01452b992d42ac06e0327ec7304ab791.ppt
- Количество слайдов: 51

Pick a bug, any bug. Comprehensive Aligned Sequence Construction for Automated Design of Effective Probes (CASCADE-P) The design and practical use of r. DNA oligonucleotide microarrays to identify microbes in complex samples Todd De. Santis, Igor Dubosarskiy (Ceres), Sonya Murray, Gary Andersen Environmental Molecular Microbiology Group BBRP - LLNL

The worries of an Italian parent Does he feel accepted? Is there sufficient diversity in his lower G. I. bacterial community? Are there archaeal organisms are in the aerosols he breathes? Is he getting enough sleep? Are the straps on his car seat irritating his neck? Enzo Salvatore De. Santis Jenny Di. Giovanni De. Santis

• Every soiled diaper sacrificed to the Diaper Genie is lost data. • But who wants to do all the work? – Culture • anaerobes • non-cultivable – Sequencing 16 S r. DNA • Need to create, clone, & process hundreds of samples • Can we create a simple, quantitative, comprehensive microbial test?

Outline • • Goals Experimental approach Why create a new 16 S r. DNA database? How do you align >65, 000 sequences? Organization of sequences into types Designing probes for each type Reassessing probe specificity as database grows Using 16 S Gene. Chip for quantitative aerosol analysis
Project Overview • Create a single Gene. Chip® capable of detecting and quantifying bacterial and/or archaeal organisms in a complex sample. • What is in a sample, as opposed to what is not in a sample. • Approach – Combinatorial power of multiple probes for sequencespecific hybridization
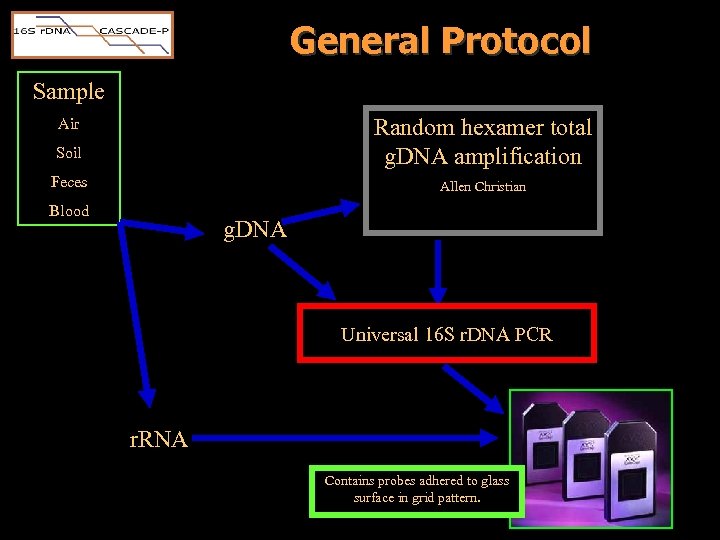
General Protocol Sample Random hexamer total g. DNA amplification Air Soil Feces Allen Christian Blood g. DNA Universal 16 S r. DNA PCR r. RNA Contains probes adhered to glass surface in grid pattern.
16 S r. RNA gene (16 S r. DNA) • Used to identify and classify organisms by gene sequence variations. • Variations have been used in design of DNA probes for the detection of: – taxonomic domains, divisions, groups … – specific organisms
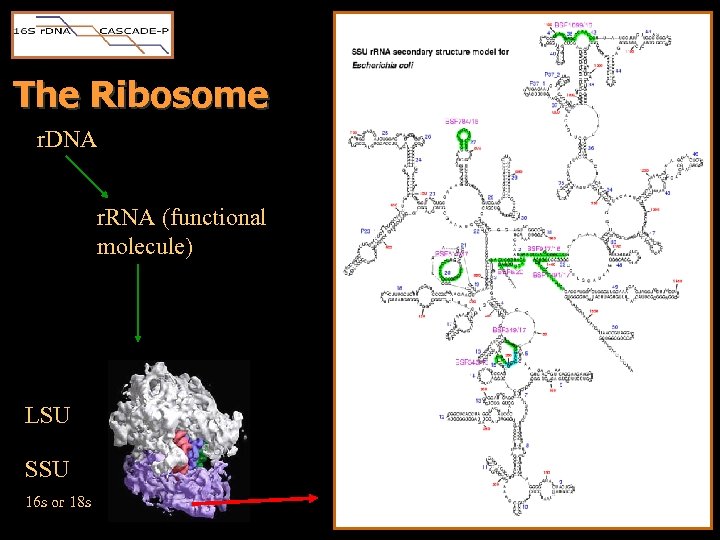
The Ribosome r. DNA r. RNA (functional molecule) LSU SSU 16 s or 18 s
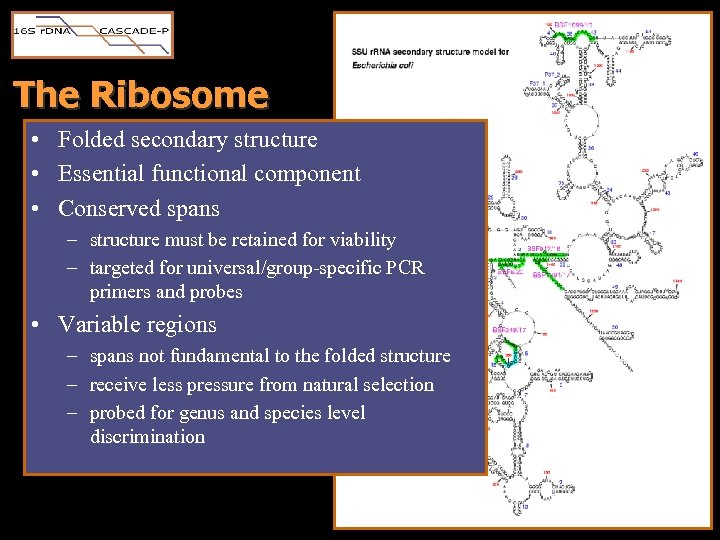
The Ribosome • Folded secondary structure • Essential functional component • Conserved spans – structure must be retained for viability – targeted for universal/group-specific PCR primers and probes • Variable regions – spans not fundamental to the folded structure – receive less pressure from natural selection – probed for genus and species level discrimination

Building upon two decades of 16 S gene cataloging • Over 75, 000 16 S records housed at NCBI. • Grows every week. • Don’t need to build a reference library for organism ID. – FAME – MS – Surface Antigen Identification
What could be amplified? • Universal 16 S PCR primers complex population of amplicons. Tom Kusmarski Variable • Must define the targets to consider as the Potential Amplicon Set or PAS. • “Give me a file of all the amplicon sequences possible if we used universal primers upon a sample containing every prokaryote. ”

Difficulties defining the PAS • Why Entrez search for “ 16 S” is insufficient: – non-16 S sequences are errantly deposited as 16 S – 16 S sequences may be concealed within longer records that cover entire operons or genomes – anti-sense strands aren't specified as such
Difficulties defining the PAS • For each sequence, need to trim away bases that won’t be amplified. • Problem: Most 16 S records are “partial”. • Can primer pattern matching within sequences allow for proper trimming? – What does it mean when a primer search fails? • Primer locus present in record but mutated, or • Primer locus outside the sequence span deposited
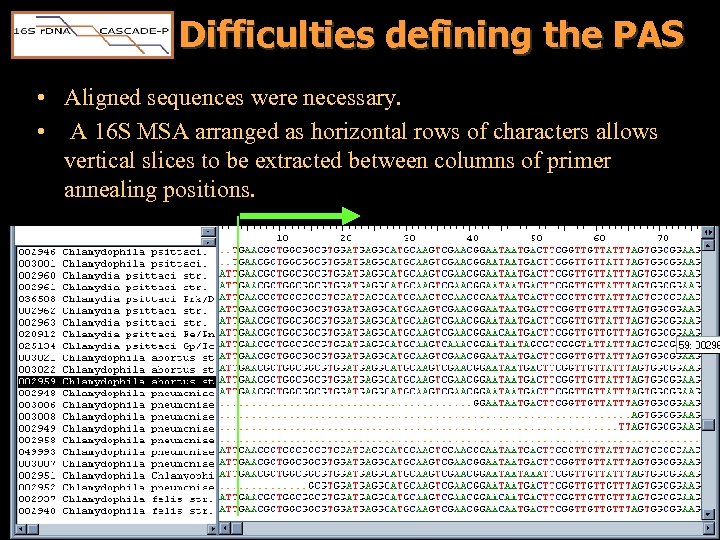
Difficulties defining the PAS • Aligned sequences were necessary. • A 16 S MSA arranged as horizontal rows of characters allows vertical slices to be extracted between columns of primer annealing positions.
Existing 16 S aligned databases • Under 20, 000 sequences among 3 databases. – Updates occurred annually or worse. – Others focused on hand-aligning complete sequences. • Structure predictions • Phylogeny assessment • Comprehensive Aligned Sequence Construction for Automated Design of Effective Probes (CASCADE-P) – need up-to-date records – need to include partial sequences • add “inertia” to region considered conserved • increase the likelihood of detecting a polymorphism • searched for unwanted cross-hybridizations with a tentative probe
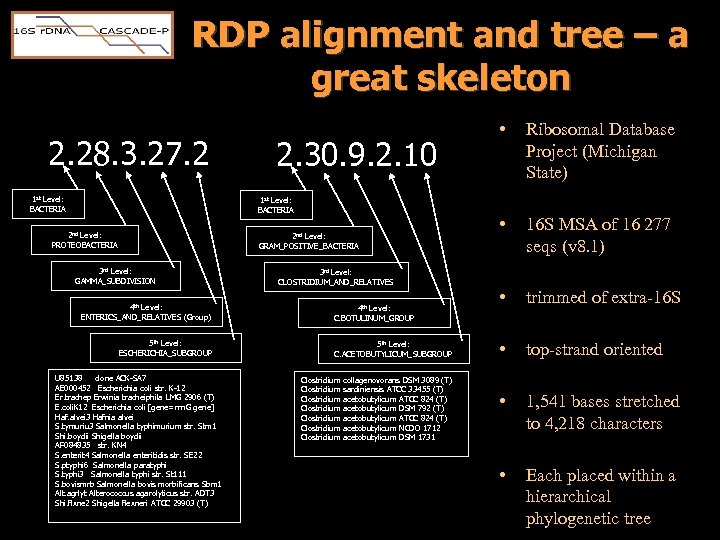
RDP alignment and tree – a great skeleton 2. 28. 3. 27. 2 1 st Level: BACTERIA 2. 30. 9. 2. 10 1 st Level: BACTERIA 2 nd Level: PROTEOBACTERIA 2 nd Level: GRAM_POSITIVE_BACTERIA 3 rd Level: GAMMA_SUBDIVISION 4 th Level: ENTERICS_AND_RELATIVES (Group) 5 th Level: ESCHERICHIA_SUBGROUP U 85138 clone ACK-SA 7 AE 000452 Escherichia coli str. K-12 Er. trachep Erwinia tracheiphila LMG 2906 (T) E. coli. K 12 Escherichia coli [gene= rrn. G gene] Haf. alvei 3 Hafnia alvei S. tymuriu 3 Salmonella typhimurium str. Stm 1 Shi. boydii Shigella boydii AF 084835 str. KN 4 S. enterit 4 Salmonella enteritidis str. SE 22 S. ptyphi 6 Salmonella paratyphi S. typhi 3 Salmonella typhi str. St 111 S. bovismrb Salmonella bovis morbificans Sbm 1 Alt. agrlyt Alterococcus agarolyticus str. ADT 3 Shi. flxne 2 Shigella flexneri ATCC 29903 (T) • Ribosomal Database Project (Michigan State) • 16 S MSA of 16 277 seqs (v 8. 1) • trimmed of extra-16 S • top-strand oriented • 1, 541 bases stretched to 4, 218 characters • Each placed within a hierarchical phylogenetic tree 3 rd Level: CLOSTRIDIUM_AND_RELATIVES 4 th Level: C. BOTULINUM_GROUP 5 th Level: C. ACETOBUTYLICUM_SUBGROUP Clostridium collagenovorans DSM 3089 (T) Clostridium sardiniensis ATCC 33455 (T) Clostridium acetobutylicum ATCC 824 (T) Clostridium acetobutylicum DSM 792 (T) Clostridium acetobutylicum ATCC 824 (T) Clostridium acetobutylicum NCDO 1712 Clostridium acetobutylicum DSM 1731
Sequence pre-processing • Download ‘ 16 S’ candidates via “ESearch” • BLAST compare to 16 S/18 S RDP “standards”. – Candidates were rejected if • the longest match length was <300 base pairs • the highest scoring BLAST subject was eukaryotic • candidate matched sequences in two or more RDP terminal tree branches equally well – Phylocode assigned from top HSP
Sequence pre-processing
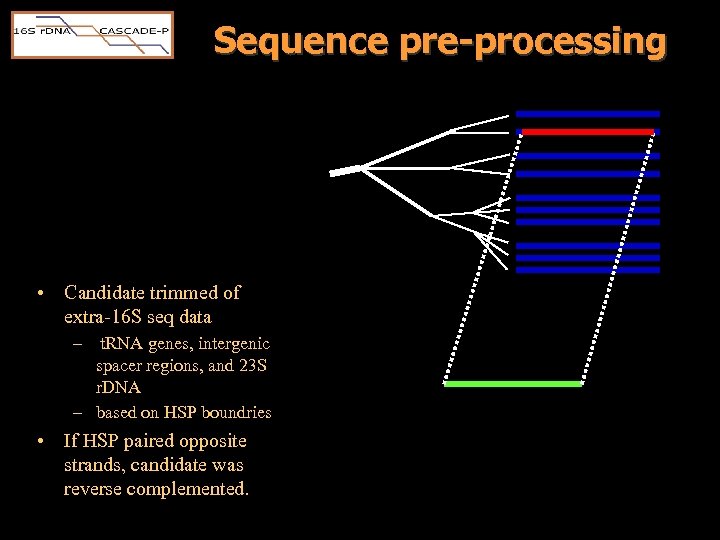
Sequence pre-processing • Candidate trimmed of extra-16 S seq data – t. RNA genes, intergenic spacer regions, and 23 S r. DNA – based on HSP boundries • If HSP paired opposite strands, candidate was reverse complemented.
Sequence pre-processing
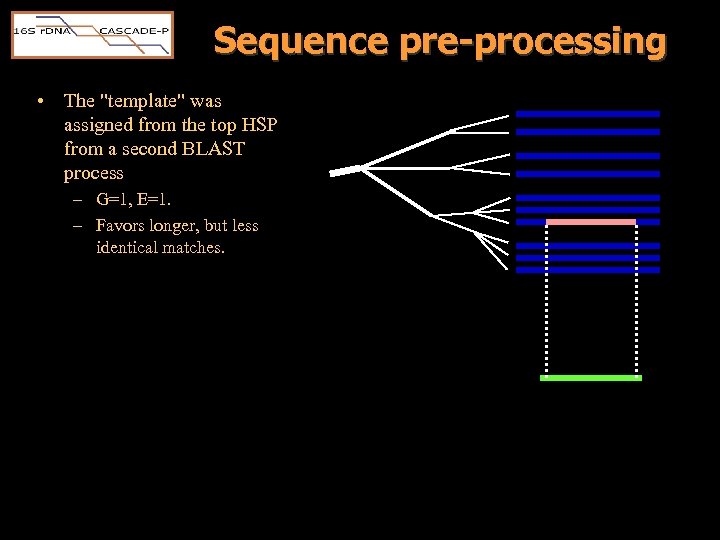
Sequence pre-processing • The "template" was assigned from the top HSP from a second BLAST process – G=1, E=1. – Favors longer, but less identical matches.

Prokaryotic Multiple Sequence Alignment prok. MSA • Essentially, the prok. MSA was a merger built serially by aligning each candidate to its closest relative in the RDP tree. • Two Steps – Align 0 • Publicly available, pair-wise SW aligner • Candidate expansion – NAST • Nearest Alignment Space Termination • Novel algorithm • Candidate compression
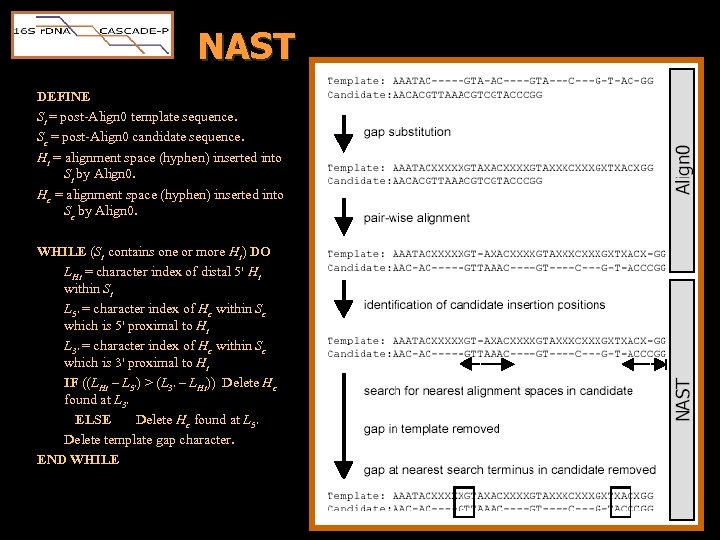
NAST DEFINE St = post-Align 0 template sequence. Sc = post-Align 0 candidate sequence. Ht = alignment space (hyphen) inserted into St by Align 0. Hc = alignment space (hyphen) inserted into Sc by Align 0. WHILE (St contains one or more Ht) DO LHt = character index of distal 5' Ht within St L 5' = character index of Hc within Sc which is 5' proximal to Ht L 3' = character index of Hc within Sc which is 3' proximal to Ht IF ((LHt – L 5') > (L 3' – LHt)) Delete Hc found at L 3' ELSE Delete Hc found at L 5' Delete template gap character. END WHILE
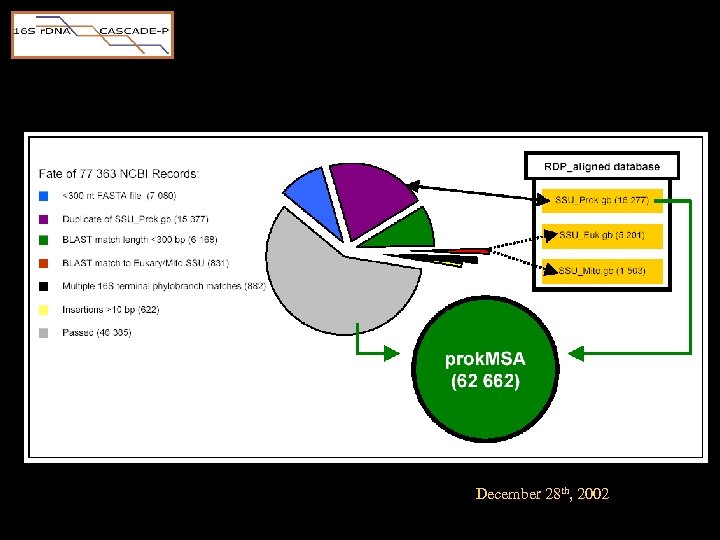
December 28 th, 2002

Operational Taxonomic Units • We did not desire to design probes for each sequence. – Many sequences are nearly identical. – Desired 20 probe per sequence: • 60, 000 seqs * ( 20 probes + 20 probes) = • 2. 4 million probes (not possible, yet) • We did desire to design probes for each type of sequence. • Need to group sequences into types amenable to probe design.

Operational Taxonomic Units • Avoid groupings based on historical nomenclature. • Sequence-dependent classification by transitive similarity clustering at 98%. if x R y & y R z x R z • Create groupings into Operational Taxonomic Units (OTU). • Each sequence must be in exactly 1 OTU
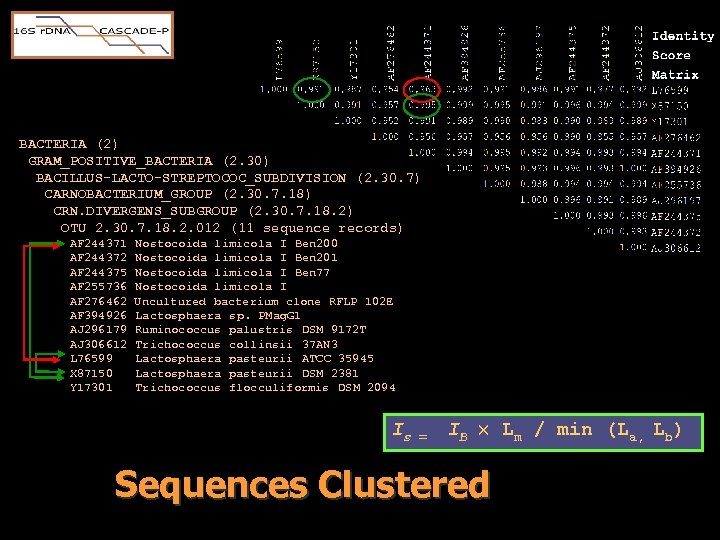
BACTERIA (2) GRAM_POSITIVE_BACTERIA (2. 30) BACILLUS-LACTO-STREPTOCOC_SUBDIVISION (2. 30. 7) CARNOBACTERIUM_GROUP (2. 30. 7. 18) CRN. DIVERGENS_SUBGROUP (2. 30. 7. 18. 2) OTU 2. 30. 7. 18. 2. 012 (11 sequence records) AF 244371 AF 244372 AF 244375 AF 255736 AF 276462 AF 394926 AJ 296179 AJ 306612 L 76599 X 87150 Y 17301 Nostocoida limicola I Ben 200 Nostocoida limicola I Ben 201 Nostocoida limicola I Ben 77 Nostocoida limicola I Uncultured bacterium clone RFLP 102 E Lactosphaera sp. PMag. G 1 Ruminococcus palustris DSM 9172 T Trichococcus collinsii 37 AN 3 Lactosphaera pasteurii ATCC 35945 Lactosphaera pasteurii DSM 2381 Trichococcus flocculiformis DSM 2094 Is = IB Lm / min (La, Lb) Sequences Clustered
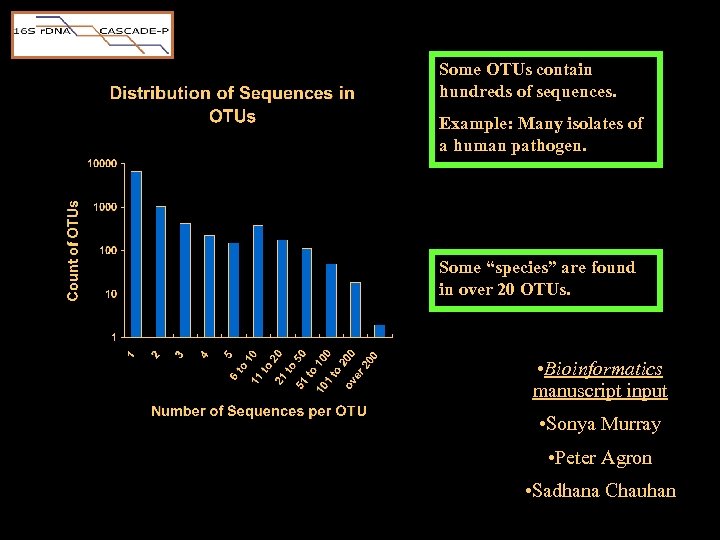
Some OTUs contain hundreds of sequences. Example: Many isolates of a human pathogen. Some “species” are found in over 20 OTUs. • Bioinformatics manuscript input • Sonya Murray • Peter Agron • Sadhana Chauhan
http: //greengenes. llnl. gov/16 S • Comprehensive Aligned Sequence Construction for Automated Design of Effective Probes • Igor Dubosarskiy – Java implementations • Tim Harsch – RDBMS consultations • Lisa Corsetti – Apache module management • Kevin Melissare – Graphics
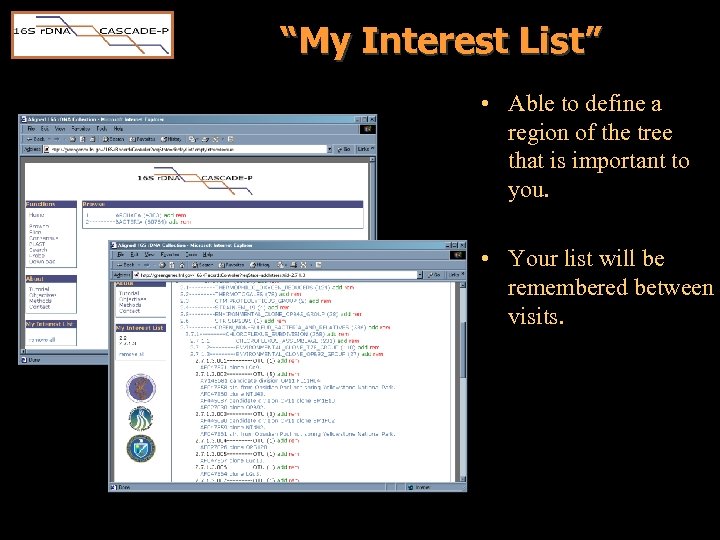
“My Interest List” • Able to define a region of the tree that is important to you. • Your list will be remembered between visits.
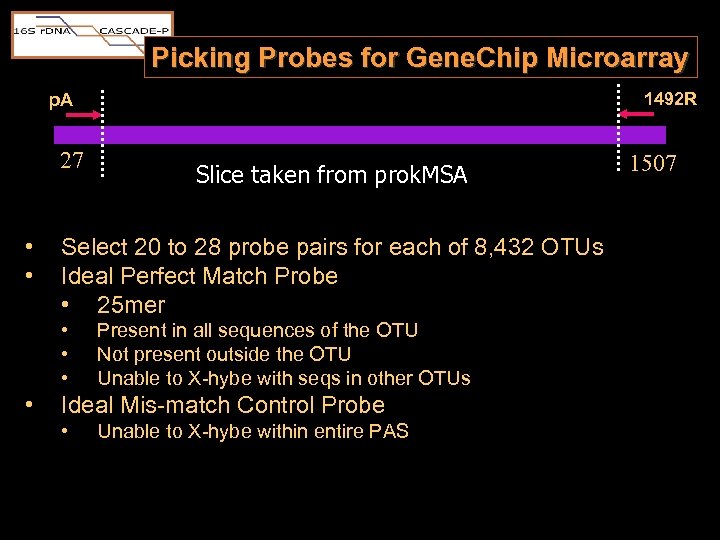
Picking Probes for Gene. Chip Microarray 1492 R p. A 27 • • Select 20 to 28 probe pairs for each of 8, 432 OTUs Ideal Perfect Match Probe • 25 mer • • Slice taken from prok. MSA Present in all sequences of the OTU Not present outside the OTU Unable to X-hybe with seqs in other OTUs Ideal Mis-match Control Probe • Unable to X-hybe within entire PAS 1507

Scoring Probe Candidates • Score is calculated for each potential probe pair. • Product of 3 factors: – Locus Specific Prevalence Factor – Perfect. Match X-hybe Factor • The closer the tree distance, the lower the factor – Mis. Match X-hybe Factor • 3 bases available • use base which produces highest factor

22/22 25/25 20/25 26 sequences Locus Specific Prevalence Scoring OTU composed of
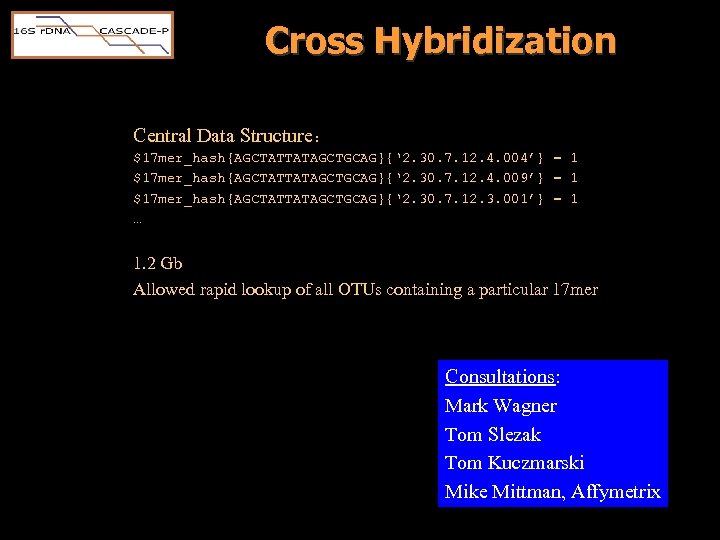
Cross Hybridization Central Data Structure: $17 mer_hash{AGCTATTATAGCTGCAG}{‘ 2. 30. 7. 12. 4. 004’} = 1 $17 mer_hash{AGCTATTATAGCTGCAG}{‘ 2. 30. 7. 12. 4. 009’} = 1 $17 mer_hash{AGCTATTATAGCTGCAG}{‘ 2. 30. 7. 12. 3. 001’} = 1 … 1. 2 Gb Allowed rapid lookup of all OTUs containing a particular 17 mer Consultations: Mark Wagner Tom Slezak Tom Kuczmarski Mike Mittman, Affymetrix
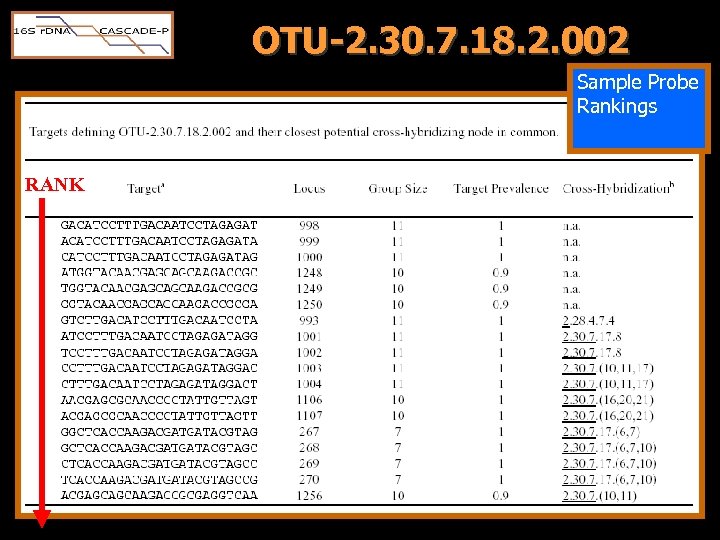
OTU-2. 30. 7. 18. 2. 002 Sample Probe Rankings RANK

S. aureus spike Combinatorial scoring of “Probe Sets” are able to categorize mixed samples. Art Koybayashi – Simulation package B. anthracis spike OTU 2. 30. 7. 12. 1. 013 * 2. 30. 7. 12. 1. 014 2. 30. 7. 12. 1. 015 2. 30. 7. 12. 1. 016 2. 30. 7. 12. 1. 017 2. 30. 7. 12. 2. 002 2. 30. 7. 12. 2. 003 2. 30. 7. 12. 2. 005 2. 30. 7. 12. 2. 006 2. 30. 7. 12. 2. 007 2. 30. 7. 12. 2. 008 2. 30. 7. 12. 3. 001 2. 30. 7. 12. 3. 002 2. 30. 7. 12. 3. 003 2. 30. 7. 12. 3. 004 2. 30. 7. 12. 3. 005 2. 30. 7. 12. 3. 006 2. 30. 7. 12. 3. 007 2. 30. 7. 12. 3. 008 2. 30. 7. 12. 3. 009 2. 30. 7. 12. 3. 010 2. 30. 7. 12. 4. 001 2. 30. 7. 12. 4. 004 * 2. 30. 7. 12. 4. 005 2. 30. 7. 12. 4. 006 2. 30. 7. 12. 4. 007 2. 30. 7. 12. 4. 008 % pos pairs 100 46 – 57 54 - 61 39 – 54 18 11 14 14 – 32 18 – 32 21 – 25 14 – 29 7 – 25 8 4 7 – 11 4 – 14 11 14 – 29 7 4 – 11 0 - 4 21 – 36 100 0 – 11 29 – 54 11 – 14 11
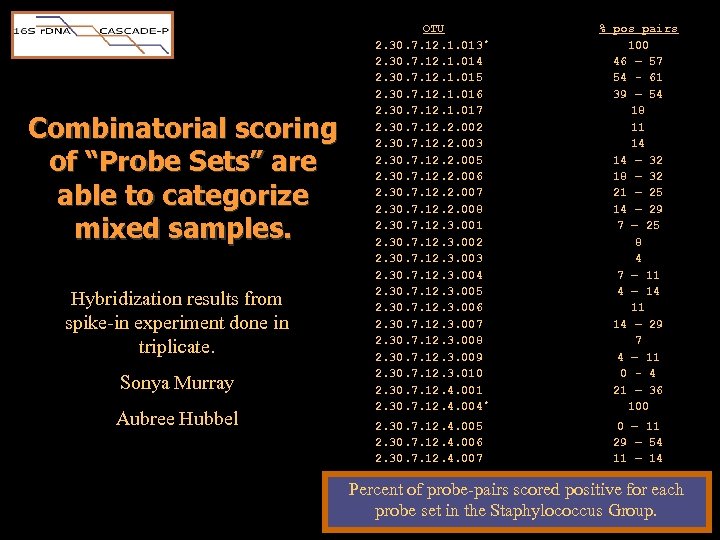
Combinatorial scoring of “Probe Sets” are able to categorize mixed samples. Hybridization results from spike-in experiment done in triplicate. Sonya Murray Aubree Hubbel OTU 2. 30. 7. 12. 1. 013 * 2. 30. 7. 12. 1. 014 2. 30. 7. 12. 1. 015 2. 30. 7. 12. 1. 016 2. 30. 7. 12. 1. 017 2. 30. 7. 12. 2. 002 2. 30. 7. 12. 2. 003 2. 30. 7. 12. 2. 005 2. 30. 7. 12. 2. 006 2. 30. 7. 12. 2. 007 2. 30. 7. 12. 2. 008 2. 30. 7. 12. 3. 001 2. 30. 7. 12. 3. 002 2. 30. 7. 12. 3. 003 2. 30. 7. 12. 3. 004 2. 30. 7. 12. 3. 005 2. 30. 7. 12. 3. 006 2. 30. 7. 12. 3. 007 2. 30. 7. 12. 3. 008 2. 30. 7. 12. 3. 009 2. 30. 7. 12. 3. 010 2. 30. 7. 12. 4. 001 2. 30. 7. 12. 4. 004 * % pos pairs 100 46 – 57 54 - 61 39 – 54 18 11 14 14 – 32 18 – 32 21 – 25 14 – 29 7 – 25 8 4 7 – 11 4 – 14 11 14 – 29 7 4 – 11 0 - 4 21 – 36 100 2. 30. 7. 12. 4. 005 0 – 11 2. 30. 7. 12. 4. 006 29 – 54 2. 30. 7. 12. 4. 007 11 – 14 2. 30. 7. 12. 4. 008 11 Percent of probe-pairs scored positive for each probe set in the Staphylococcus Group.

PAS is a moving target • Problems Opportunities – New 16 S sequence data is constantly being deposited to public databases. – Mismatch Probes can become Perfect Matches. – Phylogenetic groupings (OTUs) can change. • New transitive links may be discovered
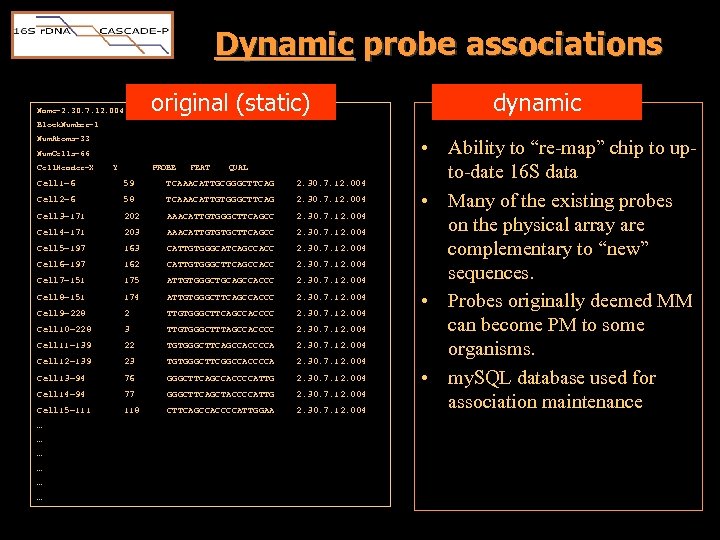
Dynamic probe associations original (static) Name=2. 30. 7. 12. 004 dynamic Block. Number=1 Num. Atoms=33 Num. Cells=66 Cell. Header=X Y PROBE FEAT QUAL Cell 1=6 59 TCAAACATTGCGGGCTTCAG 2. 30. 7. 12. 004 Cell 2=6 58 TCAAACATTGTGGGCTTCAG 2. 30. 7. 12. 004 Cell 3=171 202 AAACATTGTGGGCTTCAGCC 2. 30. 7. 12. 004 Cell 4=171 203 AAACATTGTGTGCTTCAGCC 2. 30. 7. 12. 004 Cell 5=197 163 CATTGTGGGCATCAGCCACC 2. 30. 7. 12. 004 Cell 6=197 162 CATTGTGGGCTTCAGCCACC 2. 30. 7. 12. 004 Cell 7=151 175 ATTGTGGGCTGCAGCCACCC 2. 30. 7. 12. 004 Cell 8=151 174 ATTGTGGGCTTCAGCCACCC 2. 30. 7. 12. 004 Cell 9=228 2 TTGTGGGCTTCAGCCACCCC 2. 30. 7. 12. 004 Cell 10=228 3 TTGTGGGCTTTAGCCACCCC 2. 30. 7. 12. 004 Cell 11=139 22 TGTGGGCTTCAGCCACCCCA 2. 30. 7. 12. 004 Cell 12=139 23 TGTGGGCTTCGGCCACCCCA 2. 30. 7. 12. 004 Cell 13=94 76 GGGCTTCAGCCACCCCATTG 2. 30. 7. 12. 004 Cell 14=94 77 GGGCTTCAGCTACCCCATTG 2. 30. 7. 12. 004 Cell 15=111 118 CTTCAGCCACCCCATTGGAA 2. 30. 7. 12. 004 … … … • Ability to “re-map” chip to upto-date 16 S data • Many of the existing probes on the physical array are complementary to “new” sequences. • Probes originally deemed MM can become PM to some organisms. • my. SQL database used for association maintenance

Finding groupings se q 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 A B C sequences D E F G H I J K L M N O Consider A – O to be 16 S sequences. Consider 1 – 24 to be probes already embedded on the chip. First, associate all available probes with all available sequences. Let probe similarities drive sequence groupings. 17 18 probes 19 20 21 22 23 24

Finding groupings se q 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 A B C D E F G H I J K L M N O Consider A – O to be 16 S sequences. Consider 1 – 24 to be probes already embedded on the chip. First, associate all available probes with all available sequences. Let probe similarities drive sequence groupings. 17 18 19 20 21 22 23 24

Finding groupings se q 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 A B C D E F G H I J K L M N O Consider A – O to be 16 S sequences. Consider 1 – 24 to be probes already embedded on the chip. First, associate all available probes with all available sequences. Let probe similarities drive sequence groupings. 17 18 19 20 21 22 23 24

Finding groupings se q 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 A B C D E F G H I J K L M N O Consider A – O to be 16 S sequences. Consider 1 – 24 to be probes already embedded on the chip. First, associate all available probes with all available sequences. Let probe similarities drive sequence groupings. 17 18 19 20 21 22 23 24

Finding groupings se q 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 A B C D E F G H I J K L M N O Consider A – O to be 16 S sequences. Consider 1 – 24 to be probes already embedded on the chip. First, associate all available probes with all available sequences. Let probe similarities drive sequence groupings. 17 18 19 20 21 22 23 24
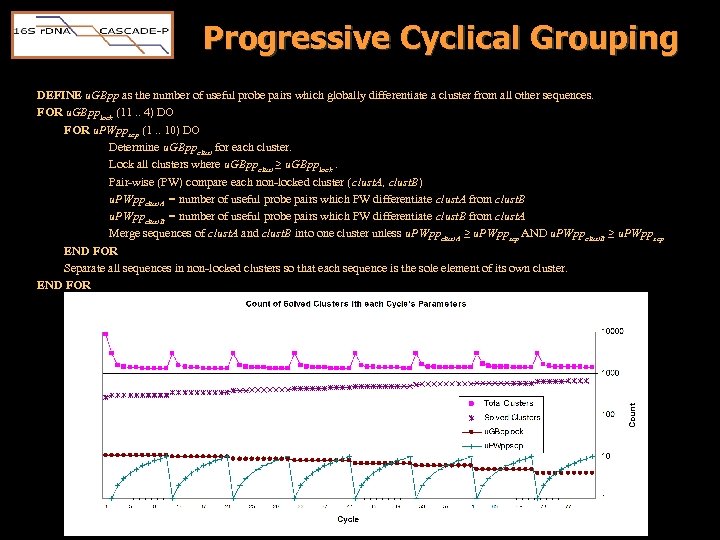
Progressive Cyclical Grouping DEFINE u. GBpp as the number of useful probe pairs which globally differentiate a cluster from all other sequences. FOR u. GBpplock (11. . 4) DO FOR u. PWppsep (1. . 10) DO Determine u. GBppclust for each cluster. Lock all clusters where u. GBppclust ≥ u. GBpplock. Pair-wise (PW) compare each non-locked cluster (clust. A, clust. B) u. PWppclust. A = number of useful probe pairs which PW differentiate clust. A from clust. B u. PWppclust. B = number of useful probe pairs which PW differentiate clust. B from clust. A Merge sequences of clust. A and clust. B into one cluster unless u. PWppclust. A ≥ u. PWppsep AND u. PWppclust. B ≥ u. PWppsep END FOR Separate all sequences in non-locked clusters so that each sequence is the sole element of its own cluster. END FOR
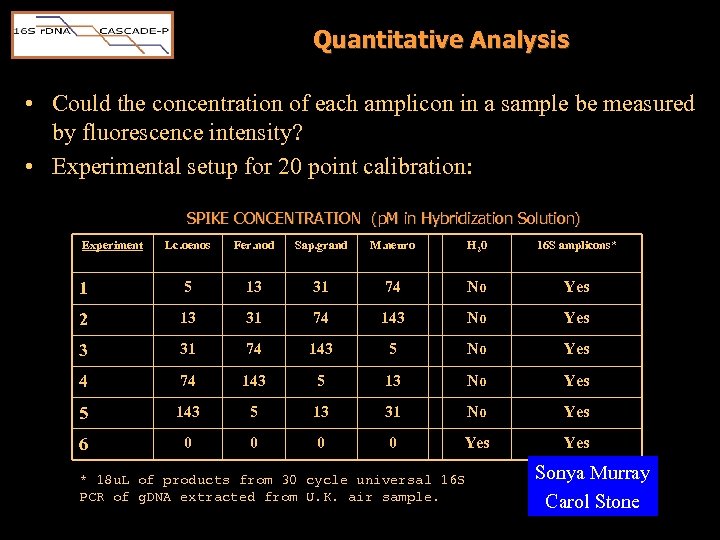
Quantitative Analysis • Could the concentration of each amplicon in a sample be measured by fluorescence intensity? • Experimental setup for 20 point calibration: SPIKE CONCENTRATION (p. M in Hybridization Solution) Experiment Lc. oenos Fer. nod Sap. grand M. neuro H 20 16 S amplicons* 1 5 13 31 74 No Yes 2 13 31 74 143 No Yes 3 31 74 143 5 No Yes 4 74 143 5 13 No Yes 5 143 5 13 31 No Yes 6 0 0 Yes * 18 u. L of products from 30 cycle universal 16 S PCR of g. DNA extracted from U. K. air sample. Sonya Murray Carol Stone
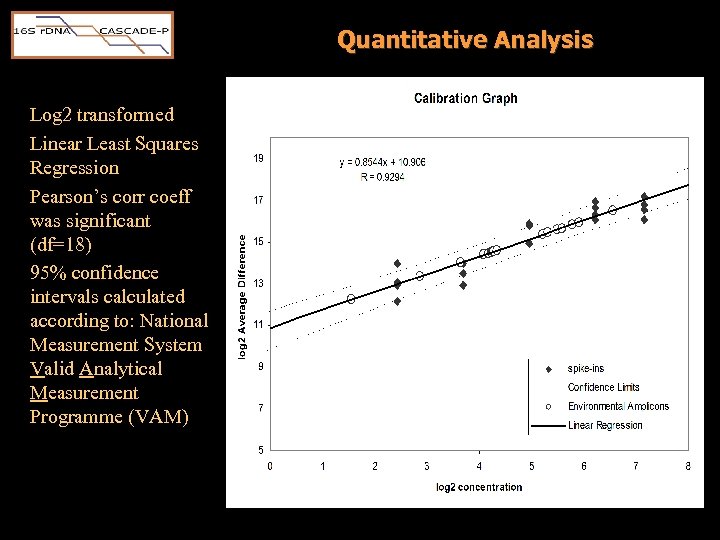
Quantitative Analysis Log 2 transformed Linear Least Squares Regression Pearson’s corr coeff was significant (df=18) 95% confidence intervals calculated according to: National Measurement System Valid Analytical Measurement Programme (VAM)
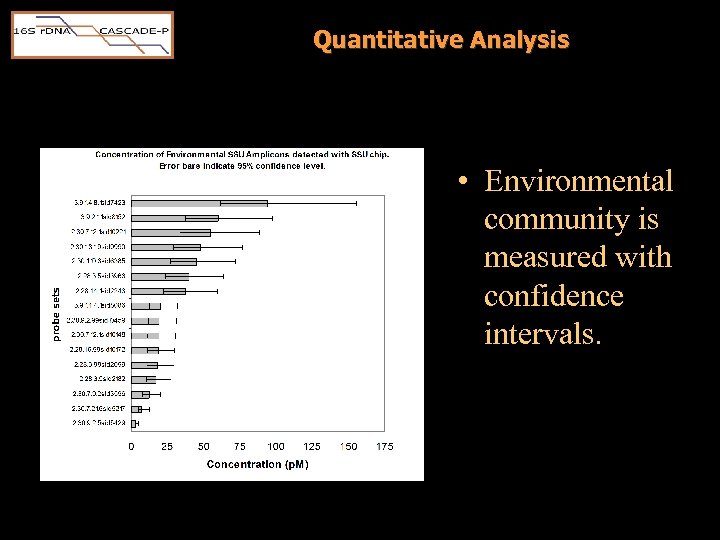
Quantitative Analysis • Environmental community is measured with confidence intervals.
Summary • prok. MSA contains 65, 000 aligned sequences and growing (largest collection). • Over 8, 000 distinct OTUs have been found. • Global probe-picking was completed. • “DOE 16 S” Gene. Chip® was manufactured. • Ability to correctly categorize spike-ins is being validated. • Detected amplicons can be quantified.

Acknowledgements • • • • Gary Andersen – Group Leader (The Tangent Terminator) Carol Stone – Sample collection, hybridization (DSLT) Aubree Hubbel – Spike synthesis Sonya Murray - Hybridizations Peter Agron – ms advise Sadhana Chauhan – ms advice Mike Mittman – probe selection constraints (Affymetrix) Art Koybayashi – Hyb simulation package, ms advice Tom Kusmarski - PAS, algorithm optimization Tom Slezak - algorithm optimization Mark Wagner - algorithm optimization Igor Dubosarskiy – Java, web front-end (Ceres) Tim Harsch - RDBMS Lisa Corsetti – Apache administration Allen Christensen – genomic sample amplification • This work was performed under the auspices of the U. S. Department of Energy by the University of California, Lawrence Livermore National Laboratory, under contract no. W-7405 -Eng-48.

01452b992d42ac06e0327ec7304ab791.ppt