9d9ee5189f6238a11fb38c73a9dd8817.ppt
- Количество слайдов: 45
 Polymerase Chain Reaction (PCR) • Polymerase chain reaction: Starting with VERY SMALL AMOUNTS OF DNA (sometimes a few molecules), one can amplify the DNA enough to detect it by electrophoresis.
Polymerase Chain Reaction (PCR) • Polymerase chain reaction: Starting with VERY SMALL AMOUNTS OF DNA (sometimes a few molecules), one can amplify the DNA enough to detect it by electrophoresis.
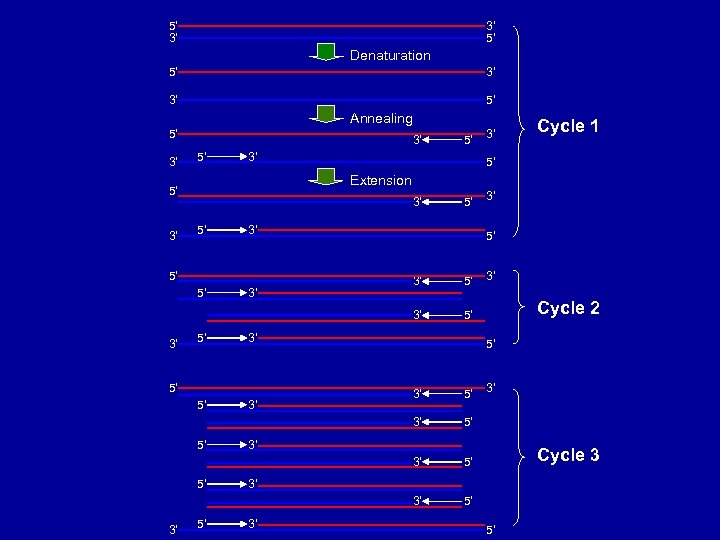 5’ 3’ 3’ 5’ Denaturation 5’ 3’ 3’ 5’ Annealing 5’ 3’ 3’ 5’ 5’ 5’ 3’ 3’ 5’ 3’ Cycle 2 5’ 3’ 5’ 3’ Cycle 1 5’ 3’ 3’ Extension 5’ 3’ 5’ 5’ 3’ Cycle 3 3’ 3’ 5’
5’ 3’ 3’ 5’ Denaturation 5’ 3’ 3’ 5’ Annealing 5’ 3’ 3’ 5’ 5’ 5’ 3’ 3’ 5’ 3’ Cycle 2 5’ 3’ 5’ 3’ Cycle 1 5’ 3’ 3’ Extension 5’ 3’ 5’ 5’ 3’ Cycle 3 3’ 3’ 5’
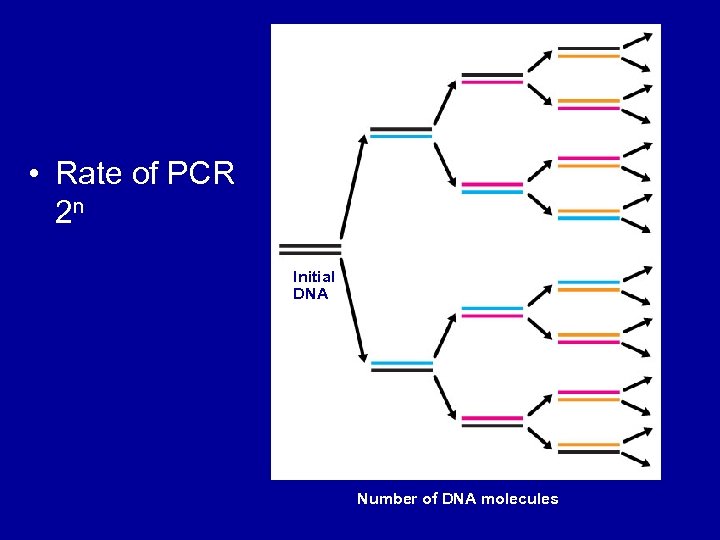 • Rate of PCR 2 n Initial DNA 1 2 4 Number of DNA molecules 8
• Rate of PCR 2 n Initial DNA 1 2 4 Number of DNA molecules 8
 Copies of DNA=2 N
Copies of DNA=2 N
 RT-PCR • Polymerase chain reaction amplification of c. DNA can also be used to detect specific transcripts in a RNA sample. • In this procedure, known as RT-PCR, reverse transcriptase is used to copy all of the m. RNAs in an RNA sample into c. DNA. • Usually, oligo d. T molecules, that anneal to the poly A tails of the m. RNA, are used as primers. • This single stranded c. DNA can then be amplified by PCR using primers that anneal to a specific transcript sequence.
RT-PCR • Polymerase chain reaction amplification of c. DNA can also be used to detect specific transcripts in a RNA sample. • In this procedure, known as RT-PCR, reverse transcriptase is used to copy all of the m. RNAs in an RNA sample into c. DNA. • Usually, oligo d. T molecules, that anneal to the poly A tails of the m. RNA, are used as primers. • This single stranded c. DNA can then be amplified by PCR using primers that anneal to a specific transcript sequence.
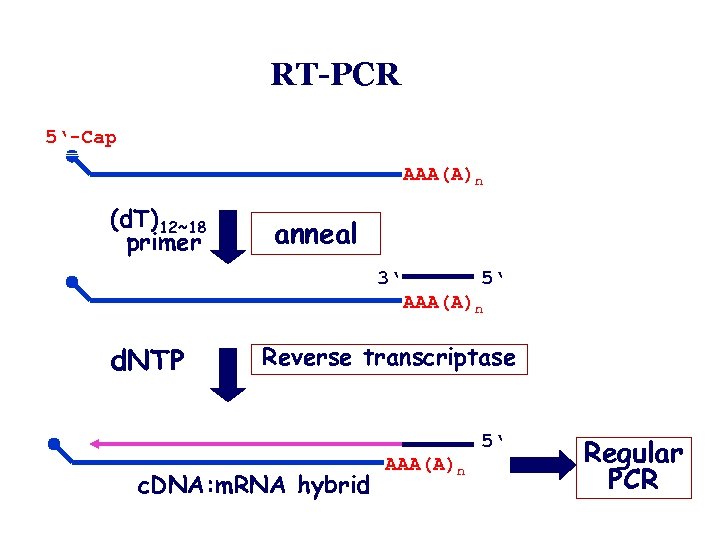 RT-PCR 5‘-Cap m. RNA (d. T)12~18 primer AAA(A)n anneal 3‘ 5‘ AAA(A)n d. NTP Reverse transcriptase 5‘ c. DNA: m. RNA hybrid AAA(A)n Regular PCR
RT-PCR 5‘-Cap m. RNA (d. T)12~18 primer AAA(A)n anneal 3‘ 5‘ AAA(A)n d. NTP Reverse transcriptase 5‘ c. DNA: m. RNA hybrid AAA(A)n Regular PCR
 • The amplified DNA fragments that are produced can by analyzed by agarose gel electrophoresis. • The amount of amplified fragment produced is proportional to the amount of target m. RNA in the original RNA sample. • Although less quantitative than Northern blots, RT-PCR is extremely sensitive and can be used to detect very rare m. RNA species.
• The amplified DNA fragments that are produced can by analyzed by agarose gel electrophoresis. • The amount of amplified fragment produced is proportional to the amount of target m. RNA in the original RNA sample. • Although less quantitative than Northern blots, RT-PCR is extremely sensitive and can be used to detect very rare m. RNA species.
 How do we accurately quantify the amount of DNA? Real-time PCR
How do we accurately quantify the amount of DNA? Real-time PCR
 Real Time PCR
Real Time PCR
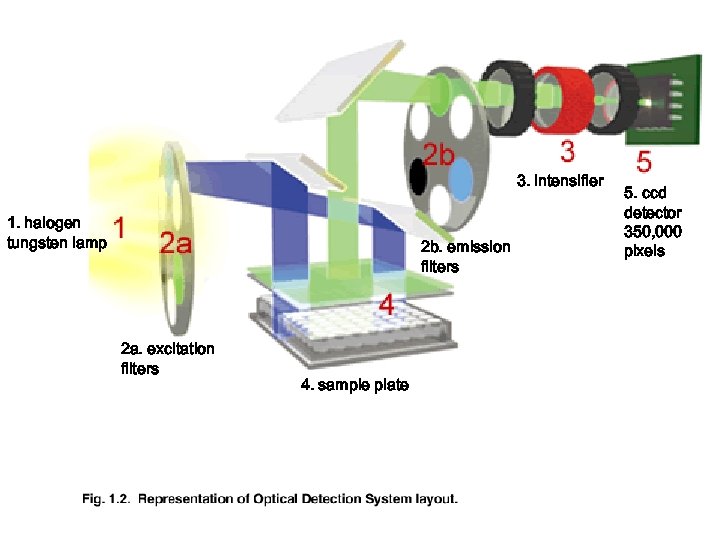 3. intensifier 1. halogen tungsten lamp 2 b. emission filters 2 a. excitation filters 4. sample plate 5. ccd detector 350, 000 pixels
3. intensifier 1. halogen tungsten lamp 2 b. emission filters 2 a. excitation filters 4. sample plate 5. ccd detector 350, 000 pixels
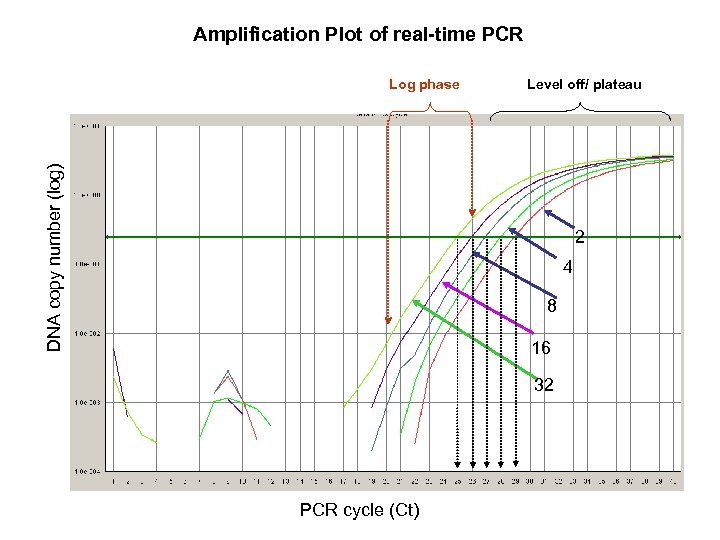 Amplification Plot of real-time PCR DNA copy number (log) Log phase Level off/ plateau 2 4 8 16 32 PCR cycle (Ct)
