79d474ae8e1cf3bbff9a8a02fca96309.ppt
- Количество слайдов: 39
 Physicochemical Methods for Protein Function Prediction Mary Jo Ondrechen Dept of Chemistry & Chemical Biology Northeastern University Boston, MA 02115
Physicochemical Methods for Protein Function Prediction Mary Jo Ondrechen Dept of Chemistry & Chemical Biology Northeastern University Boston, MA 02115
 THEMATICS Genomics and proteomics n About titration curves n Method for active site location and characterization n Examples n Future directions and conclusions n
THEMATICS Genomics and proteomics n About titration curves n Method for active site location and characterization n Examples n Future directions and conclusions n
 The post-genomic path n n n Genome sequence Protein structure Protein function Active site location and characterization, drug design, understanding protein function, normal and disease processes
The post-genomic path n n n Genome sequence Protein structure Protein function Active site location and characterization, drug design, understanding protein function, normal and disease processes
 PROTEOMICS Structural genomics – rapidly discovering new protein structures, many of unknown function The Next Frontier: Characterizing the 106 proteins for which genes hold the code.
PROTEOMICS Structural genomics – rapidly discovering new protein structures, many of unknown function The Next Frontier: Characterizing the 106 proteins for which genes hold the code.
 Predicting Protein Function Protein structure and protein function are not well correlated. Need – methods to predict function from structure (or sequence). THEMATICS “Theoretical Microscopic Titration Curves” – a reliable way to locate and characterize enzyme active sites.
Predicting Protein Function Protein structure and protein function are not well correlated. Need – methods to predict function from structure (or sequence). THEMATICS “Theoretical Microscopic Titration Curves” – a reliable way to locate and characterize enzyme active sites.
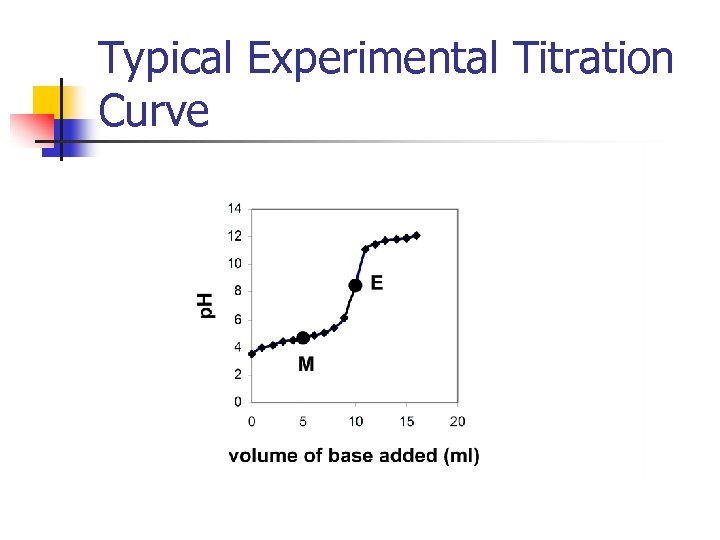 Typical Experimental Titration Curve
Typical Experimental Titration Curve
