification of domains by mass spectrometry
Domain. Hunter. TM Industrialized Domain Mapping • Partial proteolysis in 96 well plates • Optimized set of proteases • Low protein requirement • No SDS-PAGE • No N-terminal sequencing • Direct identification of domains by mass spectrometry
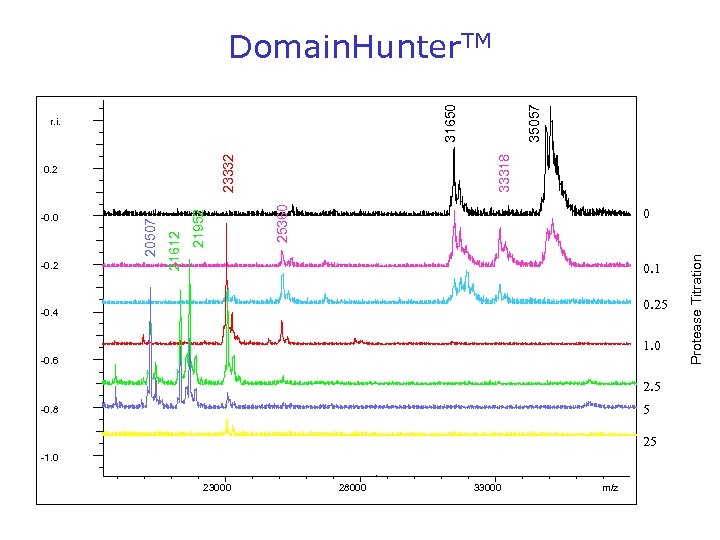 31650 25360 23332 0 0. 1 0. 25 -0. 4 1. 0 -0. 6 2. 5 5 -0. 8 25 -1. 0 23000 28000 33000 m/z Protease Titration 21952 21612 20507 0. 2 -0. 2 33318 r. i. -0. 0 35057 Domain. Hunter. TM
31650 25360 23332 0 0. 1 0. 25 -0. 4 1. 0 -0. 6 2. 5 5 -0. 8 25 -1. 0 23000 28000 33000 m/z Protease Titration 21952 21612 20507 0. 2 -0. 2 33318 r. i. -0. 0 35057 Domain. Hunter. TM
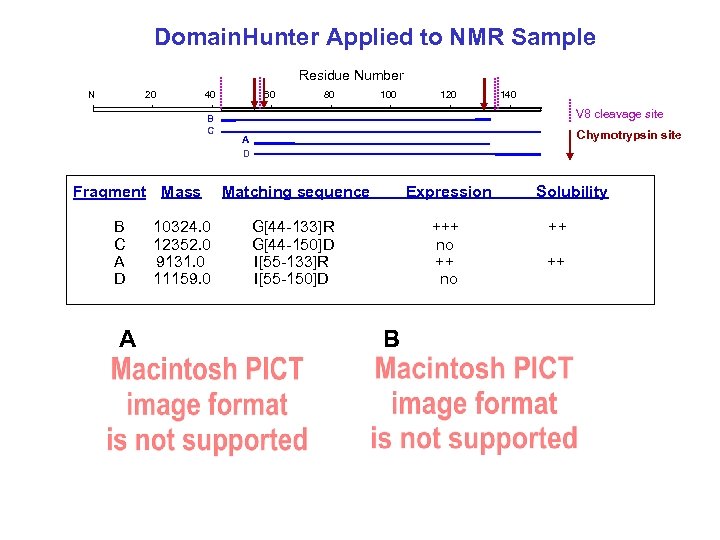 Domain. Hunter Applied to NMR Sample Residue Number N 20 40 B C Fragment Mass B C A D A 10324. 0 12352. 0 9131. 0 11159. 0 60 80 100 120 140 V 8 cleavage site Chymotrypsin site A D Matching sequence Expression G[44 -133]R G[44 -150]D I[55 -133]R I[55 -150]D +++ no B Solubility ++ ++
Domain. Hunter Applied to NMR Sample Residue Number N 20 40 B C Fragment Mass B C A D A 10324. 0 12352. 0 9131. 0 11159. 0 60 80 100 120 140 V 8 cleavage site Chymotrypsin site A D Matching sequence Expression G[44 -133]R G[44 -150]D I[55 -133]R I[55 -150]D +++ no B Solubility ++ ++
 Structural Proteomics MTH 40 MTH 1615 MTH 152 MTH 1184 MTH 1175 MTH 538 MTH 150 MTH 1790 MTH 129 MTH 1699 Nat. Str. Biol. Oct/Nov 2000 MTH 1048
Structural Proteomics MTH 40 MTH 1615 MTH 152 MTH 1184 MTH 1175 MTH 538 MTH 150 MTH 1790 MTH 129 MTH 1699 Nat. Str. Biol. Oct/Nov 2000 MTH 1048
 5 more done 3 more soon
5 more done 3 more soon
 Molecular biology for crystallization and for large-scale studies 1. Basic steps in creating expression vectors for E. coli 2. Practical tips for making fewer mistakes 3. Application of methods to higher-throughput 4. Alternate expression systems 5. Some results
Molecular biology for crystallization and for large-scale studies 1. Basic steps in creating expression vectors for E. coli 2. Practical tips for making fewer mistakes 3. Application of methods to higher-throughput 4. Alternate expression systems 5. Some results
 E coli is the first choice……why? • Cost effective • Easy to grow • Abundance of expertise and reagents • Easy to incorporate selenomethionine • High yield • Rapid doubling time and rapid scale-up
E coli is the first choice……why? • Cost effective • Easy to grow • Abundance of expertise and reagents • Easy to incorporate selenomethionine • High yield • Rapid doubling time and rapid scale-up
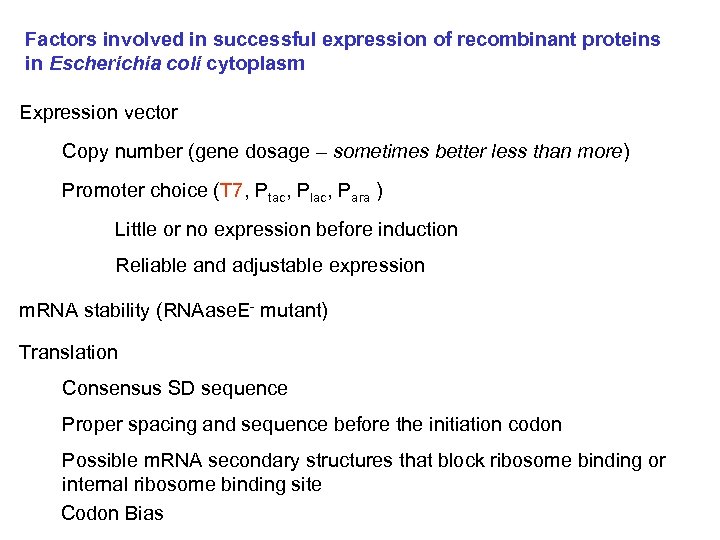 Factors involved in successful expression of recombinant proteins in Escherichia coli cytoplasm Expression vector Copy number (gene dosage – sometimes better less than more) Promoter choice (T 7, Ptac, Plac, Para ) Little or no expression before induction Reliable and adjustable expression m. RNA stability (RNAase. E- mutant) Translation Consensus SD sequence Proper spacing and sequence before the initiation codon Possible m. RNA secondary structures that block ribosome binding or internal ribosome binding site Codon Bias
Factors involved in successful expression of recombinant proteins in Escherichia coli cytoplasm Expression vector Copy number (gene dosage – sometimes better less than more) Promoter choice (T 7, Ptac, Plac, Para ) Little or no expression before induction Reliable and adjustable expression m. RNA stability (RNAase. E- mutant) Translation Consensus SD sequence Proper spacing and sequence before the initiation codon Possible m. RNA secondary structures that block ribosome binding or internal ribosome binding site Codon Bias
 But which E coli? BL 21(DE 3) F- omp. T hsd. SB (r. B-, m. B-), gal, dcm, (DE 3) BL 21 -Star(DE 3) F- omp. T hsd. SB (r. B-, m. B-), gal, dcm, rne 131, (DE 3) BL 21 -Gold(DE 3) F- omp. T hsd. S (r. B- m. B-) dcm+ Tetr gal end. A (DE 3) Tuner(DE 3) F- omp. T hsd. SB (r. B- m. B-) gal dcm lac. Y 1 (DE 3)
But which E coli? BL 21(DE 3) F- omp. T hsd. SB (r. B-, m. B-), gal, dcm, (DE 3) BL 21 -Star(DE 3) F- omp. T hsd. SB (r. B-, m. B-), gal, dcm, rne 131, (DE 3) BL 21 -Gold(DE 3) F- omp. T hsd. S (r. B- m. B-) dcm+ Tetr gal end. A (DE 3) Tuner(DE 3) F- omp. T hsd. SB (r. B- m. B-) gal dcm lac. Y 1 (DE 3)
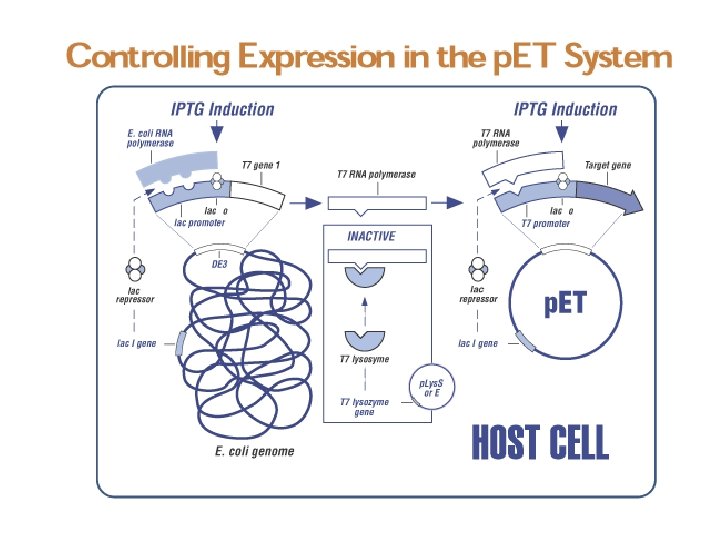
 Conventional cloning approach 1. Select vector of choice 2. Restriction digest the vector 3. PCR the insert 4. Restriction digest the insert 5. Ligate the vector and insert 6. Transform and plate 7. Pick colonies and screen for insert 8. Screen positive clones for protein expression 9. Sequence positive clones
Conventional cloning approach 1. Select vector of choice 2. Restriction digest the vector 3. PCR the insert 4. Restriction digest the insert 5. Ligate the vector and insert 6. Transform and plate 7. Pick colonies and screen for insert 8. Screen positive clones for protein expression 9. Sequence positive clones
 Which vector/tag? 1. T 7 RNA polymerase-based systems is overwhelming choice - Highly specific - High yields - Exquisitely controlled 2. Choice of vector - Restriction sites (are there internal sites in gene? ) - Are there many possible sites? - Are the enzymes commonly available? - Do the enzymes cut near ends of DNA fragments? 3. Which tag? - Relatively little data on which generates best proteins for crystallization - His-tag, GST, MBP all are effective at purification - His tag offers advantage of being able to screen +/- tag for crystals (double bang for the buck) - Make sure there is a protease site to remove tag
Which vector/tag? 1. T 7 RNA polymerase-based systems is overwhelming choice - Highly specific - High yields - Exquisitely controlled 2. Choice of vector - Restriction sites (are there internal sites in gene? ) - Are there many possible sites? - Are the enzymes commonly available? - Do the enzymes cut near ends of DNA fragments? 3. Which tag? - Relatively little data on which generates best proteins for crystallization - His-tag, GST, MBP all are effective at purification - His tag offers advantage of being able to screen +/- tag for crystals (double bang for the buck) - Make sure there is a protease site to remove tag
 Practical issues with cloning 1. Choice of protease? ? ? - Thrombin (more difficult to get but highly effective) - TEV, recombinant with his-tag, stable mutant with less autoproteolysis activity (Waugh), needs calcium, finicky - Factor X, enterokinase…. . avoid
Practical issues with cloning 1. Choice of protease? ? ? - Thrombin (more difficult to get but highly effective) - TEV, recombinant with his-tag, stable mutant with less autoproteolysis activity (Waugh), needs calcium, finicky - Factor X, enterokinase…. . avoid
 “I can’t use thrombin, it digests my protein”
“I can’t use thrombin, it digests my protein”
 Practical issues with cloning Restrict the plasmid - Double digestion often leave one end undigested, which in turn results in high background due to re-ligation - Phosphatase treatment and gel purification of large prep makes life much easier in long run - Optimize system to get no background
Practical issues with cloning Restrict the plasmid - Double digestion often leave one end undigested, which in turn results in high background due to re-ligation - Phosphatase treatment and gel purification of large prep makes life much easier in long run - Optimize system to get no background
 Practical issues with cloning PCR the insert - For HTP studies need to optimize condition for genome or clone - Order primers from reputable supplier (most common problem is in deprotecting oligos) - Have someone else double-check primer sequence - Order primers with requisite overhang (be over-cautious) - Use error-correcting polymerase
Practical issues with cloning PCR the insert - For HTP studies need to optimize condition for genome or clone - Order primers from reputable supplier (most common problem is in deprotecting oligos) - Have someone else double-check primer sequence - Order primers with requisite overhang (be over-cautious) - Use error-correcting polymerase
 Practical issues with cloning Digest the PCR insert - Make sure that there are no internal sites - Purify the restricted product
Practical issues with cloning Digest the PCR insert - Make sure that there are no internal sites - Purify the restricted product
 Practical issues with cloning Ligation and transformation - If vector control background is low, and PCR product is purified, then should be no problem - Use highly competent cells
Practical issues with cloning Ligation and transformation - If vector control background is low, and PCR product is purified, then should be no problem - Use highly competent cells
 Practical issues with cloning Screen for positive clones - PCR screen from colony - Screen by protein expression - Make note of expression, as well as solubility
Practical issues with cloning Screen for positive clones - PCR screen from colony - Screen by protein expression - Make note of expression, as well as solubility
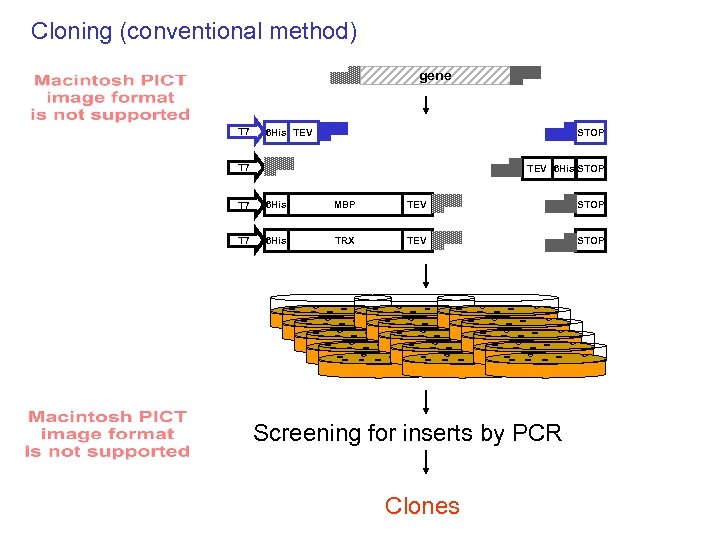 Cloning (conventional method) gene T 7 6 His TEV STOP T 7 TEV 6 His STOP T 7 6 His MBP TEV STOP T 7 6 His TRX TEV STOP Screening for inserts by PCR Clones
Cloning (conventional method) gene T 7 6 His TEV STOP T 7 TEV 6 His STOP T 7 6 His MBP TEV STOP T 7 6 His TRX TEV STOP Screening for inserts by PCR Clones
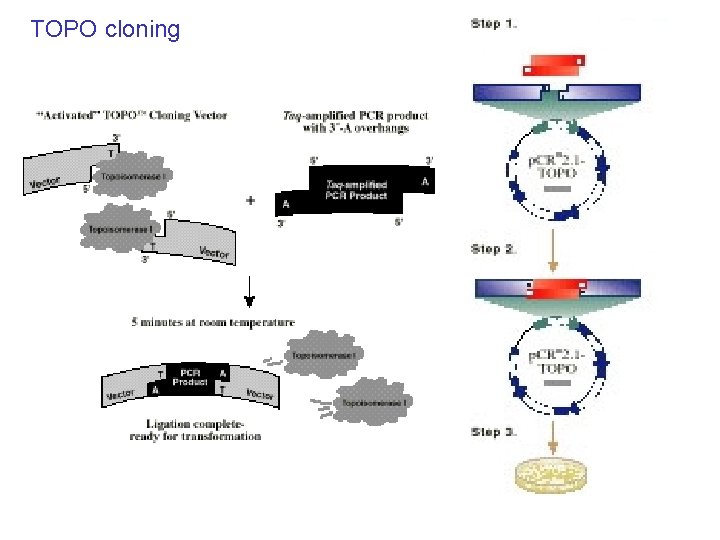 TOPO cloning
TOPO cloning
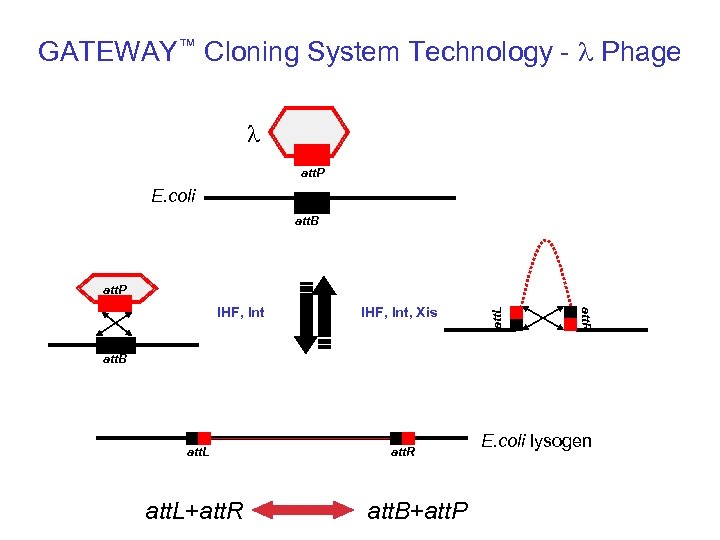 GATEWAY™ Cloning System Technology - l Phage l att. P E. coli att. B IHF, Int, Xis att. R IHF, Int att. L att. P att. B att. L+att. R att. B+att. P E. coli lysogen
GATEWAY™ Cloning System Technology - l Phage l att. P E. coli att. B IHF, Int, Xis att. R IHF, Int att. L att. P att. B att. L+att. R att. B+att. P E. coli lysogen
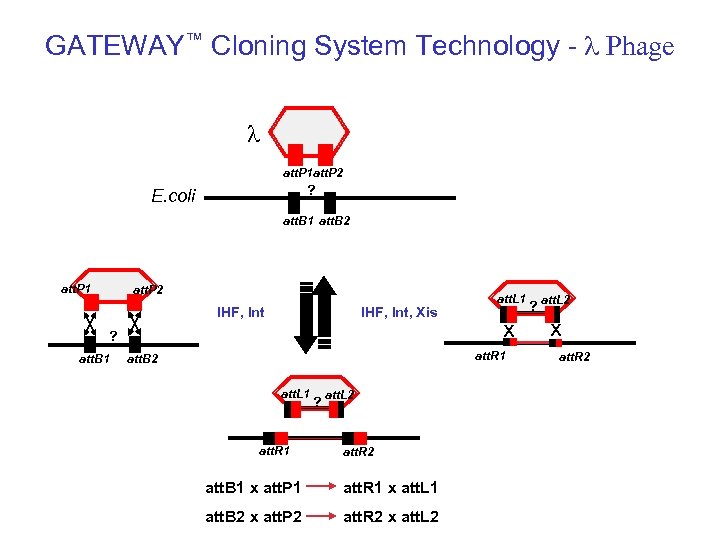 GATEWAY™ Cloning System Technology - l Phage l att. P 1 att. P 2 att. P ? E. coli att. B 2 att. B 1 att. B att. P 1 att. P 2 IHF, Int, Xis ? att. B 1 att. L 1 x att. R 1 att. B 2 att. L 1 att. R 1 ? att. L 2 att. R 2 att. B 1 x att. P 1 att. R 1 x att. L 1 att. B 2 x att. P 2 att. R 2 x att. L 2 ? att. L 2 x att. R 2
GATEWAY™ Cloning System Technology - l Phage l att. P 1 att. P 2 att. P ? E. coli att. B 2 att. B 1 att. B att. P 1 att. P 2 IHF, Int, Xis ? att. B 1 att. L 1 x att. R 1 att. B 2 att. L 1 att. R 1 ? att. L 2 att. R 2 att. B 1 x att. P 1 att. R 1 x att. L 1 att. B 2 x att. P 2 att. R 2 x att. L 2 ? att. L 2 x att. R 2
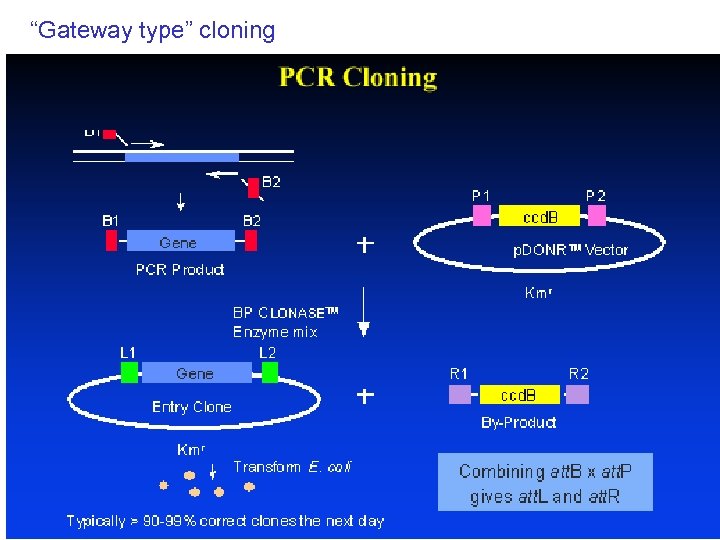 “Gateway type” cloning
“Gateway type” cloning
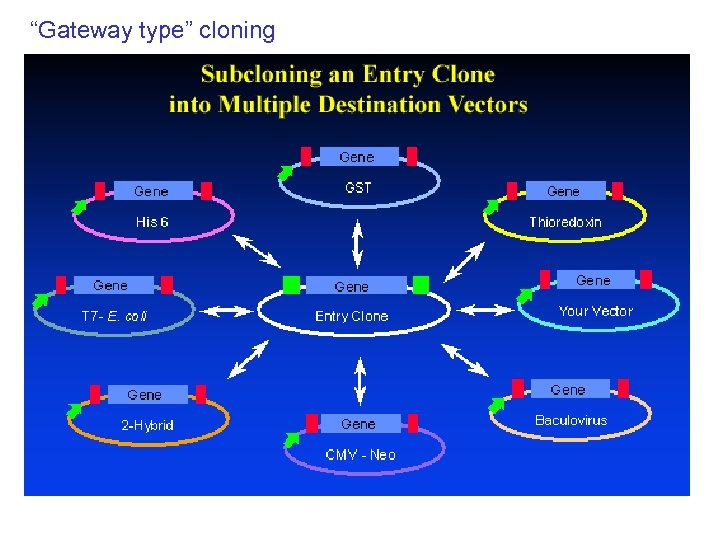 “Gateway type” cloning
“Gateway type” cloning