Amplification Plot of real-time PCR DNA copy number (log) Log phase Level off/ plateau 2 4 8 16 32 PCR cycle (Ct)
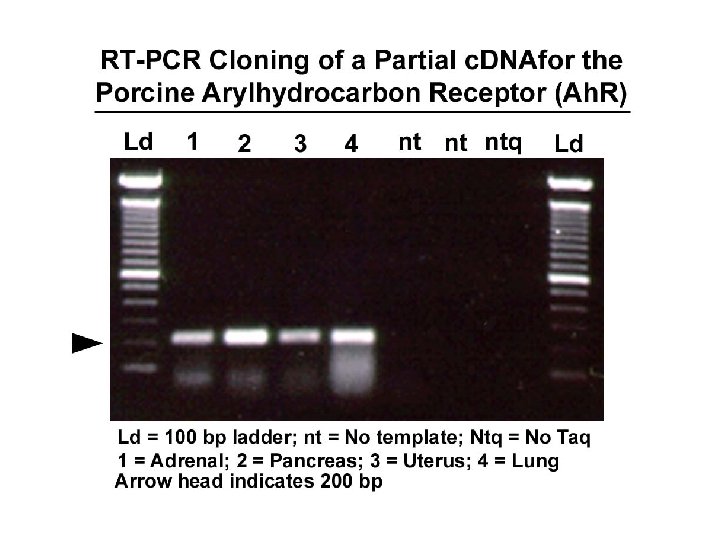
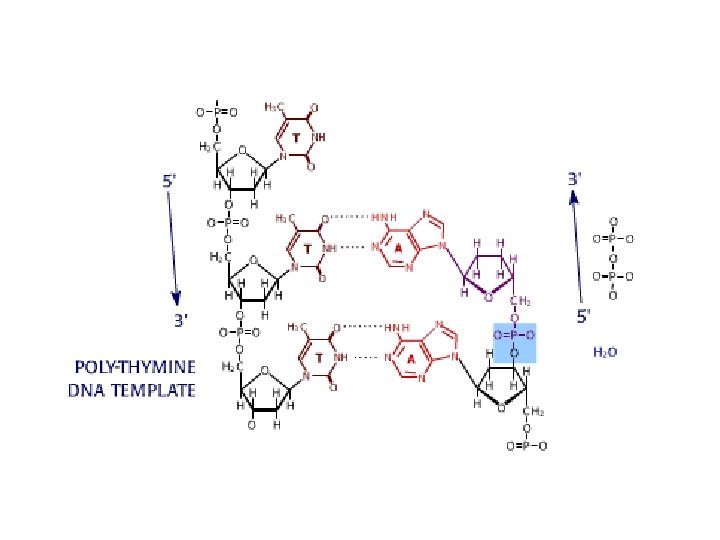
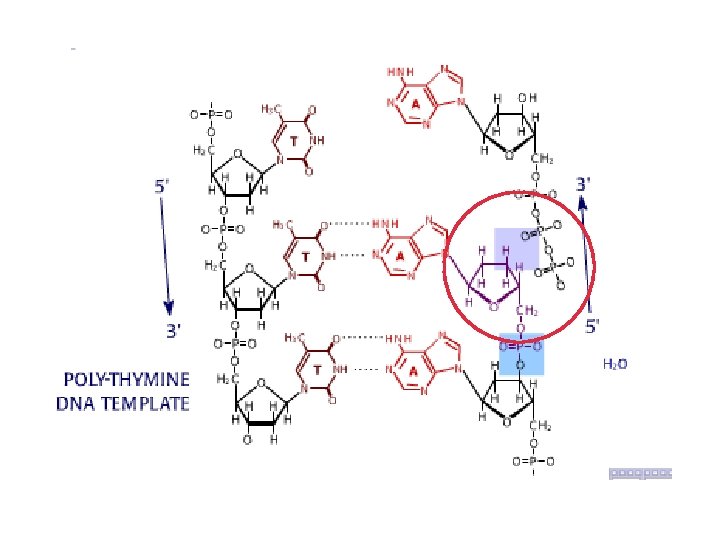
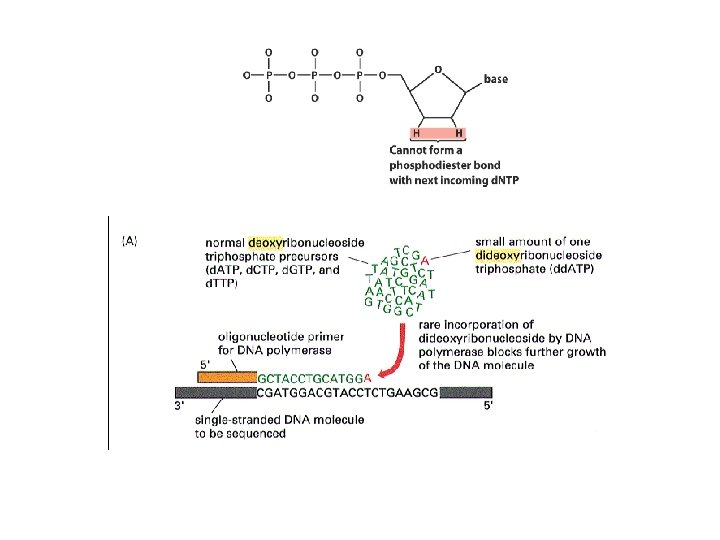
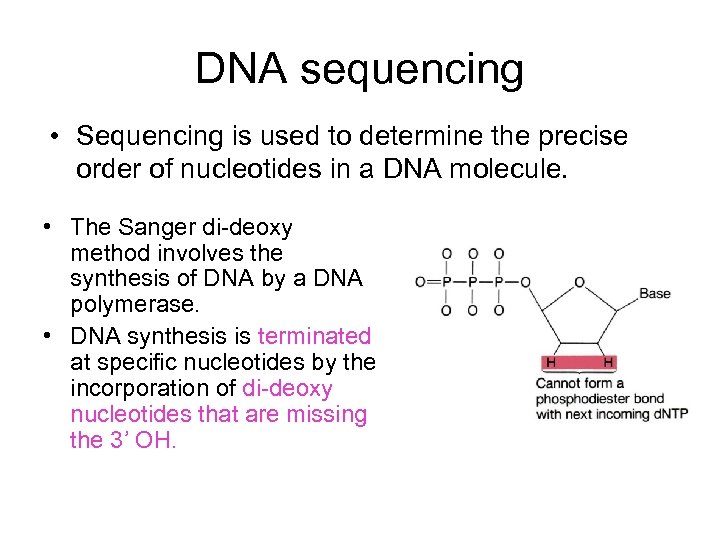 DNA sequencing • Sequencing is used to determine the precise order of nucleotides in a DNA molecule. • The Sanger di-deoxy method involves the synthesis of DNA by a DNA polymerase. • DNA synthesis is terminated at specific nucleotides by the incorporation of di-deoxy nucleotides that are missing the 3’ OH.
DNA sequencing • Sequencing is used to determine the precise order of nucleotides in a DNA molecule. • The Sanger di-deoxy method involves the synthesis of DNA by a DNA polymerase. • DNA synthesis is terminated at specific nucleotides by the incorporation of di-deoxy nucleotides that are missing the 3’ OH.
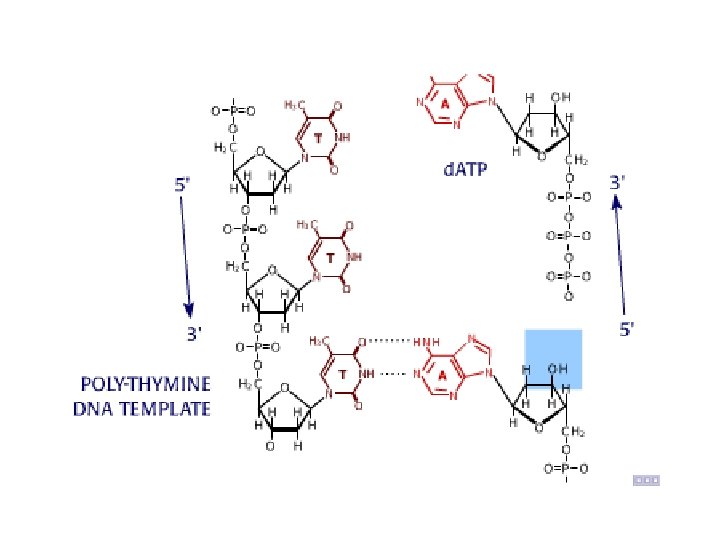
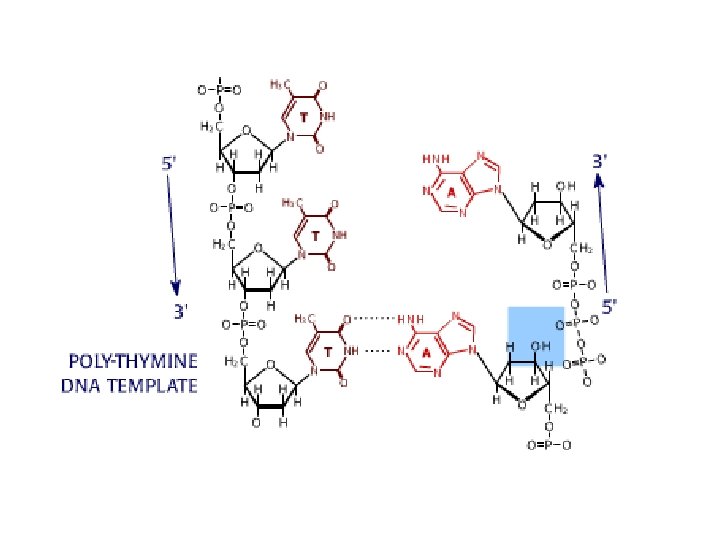
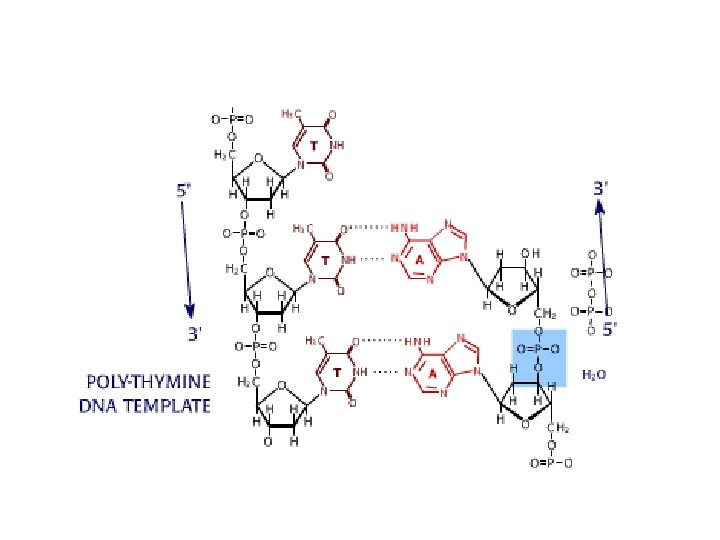
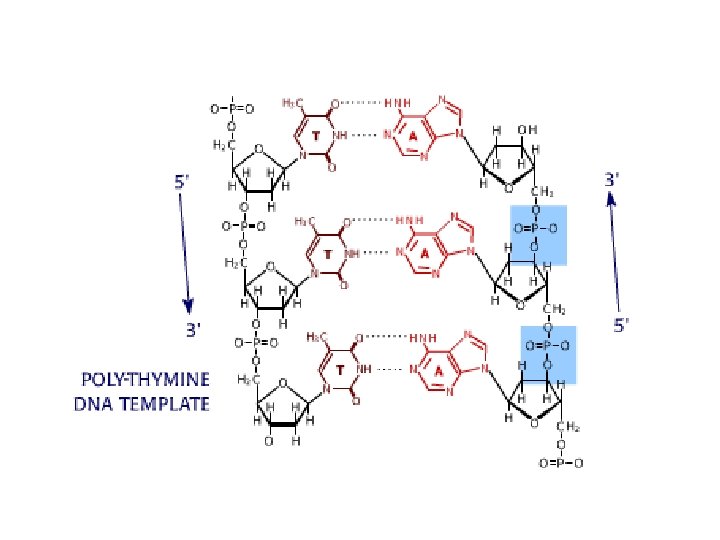
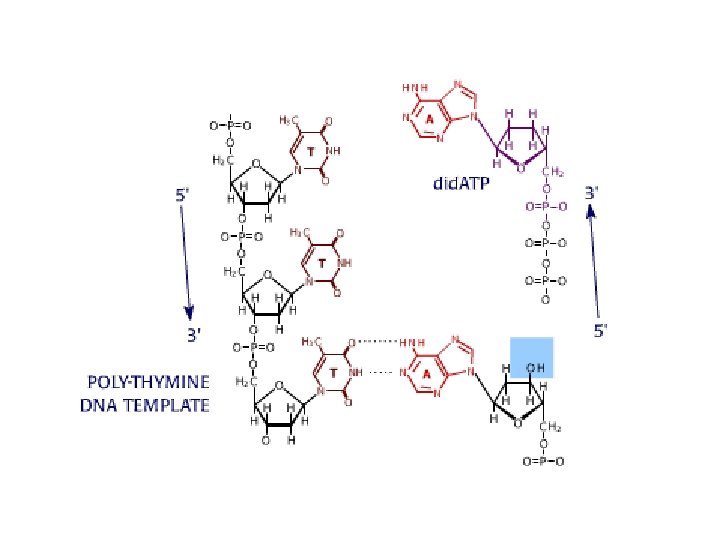
 Sequence analysis • Four different reactions produce DNA fragments that are terminated (randomly) at each of the four nucleotides. • These samples are resolved by electrophoresis. • The shortest fragments, those terminated closest to the primer, run faster than the longer fragments.
Sequence analysis • Four different reactions produce DNA fragments that are terminated (randomly) at each of the four nucleotides. • These samples are resolved by electrophoresis. • The shortest fragments, those terminated closest to the primer, run faster than the longer fragments.