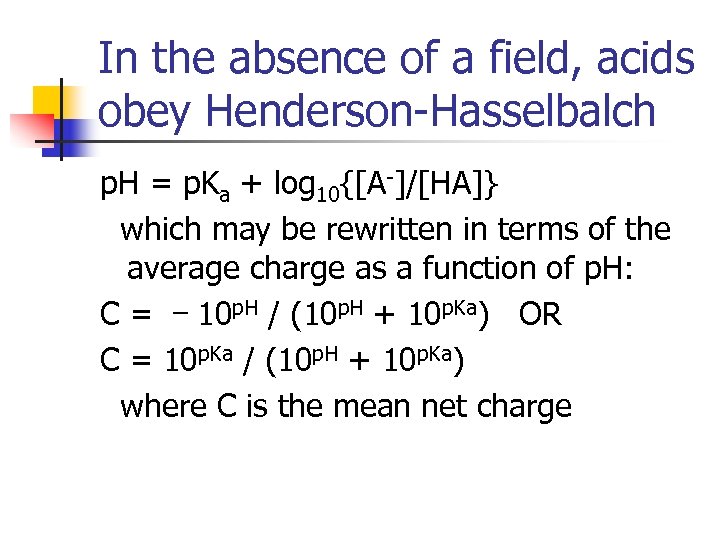 In the absence of a field, acids obey Henderson-Hasselbalch p. H = p. Ka + log 10{[A-]/[HA]} which may be rewritten in terms of the average charge as a function of p. H: _ C = 10 p. H / (10 p. H + 10 p. Ka) OR C = 10 p. Ka / (10 p. H + 10 p. Ka) where C is the mean net charge
In the absence of a field, acids obey Henderson-Hasselbalch p. H = p. Ka + log 10{[A-]/[HA]} which may be rewritten in terms of the average charge as a function of p. H: _ C = 10 p. H / (10 p. H + 10 p. Ka) OR C = 10 p. Ka / (10 p. H + 10 p. Ka) where C is the mean net charge
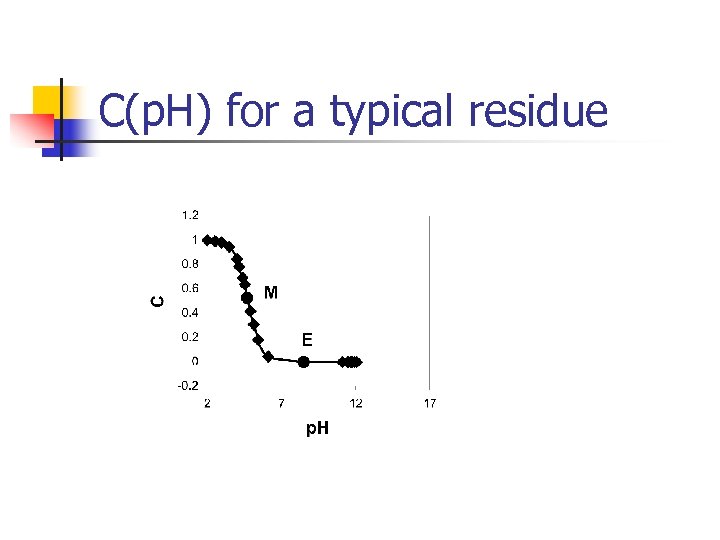 C(p. H) for a typical residue
C(p. H) for a typical residue
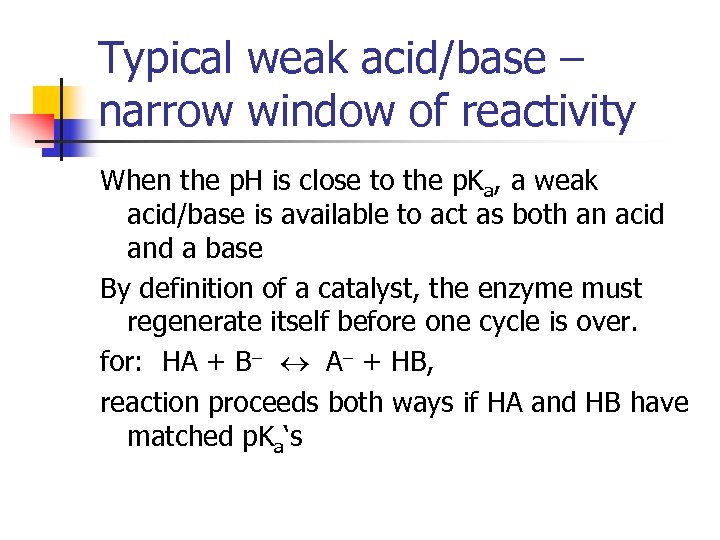 Typical weak acid/base – narrow window of reactivity When the p. H is close to the p. Ka, a weak acid/base is available to act as both an acid and a base By definition of a catalyst, the enzyme must regenerate itself before one cycle is over. for: HA + B A + HB, reaction proceeds both ways if HA and HB have matched p. Ka‘s
Typical weak acid/base – narrow window of reactivity When the p. H is close to the p. Ka, a weak acid/base is available to act as both an acid and a base By definition of a catalyst, the enzyme must regenerate itself before one cycle is over. for: HA + B A + HB, reaction proceeds both ways if HA and HB have matched p. Ka‘s
 A common catalysis - st 1 step in enzyme Deprotonate a C-H bond: C-H + B: C + HB+ What is required of B? n n It must be a strong enough base It must be deprotonated at neutral p. H Mutually contradictory requirements (for a Henderson-Hasselbalch acid/base)
A common catalysis - st 1 step in enzyme Deprotonate a C-H bond: C-H + B: C + HB+ What is required of B? n n It must be a strong enough base It must be deprotonated at neutral p. H Mutually contradictory requirements (for a Henderson-Hasselbalch acid/base)
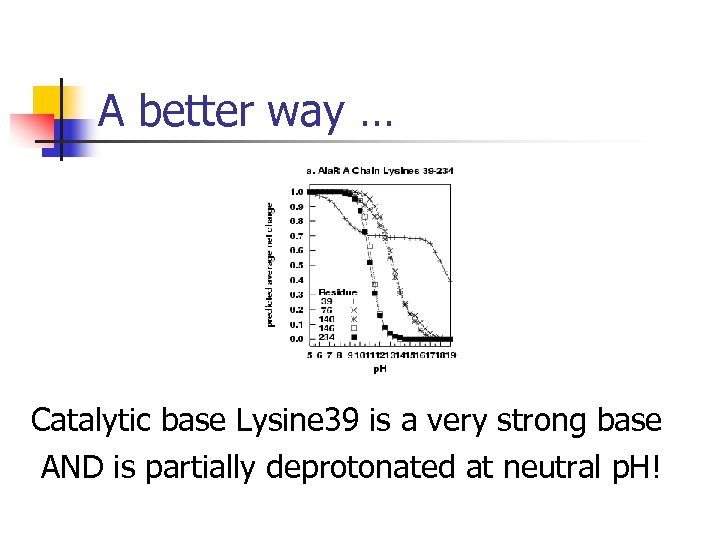 A better way … Catalytic base Lysine 39 is a very strong base AND is partially deprotonated at neutral p. H!
A better way … Catalytic base Lysine 39 is a very strong base AND is partially deprotonated at neutral p. H!
 Perturbed titration curves n n Enable residue to act as acid/base over a wide p. H range Precise p. H adjustment not needed Precise p. Ka match not required Enable residue to have right mix of chemical properties: n n acid (or base) strength right protonation state at neutrality
Perturbed titration curves n n Enable residue to act as acid/base over a wide p. H range Precise p. H adjustment not needed Precise p. Ka match not required Enable residue to have right mix of chemical properties: n n acid (or base) strength right protonation state at neutrality
 Perturbed curves Have been noticed before (in titration curves obtained computationally for proteins) We now understand are significant: n n Are markers of chemical reactivity Can be used to locate active site M. J. Ondrechen, J. G. Clifton and D. Ringe, Proc. Natl. Acad Sci USA 98, 12473 -12478 (2001)
Perturbed curves Have been noticed before (in titration curves obtained computationally for proteins) We now understand are significant: n n Are markers of chemical reactivity Can be used to locate active site M. J. Ondrechen, J. G. Clifton and D. Ringe, Proc. Natl. Acad Sci USA 98, 12473 -12478 (2001)
 THEMATICS Theoretical Microscopic Titration Curves n Conceptually simple n Require a known structure n Can be computed n Highly reliable identifier of active site n Characterize enzyme active site
THEMATICS Theoretical Microscopic Titration Curves n Conceptually simple n Require a known structure n Can be computed n Highly reliable identifier of active site n Characterize enzyme active site
 Complementary to other methods THEMATICS complements well other methods that predict, or provide clues about, function: Evolutionary history; sequence relationships; sequence homology; domain fusion; conservation of gene position; gene coinheritance; geometric motif search; cleft search; small molecule docking; energetics; flexibility Characterize function by chemical reactivity
Complementary to other methods THEMATICS complements well other methods that predict, or provide clues about, function: Evolutionary history; sequence relationships; sequence homology; domain fusion; conservation of gene position; gene coinheritance; geometric motif search; cleft search; small molecule docking; energetics; flexibility Characterize function by chemical reactivity
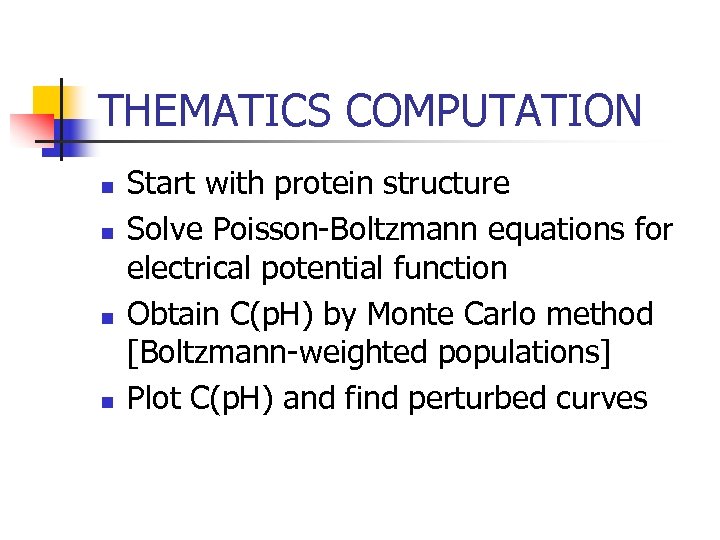 THEMATICS COMPUTATION n n Start with protein structure Solve Poisson-Boltzmann equations for electrical potential function Obtain C(p. H) by Monte Carlo method [Boltzmann-weighted populations] Plot C(p. H) and find perturbed curves
THEMATICS COMPUTATION n n Start with protein structure Solve Poisson-Boltzmann equations for electrical potential function Obtain C(p. H) by Monte Carlo method [Boltzmann-weighted populations] Plot C(p. H) and find perturbed curves
 Which curves are perturbed? n n Visual inspection Mathematical analysis (H. Yang) n n Statistical analysis Fit to parametrized sigmoid function Neural networks / Support Vector Machines (W. Tong) Only small fraction (~3 -7%) of all ionizable residues are perturbed n
Which curves are perturbed? n n Visual inspection Mathematical analysis (H. Yang) n n Statistical analysis Fit to parametrized sigmoid function Neural networks / Support Vector Machines (W. Tong) Only small fraction (~3 -7%) of all ionizable residues are perturbed n
 Ionizable residues Arg Asp Cys Glu His Lys Tyr + termini A cluster of two or more perturbed residues in physical proximity is a reliable predictor of active site location Success in finding active site is not particularly sensitive to selection criteria
Ionizable residues Arg Asp Cys Glu His Lys Tyr + termini A cluster of two or more perturbed residues in physical proximity is a reliable predictor of active site location Success in finding active site is not particularly sensitive to selection criteria
 THEMATICS – A unique predictive tool for Proteomics n n n Gives chemical information Indicates why a particular residue may be involved in catalysis Highly reliable for identifying active sites Conceptually simple Computationally (relatively) fast
THEMATICS – A unique predictive tool for Proteomics n n n Gives chemical information Indicates why a particular residue may be involved in catalysis Highly reliable for identifying active sites Conceptually simple Computationally (relatively) fast
 Alanine Racemase n n n Used by bacteria in cell wall construction A target for antibiotics (and for drugs to treat tuberculosis) Vitamin B 6 – dependent Active as a dimer Active site located at dimer interface
Alanine Racemase n n n Used by bacteria in cell wall construction A target for antibiotics (and for drugs to treat tuberculosis) Vitamin B 6 – dependent Active as a dimer Active site located at dimer interface
 Alanine Racemase catalysis n n Catalyzes interconversion of D-Ala and L -Ala Reaction occurs on a Schiff base intermediate (alanine + pyridoxal phosphate) First step on Schiff base: remove alpha. H atom from Ala moiety K 39 and Y 265’ are the catalytic bases
Alanine Racemase catalysis n n Catalyzes interconversion of D-Ala and L -Ala Reaction occurs on a Schiff base intermediate (alanine + pyridoxal phosphate) First step on Schiff base: remove alpha. H atom from Ala moiety K 39 and Y 265’ are the catalytic bases
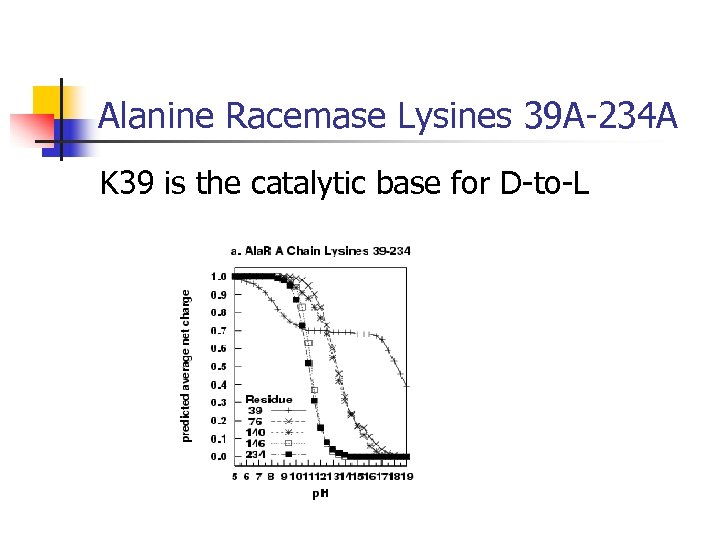 Alanine Racemase Lysines 39 A-234 A K 39 is the catalytic base for D-to-L
Alanine Racemase Lysines 39 A-234 A K 39 is the catalytic base for D-to-L
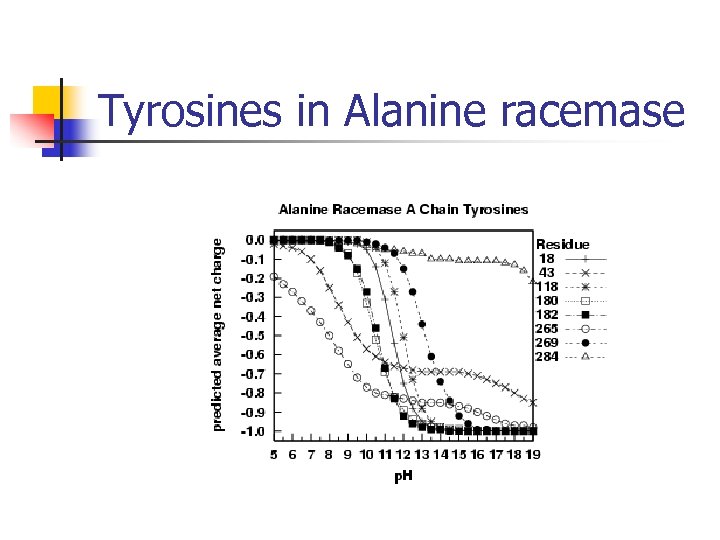 Tyrosines in Alanine racemase
Tyrosines in Alanine racemase
 Results for Alanine Racemase Full results for Alanine racemase: n [R 219, C 311, K 39, Y 43, Y 265, Y 284, Y 354, C 358], [R 366], [D 68] Bold = known active site residue Italics = “second shell” False positives – tend to be isolated
Results for Alanine Racemase Full results for Alanine racemase: n [R 219, C 311, K 39, Y 43, Y 265, Y 284, Y 354, C 358], [R 366], [D 68] Bold = known active site residue Italics = “second shell” False positives – tend to be isolated
 THEMATICS results THEMATICS has succeeded in finding the active site for several dozen proteins with a variety of folds and chemistries. Occasionally, get two or more clusters Occasionally, when visual inspection has not found a positive cluster, statistical analysis has.
THEMATICS results THEMATICS has succeeded in finding the active site for several dozen proteins with a variety of folds and chemistries. Occasionally, get two or more clusters Occasionally, when visual inspection has not found a positive cluster, statistical analysis has.
 Human Adenosine Kinase n n n Catalyzes the transfer of phosphate from ATP to a nucleoside analogue Unique fold – an - three-layer sandwich plus a smaller - two-layer domain Antiviral and anticancer drug target
Human Adenosine Kinase n n n Catalyzes the transfer of phosphate from ATP to a nucleoside analogue Unique fold – an - three-layer sandwich plus a smaller - two-layer domain Antiviral and anticancer drug target
 Human Adenosine Kinase n n n One of two proteins to date where the human observer was unable to locate the active site Statistical analysis successful in finding the active site (H. Yang) Perturbations in predicted titration curves are subtle, but statistically significant
Human Adenosine Kinase n n n One of two proteins to date where the human observer was unable to locate the active site Statistical analysis successful in finding the active site (H. Yang) Perturbations in predicted titration curves are subtle, but statistically significant
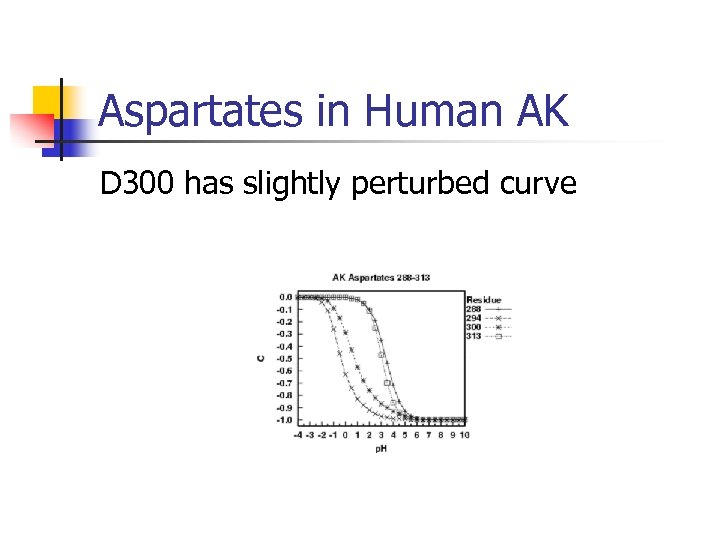 Aspartates in Human AK D 300 has slightly perturbed curve
Aspartates in Human AK D 300 has slightly perturbed curve
 Colicin E 3 – important test case n n Nuclease - cleaves a phosphodiester linkage in the RNA of the ribosome Used by e coli to kill rival bacteria Unique fold – cannot infer active site location from other RNAases Structure provided by Prof. M. Shoham (CWR) prior to publication
Colicin E 3 – important test case n n Nuclease - cleaves a phosphodiester linkage in the RNA of the ribosome Used by e coli to kill rival bacteria Unique fold – cannot infer active site location from other RNAases Structure provided by Prof. M. Shoham (CWR) prior to publication
 THEMATICS results Colicin E 3 n n [E 517, H 526, R 495, R 545, Y 519] