9caeedf66697f97ec15902f95411e626.ppt
- Количество слайдов: 26
Practical molecular biology PD Dr. Alexei Gratchev Prof. Dr. Julia Kzhyshkowska Prof. Dr. W. Kaminski
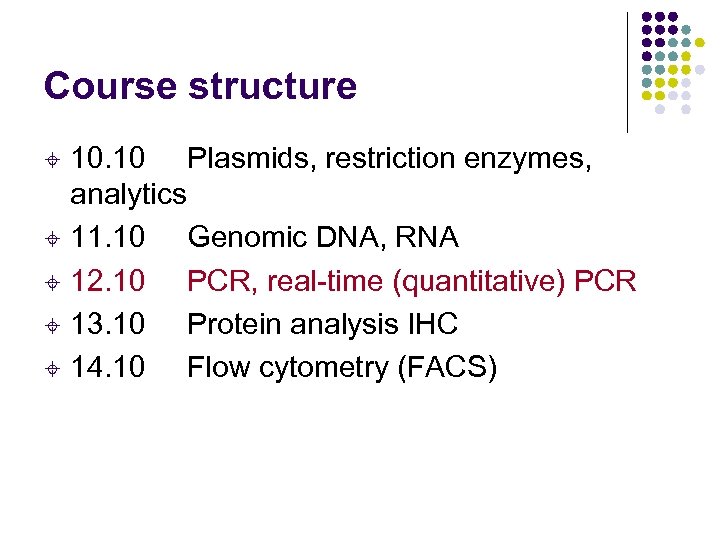
Course structure 10. 10 Plasmids, restriction enzymes, analytics ± 11. 10 Genomic DNA, RNA ± 12. 10 PCR, real-time (quantitative) PCR ± 13. 10 Protein analysis IHC ± 14. 10 Flow cytometry (FACS) ±
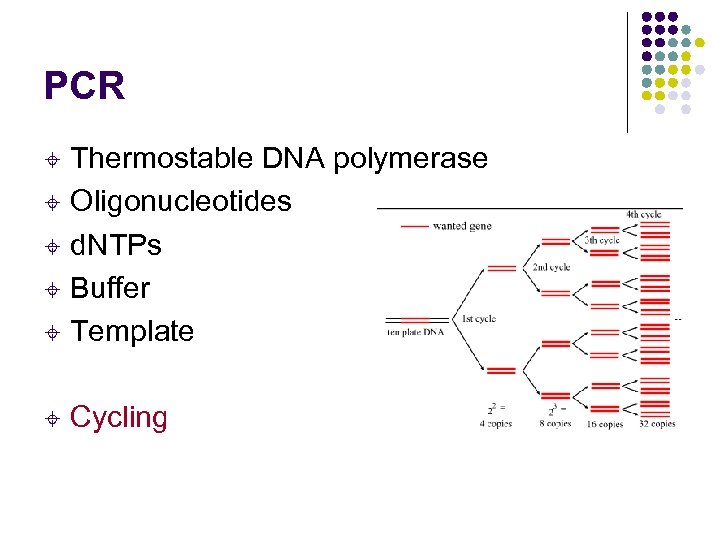
PCR Thermostable DNA polymerase ± Oligonucleotides ± d. NTPs ± Buffer ± Template ± ± Cycling

PCR Detection of pathogens ± Detection of mutations ± Person identification ± Cloning ± Mutagenesis ± and may more… ±
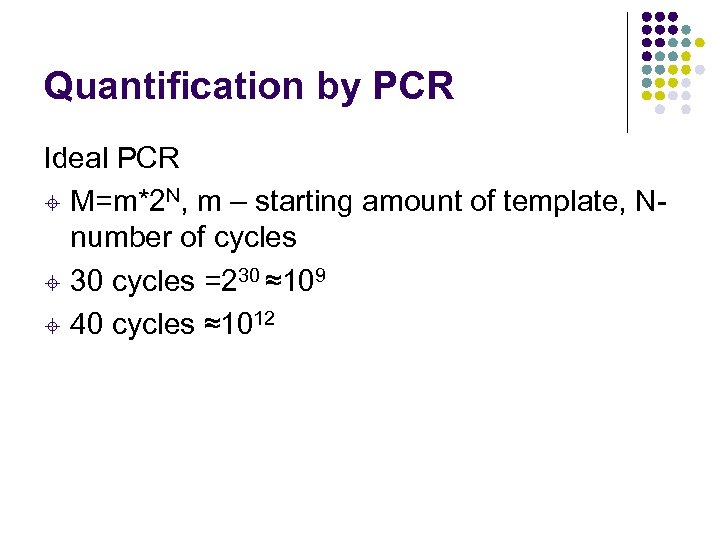
Quantification by PCR Ideal PCR ± M=m*2 N, m – starting amount of template, Nnumber of cycles ± 30 cycles =230 ≈109 ± 40 cycles ≈1012
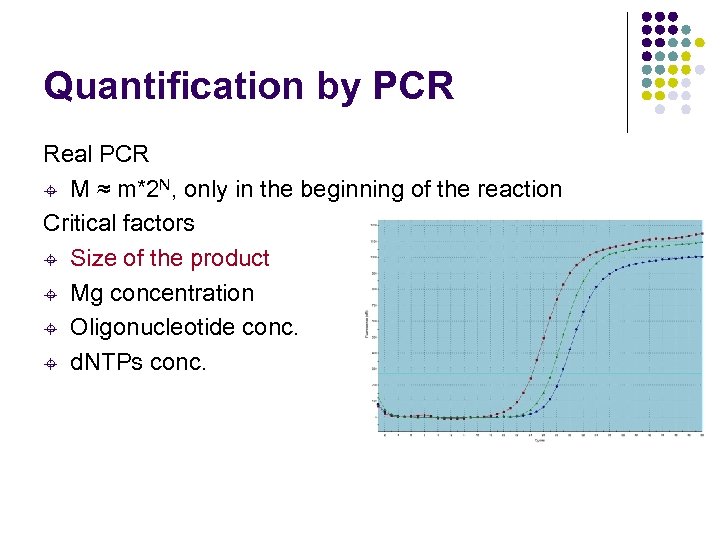
Quantification by PCR Real PCR ± M ≈ m*2 N, only in the beginning of the reaction Critical factors ± Size of the product ± Mg concentration ± Oligonucleotide conc. ± d. NTPs conc.
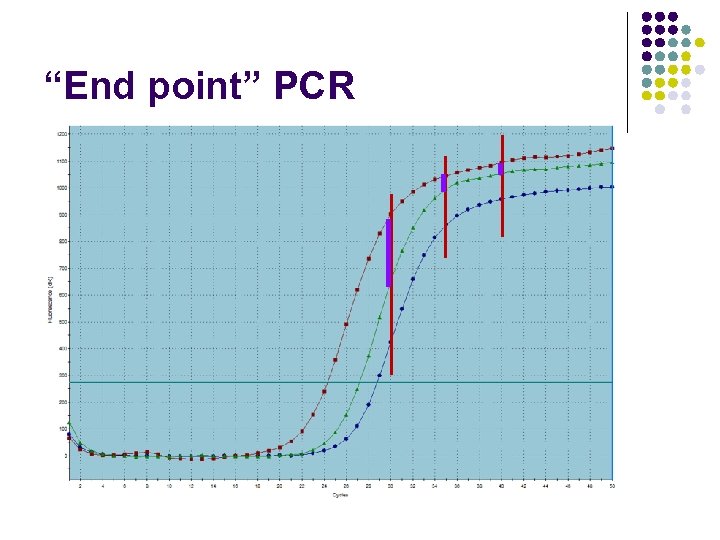
“End point” PCR
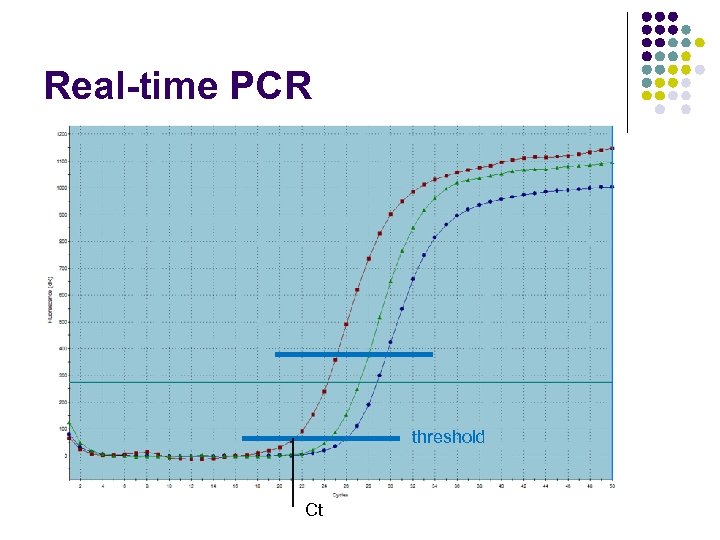
Real-time PCR threshold Ct
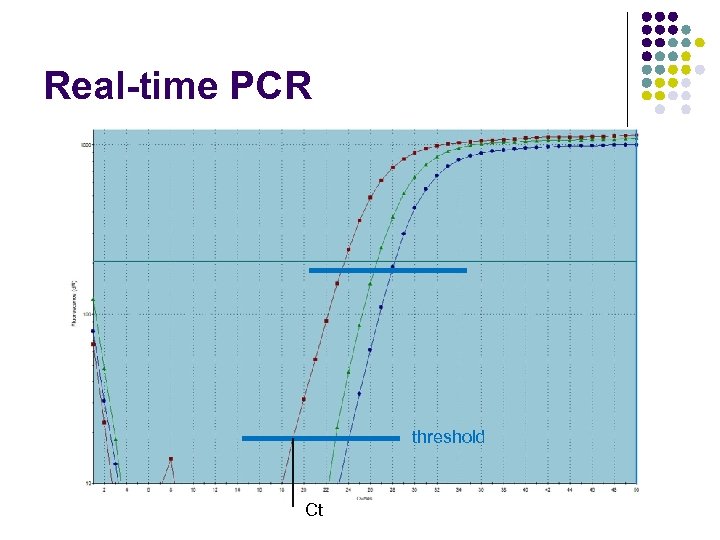
Real-time PCR threshold Ct

Quantification by PCR ± Measure the amount of the product after every cycle Determine threshold cycle (Ct) value for each sample Calculate the amount of the product ± Note: Ct can be a fraction ± ±
Real-time data collection ± Intercalating dyes ± ± ± Cheap Low specificity Can measure only one gene per tube Molecular beacons Taq. Man® probes ± ± Highly specific Several genes can be measured in one tube (Multiplex PCR) Expensive Multiplex PCR is hard to optimize
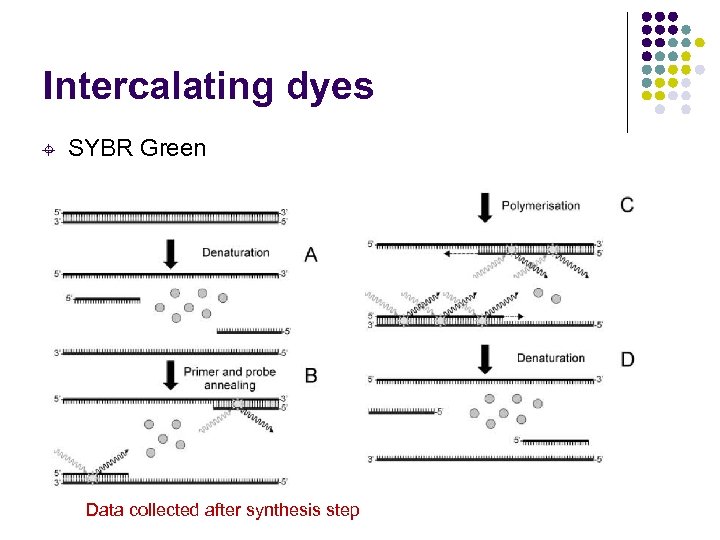
Intercalating dyes ± SYBR Green Data collected after synthesis step
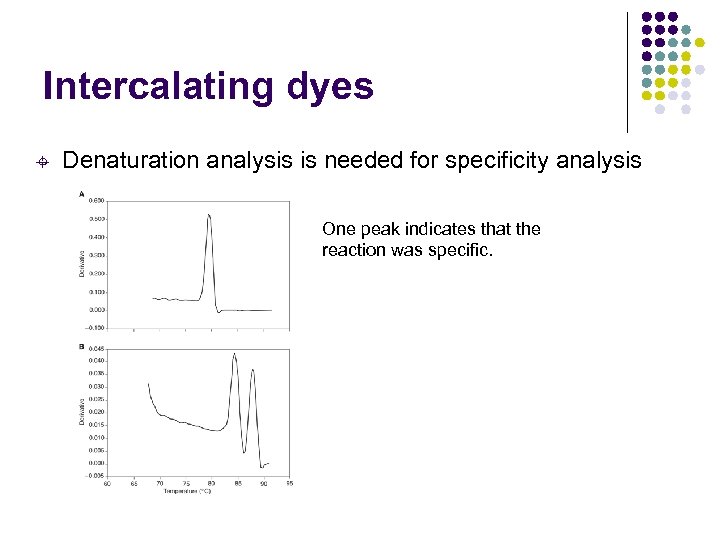
Intercalating dyes ± Denaturation analysis is needed for specificity analysis One peak indicates that the reaction was specific.
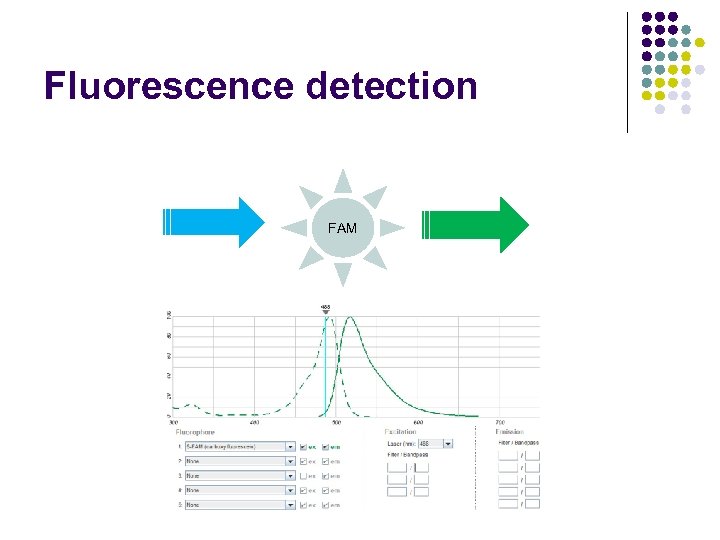
Fluorescence detection FAM
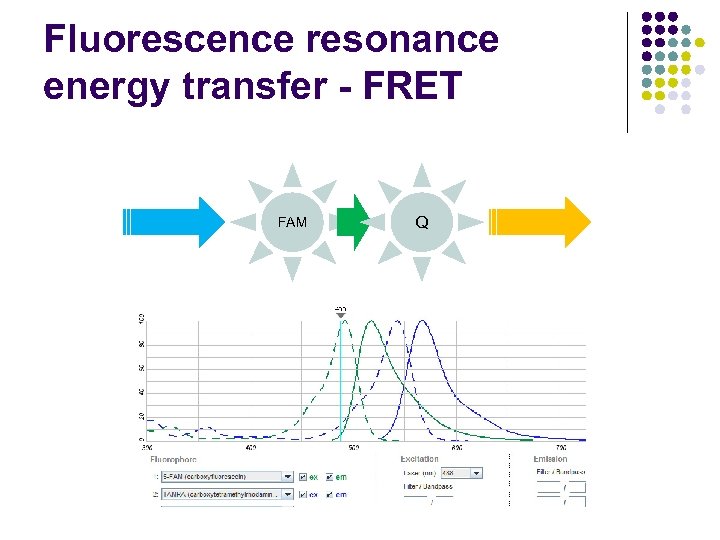
Fluorescence resonance energy transfer - FRET FAM Q
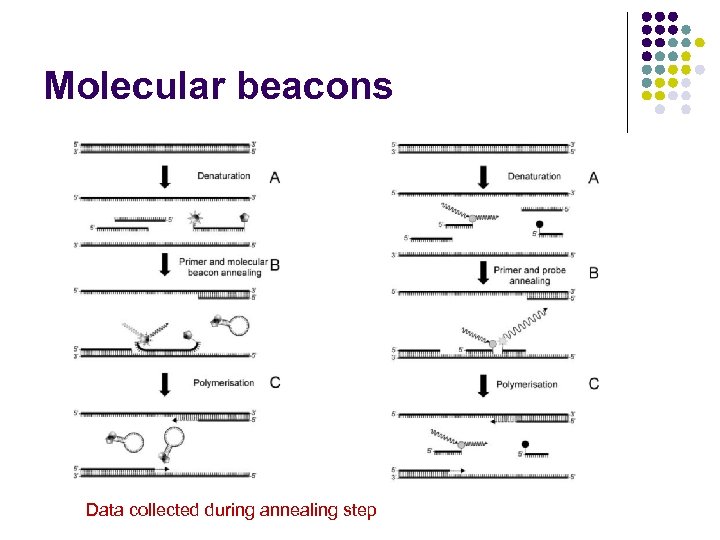
Molecular beacons Data collected during annealing step
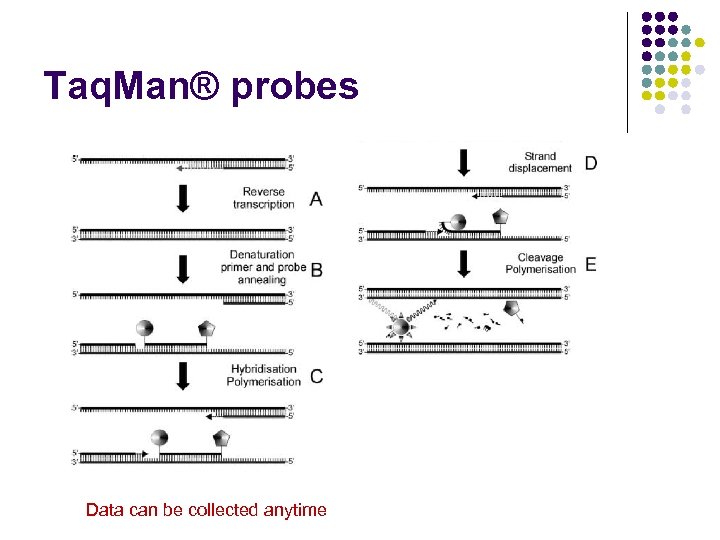
Taq. Man® probes Data can be collected anytime
Real-time PCR equipment ± Light sources ± ± ± Laser LED Array Focused halogen lamp Halogen lamp Detectors ± ± PMT (Photo Multiplier Tube) CCD camera Light source PMT
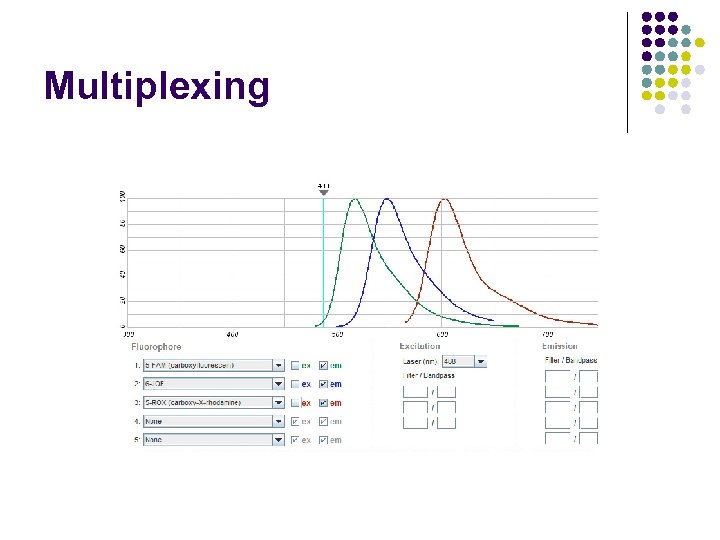
Multiplexing

Experiment planning Selection detection method ± Intercalating dye ± Molecular beacon ± Taq. Man® probe Selection of house keeping gene ± ± GAPD beta actin Selection of quantification method ± ± absolute (Standard curve) relative (dd. Ct)
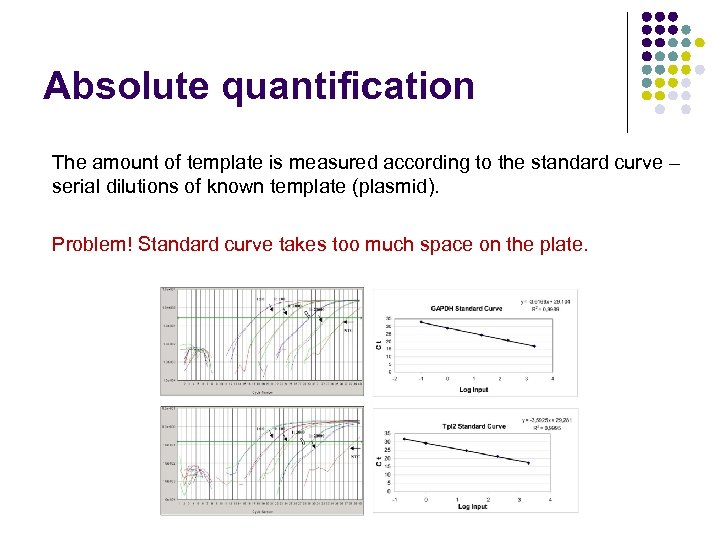
Absolute quantification The amount of template is measured according to the standard curve – serial dilutions of known template (plasmid). Problem! Standard curve takes too much space on the plate.
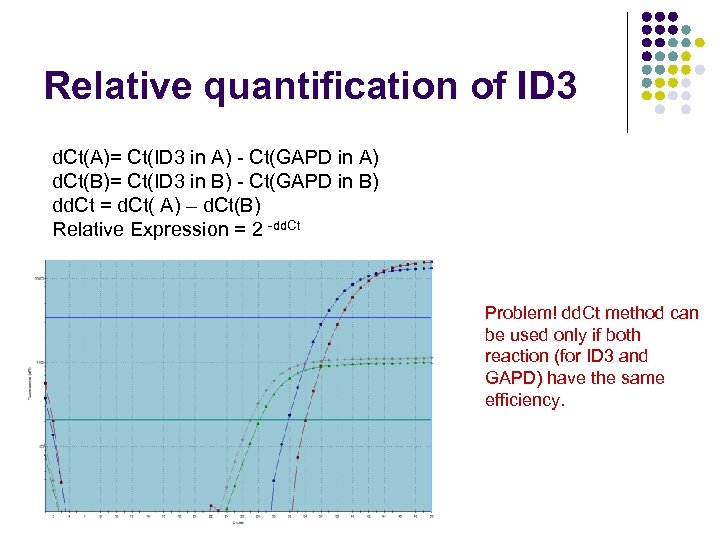
Relative quantification of ID 3 d. Ct(A)= Ct(ID 3 in A) - Ct(GAPD in A) d. Ct(B)= Ct(ID 3 in B) - Ct(GAPD in B) dd. Ct = d. Ct( A) – d. Ct(B) Relative Expression = 2 -dd. Ct Problem! dd. Ct method can be used only if both reaction (for ID 3 and GAPD) have the same efficiency.
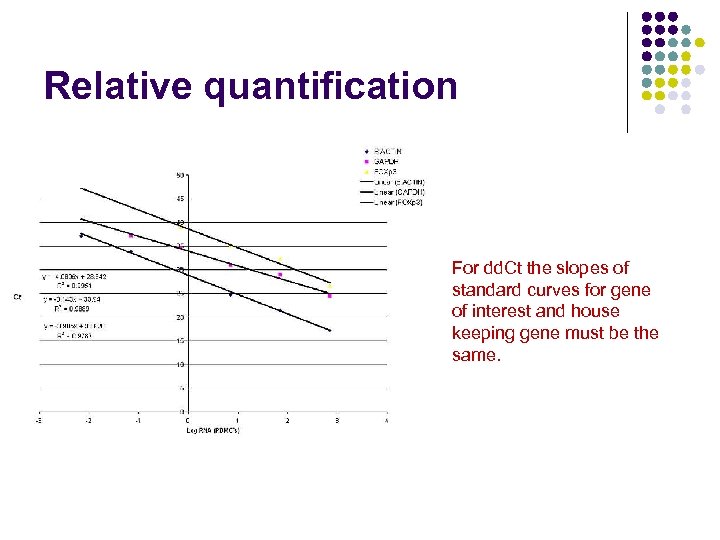
Relative quantification For dd. Ct the slopes of standard curves for gene of interest and house keeping gene must be the same.
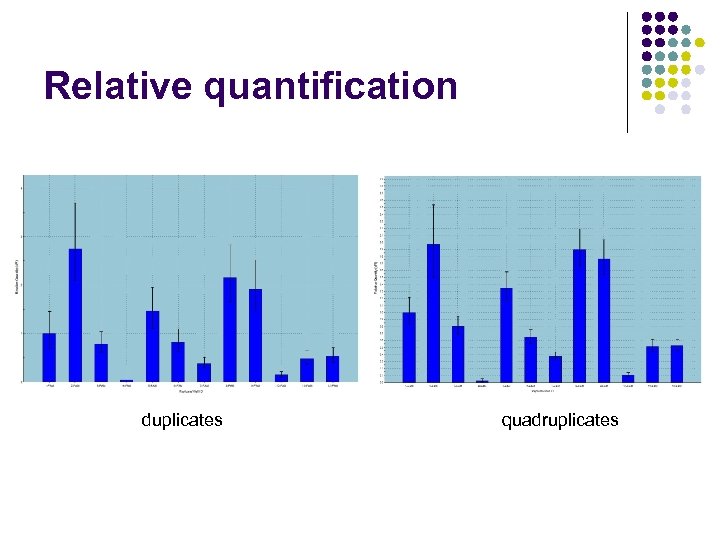
Relative quantification duplicates quadruplicates
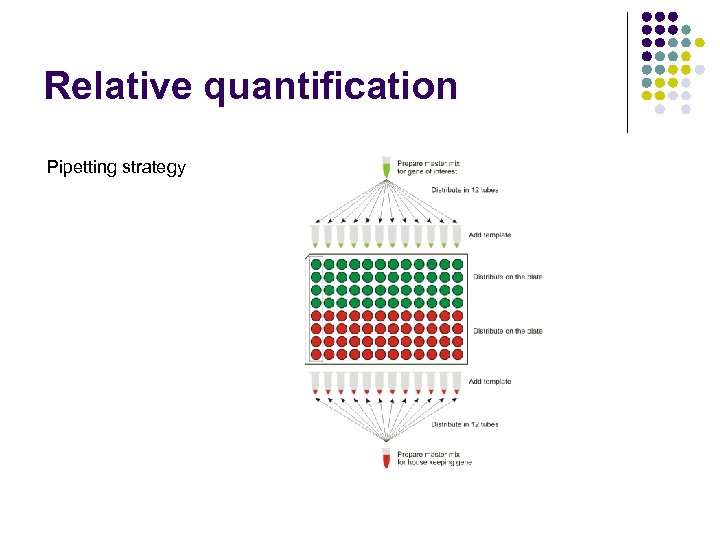
Relative quantification Pipetting strategy

Questions?
9caeedf66697f97ec15902f95411e626.ppt