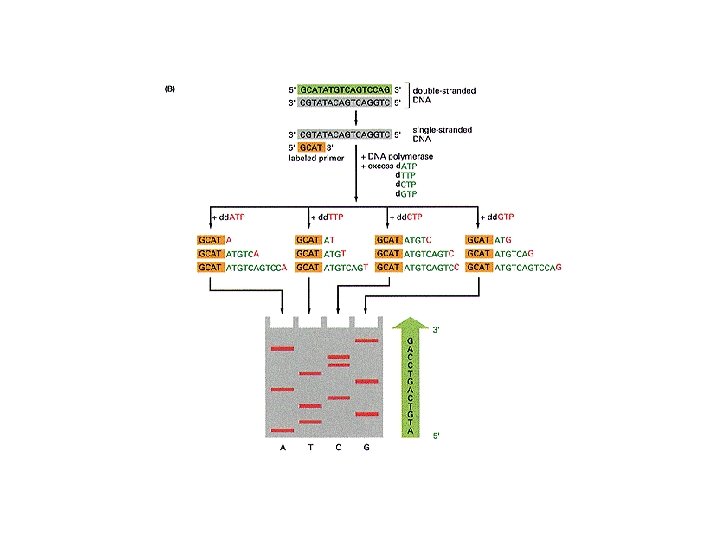
 ACGT • DNA sequencing reactions can be radioactively labeled. • Bands detected by Xray film exposure. • Sequence can be read in the 5’ to 3’ direction from the bottom of the image towards the top. A A T C T A A C G
ACGT • DNA sequencing reactions can be radioactively labeled. • Bands detected by Xray film exposure. • Sequence can be read in the 5’ to 3’ direction from the bottom of the image towards the top. A A T C T A A C G
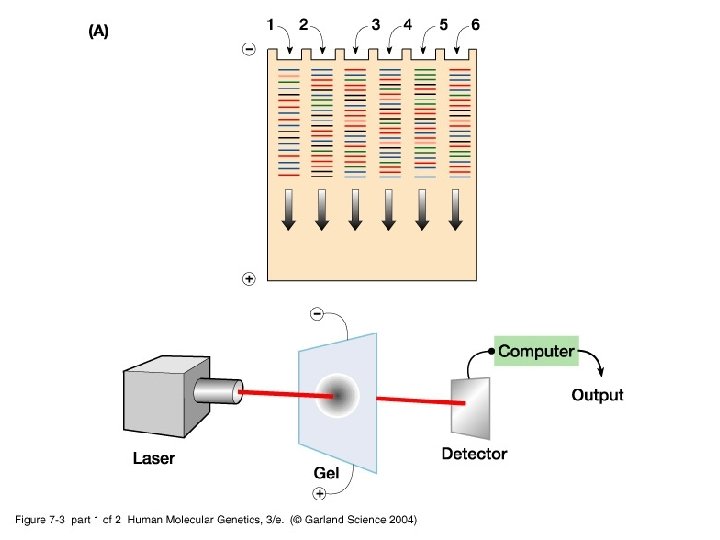
 This is great but… Wouldn’t it be great to run everything in one lane? Saves space and time, more efficient Fluorescently label the dd. NTPs so that they each have a different color…
This is great but… Wouldn’t it be great to run everything in one lane? Saves space and time, more efficient Fluorescently label the dd. NTPs so that they each have a different color…
 07_03. jpg
07_03. jpg
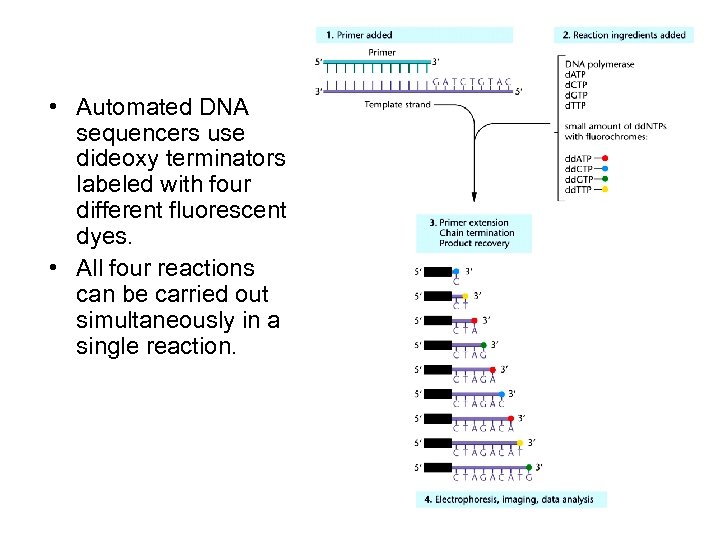 • Automated DNA sequencers use dideoxy terminators labeled with four different fluorescent dyes. • All four reactions can be carried out simultaneously in a single reaction.
• Automated DNA sequencers use dideoxy terminators labeled with four different fluorescent dyes. • All four reactions can be carried out simultaneously in a single reaction.
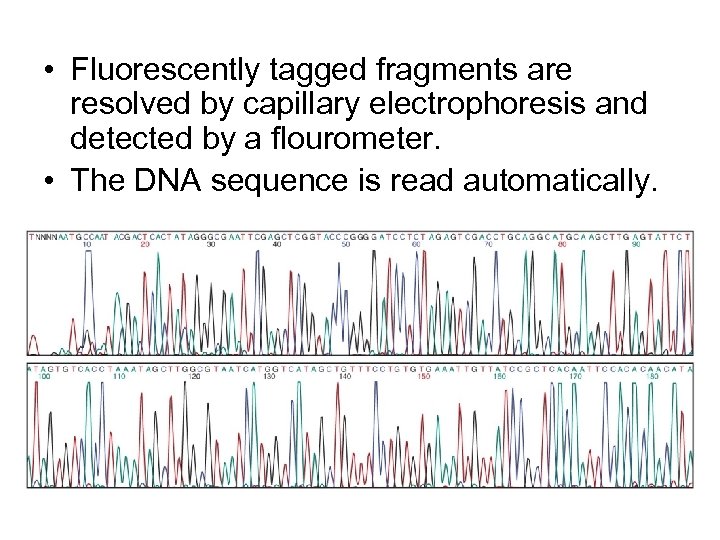 • Fluorescently tagged fragments are resolved by capillary electrophoresis and detected by a flourometer. • The DNA sequence is read automatically.
• Fluorescently tagged fragments are resolved by capillary electrophoresis and detected by a flourometer. • The DNA sequence is read automatically.
 Beckman CEQ 2000, 8 capillary ABI Prism 3730, 96 capillary
Beckman CEQ 2000, 8 capillary ABI Prism 3730, 96 capillary
 Affymetrix Gene Chip
Affymetrix Gene Chip
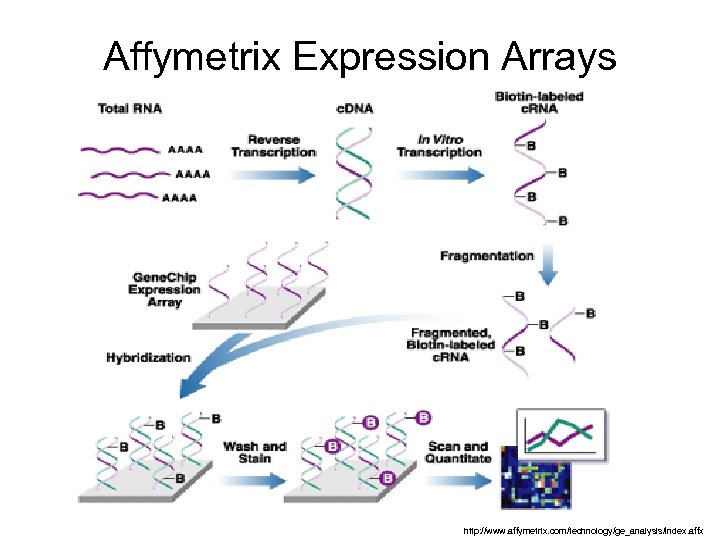 Affymetrix Expression Arrays http: //www. affymetrix. com/technology/ge_analysis/index. affx
Affymetrix Expression Arrays http: //www. affymetrix. com/technology/ge_analysis/index. affx
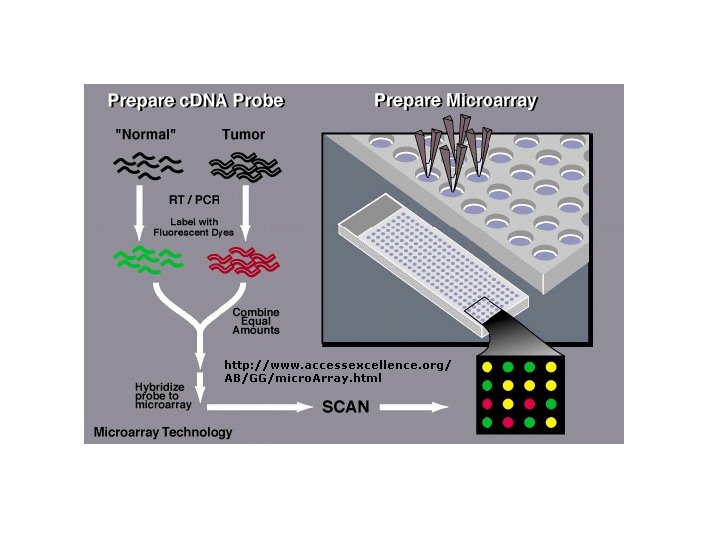
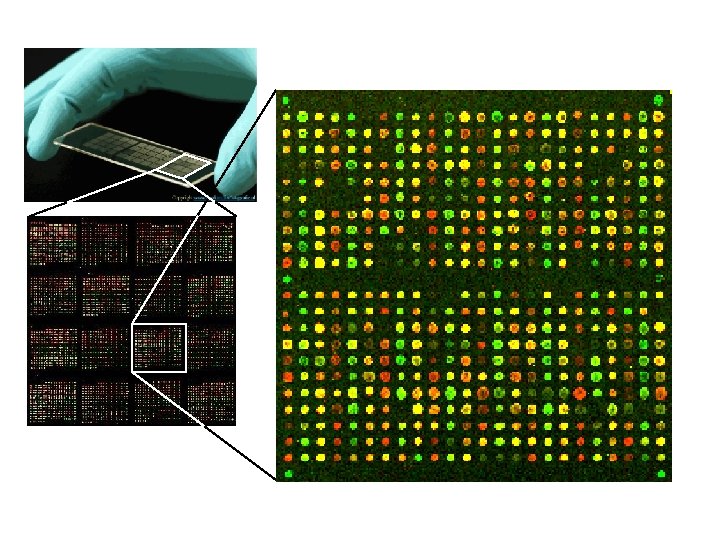
 Analysis of microarrays • Microarrays allow for the simultaneous analysis of the expression of thousands of m. RNAs. • Useful for determining changes in gene expression patterns from one sample tissue to another. • For example, microarrays have been used to study differences in gene expression in different tumor tissues.
Analysis of microarrays • Microarrays allow for the simultaneous analysis of the expression of thousands of m. RNAs. • Useful for determining changes in gene expression patterns from one sample tissue to another. • For example, microarrays have been used to study differences in gene expression in different tumor tissues.
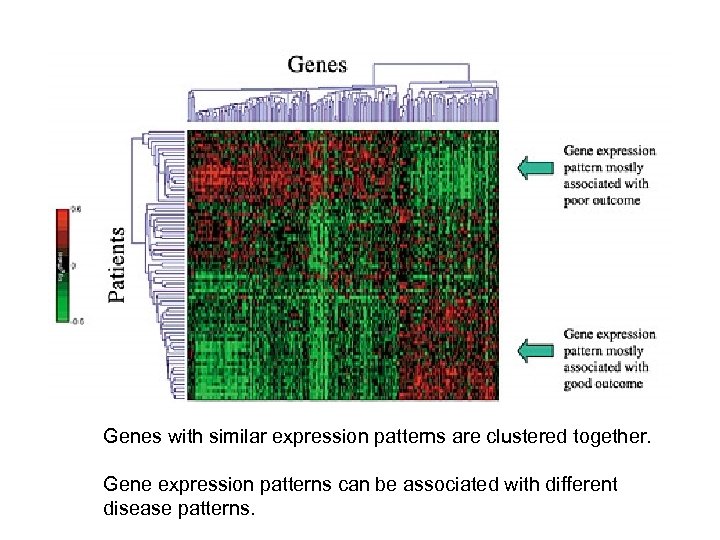 Genes with similar expression patterns are clustered together. Gene expression patterns can be associated with different disease patterns.
Genes with similar expression patterns are clustered together. Gene expression patterns can be associated with different disease patterns.
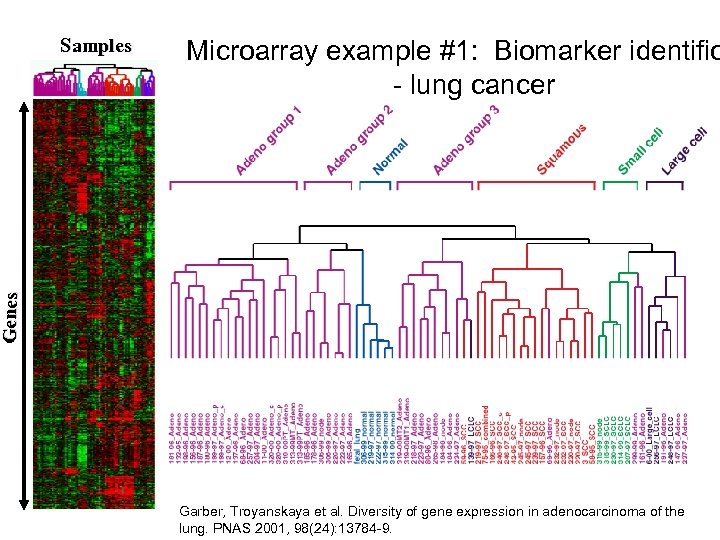 Microarray example #1: Biomarker identific - lung cancer G e ne s Samples Garber, Troyanskaya et al. Diversity of gene expression in adenocarcinoma of the lung. PNAS 2001, 98(24): 13784 -9.
Microarray example #1: Biomarker identific - lung cancer G e ne s Samples Garber, Troyanskaya et al. Diversity of gene expression in adenocarcinoma of the lung. PNAS 2001, 98(24): 13784 -9.
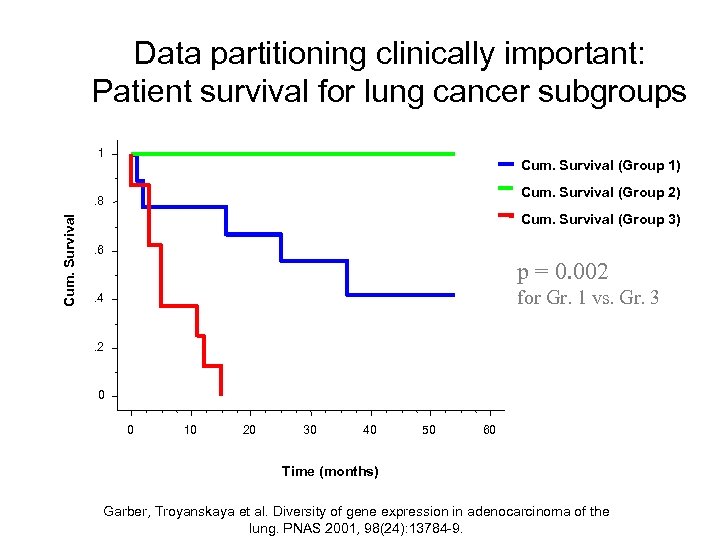 Data partitioning clinically important: Patient survival for lung cancer subgroups 1 Cum. Survival (Group 1) Cum. Survival (Group 2) Cum. Survival . 8 Cum. Survival (Group 3). 6 p = 0. 002 for Gr. 1 vs. Gr. 3 . 4 . 2 0 0 10 20 30 40 50 60 Time (months) Garber, Troyanskaya et al. Diversity of gene expression in adenocarcinoma of the lung. PNAS 2001, 98(24): 13784 -9.
Data partitioning clinically important: Patient survival for lung cancer subgroups 1 Cum. Survival (Group 1) Cum. Survival (Group 2) Cum. Survival . 8 Cum. Survival (Group 3). 6 p = 0. 002 for Gr. 1 vs. Gr. 3 . 4 . 2 0 0 10 20 30 40 50 60 Time (months) Garber, Troyanskaya et al. Diversity of gene expression in adenocarcinoma of the lung. PNAS 2001, 98(24): 13784 -9.
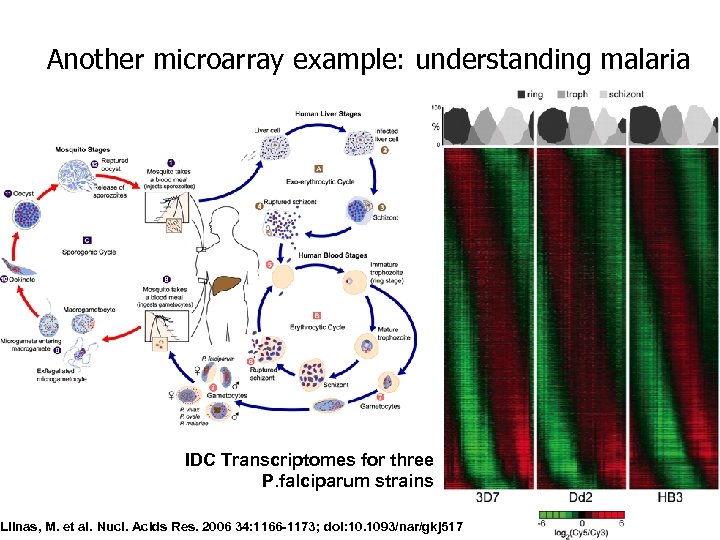 Another microarray example: understanding malaria IDC Transcriptomes for three P. falciparum strains Llinas, M. et al. Nucl. Acids Res. 2006 34: 1166 -1173; doi: 10. 1093/nar/gkj 517
Another microarray example: understanding malaria IDC Transcriptomes for three P. falciparum strains Llinas, M. et al. Nucl. Acids Res. 2006 34: 1166 -1173; doi: 10. 1093/nar/gkj 517
 Yeast 2 Hybrid Allows the genetic detection of physical interactions between proteins
Yeast 2 Hybrid Allows the genetic detection of physical interactions between proteins
 Yeast Two Hybrid Assay • • The two-hybrid system is a molecular genetic tool which facilitates the study of protein-protein interactions. If two proteins interact, then a reporter gene is transcriptionally activated. – e. g. gal 1 -lac. Z - the beta-galactosidase gene • • A colour reaction can be seen on specific media. You can use this to – Study the interaction between two proteins which you expect to interact – Find proteins (prey) which interact with a protein you have already (bait).
Yeast Two Hybrid Assay • • The two-hybrid system is a molecular genetic tool which facilitates the study of protein-protein interactions. If two proteins interact, then a reporter gene is transcriptionally activated. – e. g. gal 1 -lac. Z - the beta-galactosidase gene • • A colour reaction can be seen on specific media. You can use this to – Study the interaction between two proteins which you expect to interact – Find proteins (prey) which interact with a protein you have already (bait).
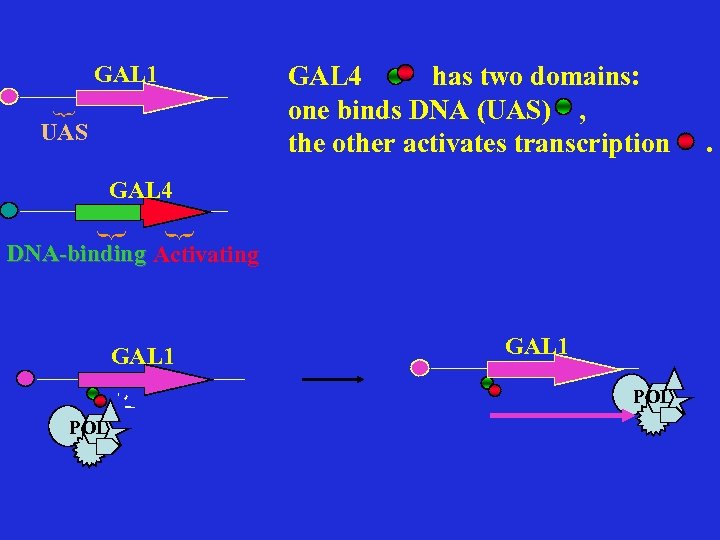 GAL 4 has two domains: one binds DNA (UAS) , the other activates transcription { GAL 1 UAS { { GAL 4 DNA-binding Activating GAL 1 POL .
GAL 4 has two domains: one binds DNA (UAS) , the other activates transcription { GAL 1 UAS { { GAL 4 DNA-binding Activating GAL 1 POL .
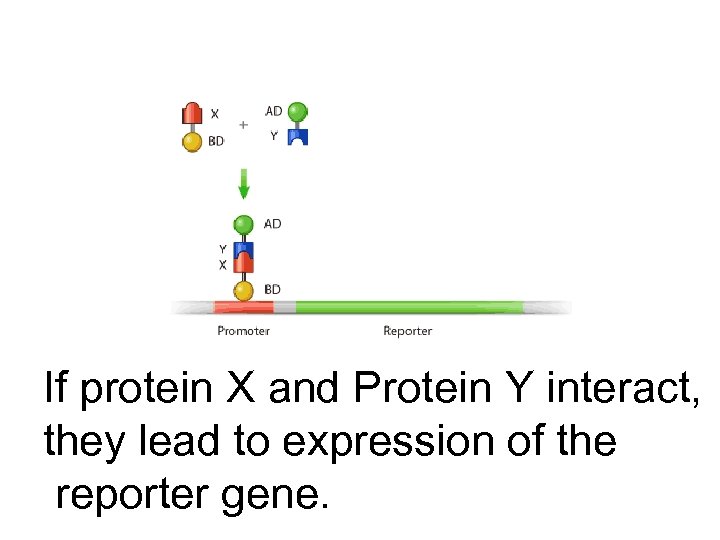 If protein X and Protein Y interact, they lead to expression of the reporter gene.
If protein X and Protein Y interact, they lead to expression of the reporter gene.
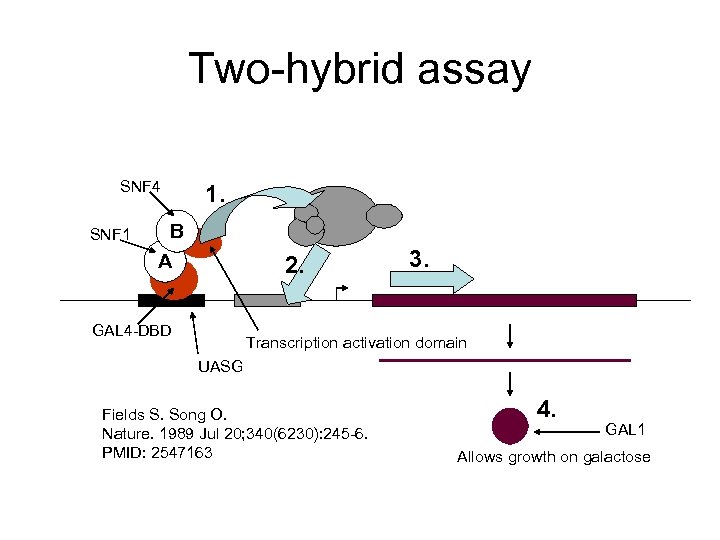 Two-hybrid assay SNF 4 SNF 1 1. B A 2. GAL 4 -DBD 3. Transcription activation domain UASG Fields S. Song O. Nature. 1989 Jul 20; 340(6230): 245 -6. PMID: 2547163 4. GAL 1 Allows growth on galactose
Two-hybrid assay SNF 4 SNF 1 1. B A 2. GAL 4 -DBD 3. Transcription activation domain UASG Fields S. Song O. Nature. 1989 Jul 20; 340(6230): 245 -6. PMID: 2547163 4. GAL 1 Allows growth on galactose
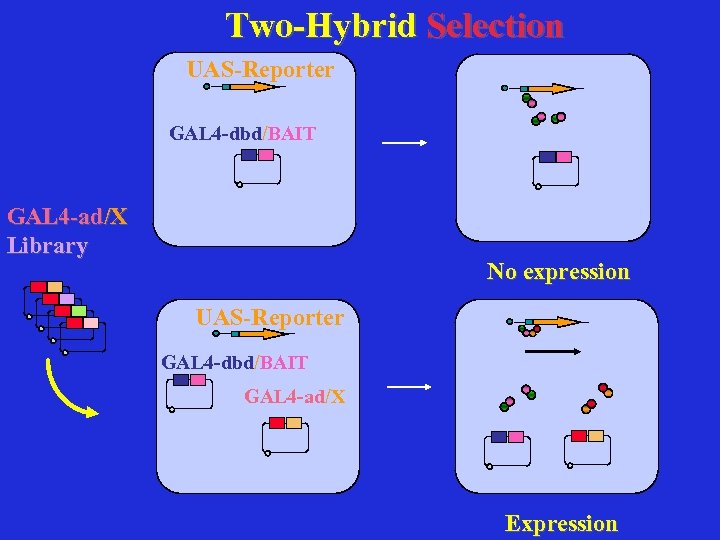 Two-Hybrid Selection UAS-Reporter GAL 4 -dbd/BAIT GAL 4 -ad/X Library No expression UAS-Reporter GAL 4 -dbd/BAIT GAL 4 -ad/X Expression
Two-Hybrid Selection UAS-Reporter GAL 4 -dbd/BAIT GAL 4 -ad/X Library No expression UAS-Reporter GAL 4 -dbd/BAIT GAL 4 -ad/X Expression