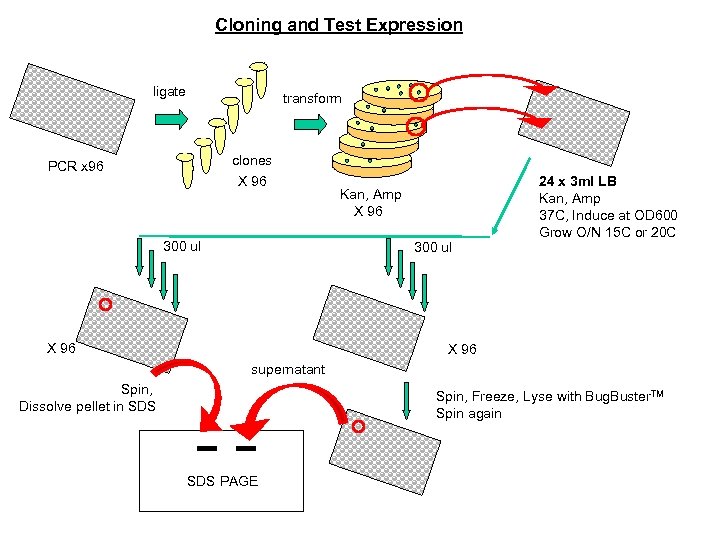 Cloning and Test Expression ligate transform clones X 96 PCR x 96 300 ul Kan, Amp X 96 300 ul X 96 24 x 3 ml LB Kan, Amp 37 C, Induce at OD 600 Grow O/N 15 C or 20 C X 96 supernatant Spin, Dissolve pellet in SDS Spin, Freeze, Lyse with Bug. Buster. TM Spin again SDS PAGE
Cloning and Test Expression ligate transform clones X 96 PCR x 96 300 ul Kan, Amp X 96 300 ul X 96 24 x 3 ml LB Kan, Amp 37 C, Induce at OD 600 Grow O/N 15 C or 20 C X 96 supernatant Spin, Dissolve pellet in SDS Spin, Freeze, Lyse with Bug. Buster. TM Spin again SDS PAGE
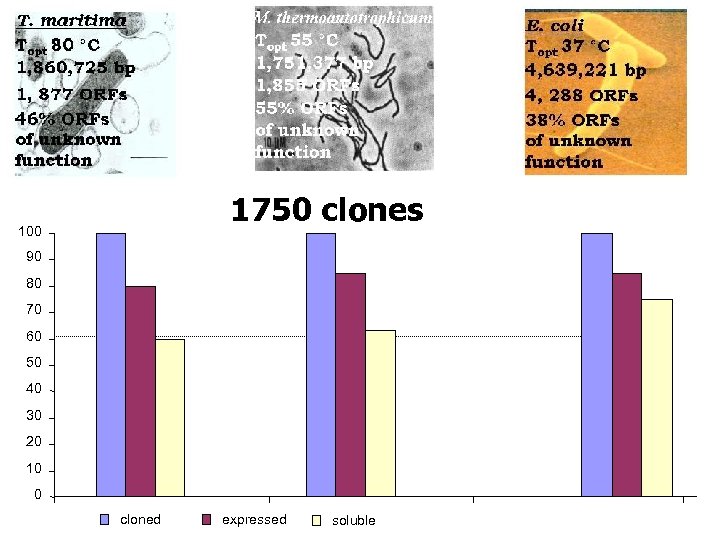 1750 clones 100 90 80 70 60 50 40 30 20 10 0 cloned expressed soluble
1750 clones 100 90 80 70 60 50 40 30 20 10 0 cloned expressed soluble
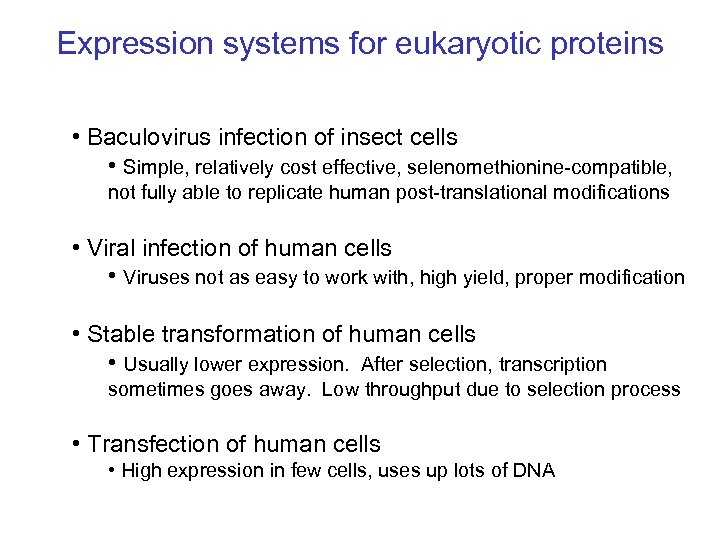 Expression systems for eukaryotic proteins • Baculovirus infection of insect cells • Simple, relatively cost effective, selenomethionine-compatible, not fully able to replicate human post-translational modifications • Viral infection of human cells • Viruses not as easy to work with, high yield, proper modification • Stable transformation of human cells • Usually lower expression. After selection, transcription sometimes goes away. Low throughput due to selection process • Transfection of human cells • High expression in few cells, uses up lots of DNA
Expression systems for eukaryotic proteins • Baculovirus infection of insect cells • Simple, relatively cost effective, selenomethionine-compatible, not fully able to replicate human post-translational modifications • Viral infection of human cells • Viruses not as easy to work with, high yield, proper modification • Stable transformation of human cells • Usually lower expression. After selection, transcription sometimes goes away. Low throughput due to selection process • Transfection of human cells • High expression in few cells, uses up lots of DNA

 Protein Purification
Protein Purification
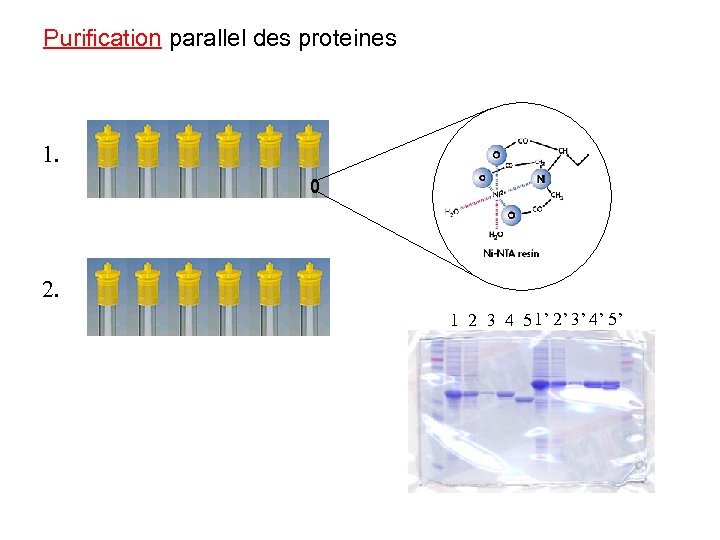 Purification parallel des proteines 1. 2. 1 2 3 4 5 1’ 2’ 3’ 4’ 5’
Purification parallel des proteines 1. 2. 1 2 3 4 5 1’ 2’ 3’ 4’ 5’
 Proteo. Max – Automated Protein Purification and Concentration System Affinium Pharmaceuticals
Proteo. Max – Automated Protein Purification and Concentration System Affinium Pharmaceuticals
 A few observations from our work
A few observations from our work
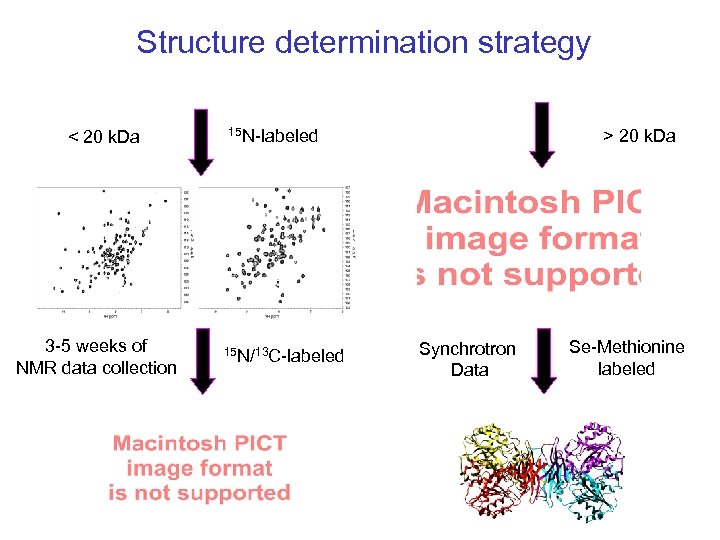 Structure determination strategy < 20 k. Da 3 -5 weeks of NMR data collection 15 N-labeled 15 N/13 C-labeled > 20 k. Da Synchrotron Data Se-Methionine labeled
Structure determination strategy < 20 k. Da 3 -5 weeks of NMR data collection 15 N-labeled 15 N/13 C-labeled > 20 k. Da Synchrotron Data Se-Methionine labeled
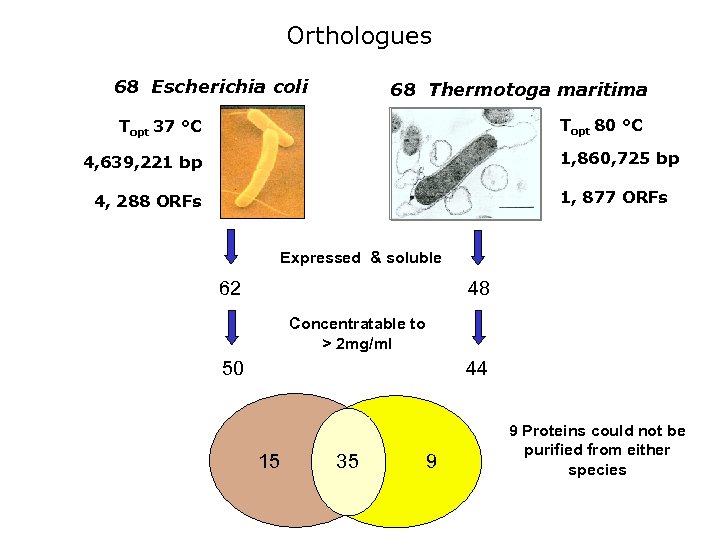 Orthologues 68 Escherichia coli 68 Thermotoga maritima Topt 80 °C Topt 37 °C 1, 860, 725 bp 4, 639, 221 bp 1, 877 ORFs 4, 288 ORFs Expressed & soluble 62 48 Concentratable to > 2 mg/ml 50 44 15 35 9 9 Proteins could not be purified from either species
Orthologues 68 Escherichia coli 68 Thermotoga maritima Topt 80 °C Topt 37 °C 1, 860, 725 bp 4, 639, 221 bp 1, 877 ORFs 4, 288 ORFs Expressed & soluble 62 48 Concentratable to > 2 mg/ml 50 44 15 35 9 9 Proteins could not be purified from either species
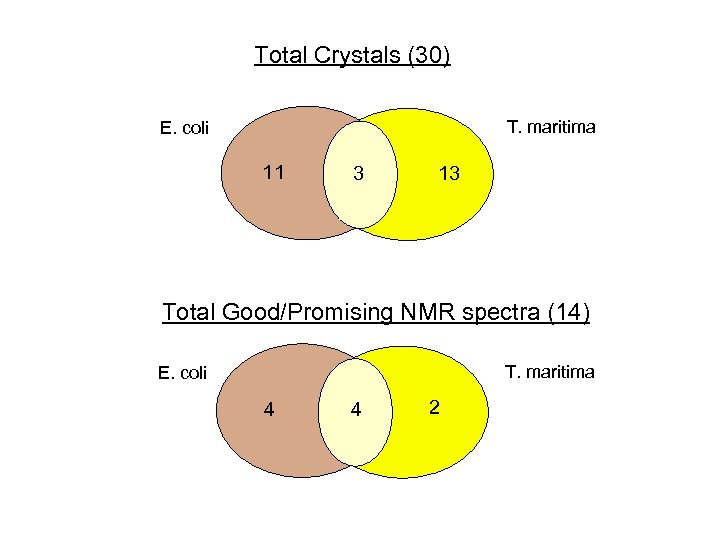 Total Crystals (30) T. maritima E. coli 11 3 13 Total Good/Promising NMR spectra (14) T. maritima E. coli 4 4 2
Total Crystals (30) T. maritima E. coli 11 3 13 Total Good/Promising NMR spectra (14) T. maritima E. coli 4 4 2
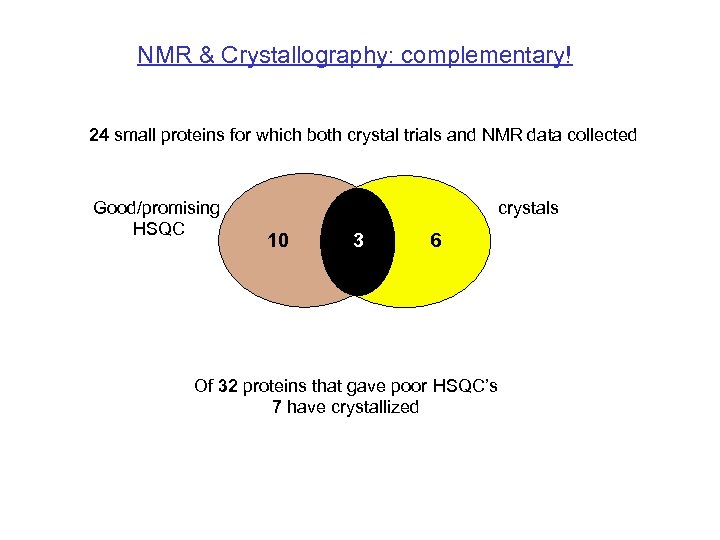 NMR & Crystallography: complementary! 24 small proteins for which both crystal trials and NMR data collected Good/promising HSQC crystals 10 3 6 Of 32 proteins that gave poor HSQC’s 7 have crystallized
NMR & Crystallography: complementary! 24 small proteins for which both crystal trials and NMR data collected Good/promising HSQC crystals 10 3 6 Of 32 proteins that gave poor HSQC’s 7 have crystallized
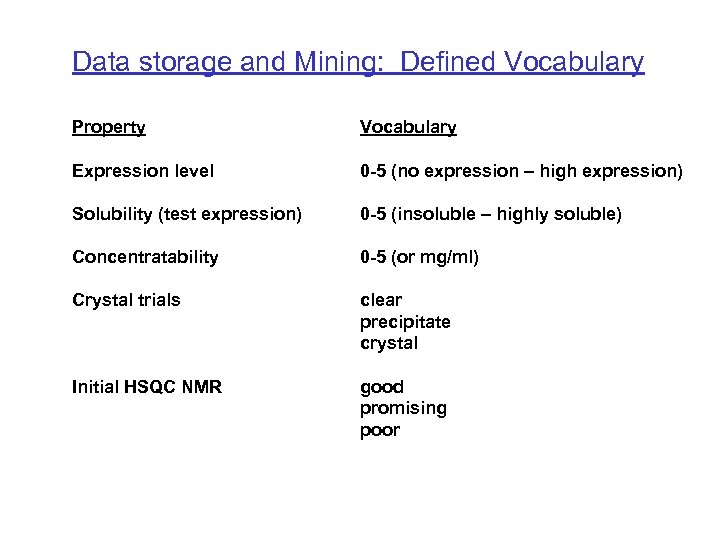 Data storage and Mining: Defined Vocabulary Property Vocabulary Expression level 0 -5 (no expression – high expression) Solubility (test expression) 0 -5 (insoluble – highly soluble) Concentratability 0 -5 (or mg/ml) Crystal trials clear precipitate crystal Initial HSQC NMR good promising poor
Data storage and Mining: Defined Vocabulary Property Vocabulary Expression level 0 -5 (no expression – high expression) Solubility (test expression) 0 -5 (insoluble – highly soluble) Concentratability 0 -5 (or mg/ml) Crystal trials clear precipitate crystal Initial HSQC NMR good promising poor
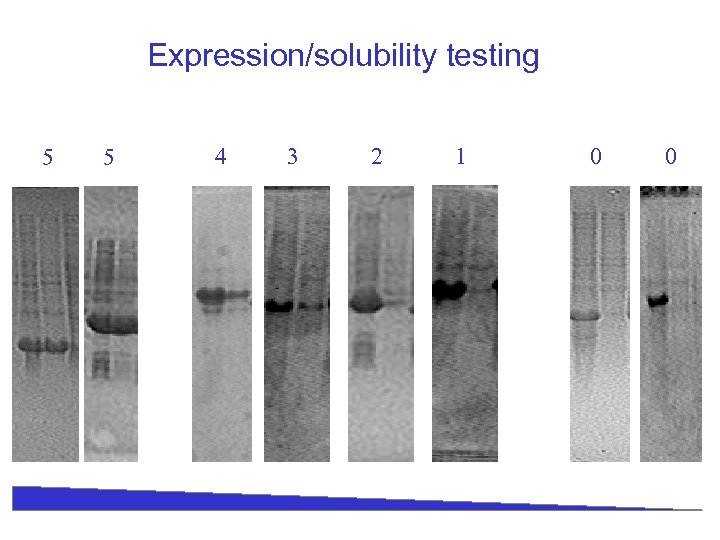 Expression/solubility testing 5 5 4 3 2 1 0 0
Expression/solubility testing 5 5 4 3 2 1 0 0
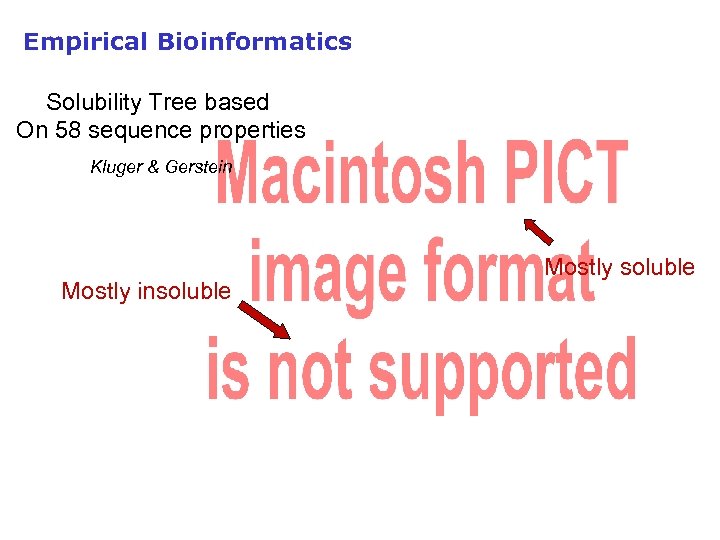 Empirical Bioinformatics Solubility Tree based On 58 sequence properties Kluger & Gerstein Mostly insoluble Mostly soluble
Empirical Bioinformatics Solubility Tree based On 58 sequence properties Kluger & Gerstein Mostly insoluble Mostly soluble
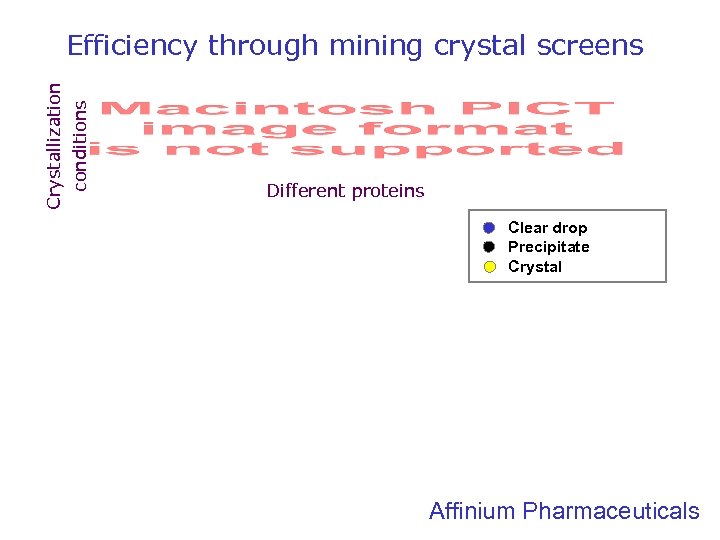 Crystallization conditions Efficiency through mining crystal screens Different proteins Clear drop Precipitate Crystal Affinium Pharmaceuticals
Crystallization conditions Efficiency through mining crystal screens Different proteins Clear drop Precipitate Crystal Affinium Pharmaceuticals
 Crystal trial: Diminishing Returns
Crystal trial: Diminishing Returns
 Collaborators on Structural Proteomics Lawrence Mc. Intosh (UBC) C. Mackereth, G. Lee Thomas Szypersky* (SUNY Buffalo) Mike Kennedy (PNNL)* J. Cort, T. Ramelot Mark Gerstein (Yale) * Yval Kluger Ning Lan Kalle Gehring (Mc. Gill) I. Ekiel G. Kozlov Dave Wishart (U. Alberta) S. Bhattacharyya Sherry Mowbray (Sweden) Liang Tong (Columbia) * John Hunt (Columbia) * Andrzej Joachimiak (ANL)* Weontae Lee (Yonsei U. ) Guy Montelione (Rutgers) * Emil Pai (U. Toronto) V. Saridakis, N. Wu *Northeast Structural Genomics Consortium *Midwest Structural Genomics Consortium
Collaborators on Structural Proteomics Lawrence Mc. Intosh (UBC) C. Mackereth, G. Lee Thomas Szypersky* (SUNY Buffalo) Mike Kennedy (PNNL)* J. Cort, T. Ramelot Mark Gerstein (Yale) * Yval Kluger Ning Lan Kalle Gehring (Mc. Gill) I. Ekiel G. Kozlov Dave Wishart (U. Alberta) S. Bhattacharyya Sherry Mowbray (Sweden) Liang Tong (Columbia) * John Hunt (Columbia) * Andrzej Joachimiak (ANL)* Weontae Lee (Yonsei U. ) Guy Montelione (Rutgers) * Emil Pai (U. Toronto) V. Saridakis, N. Wu *Northeast Structural Genomics Consortium *Midwest Structural Genomics Consortium


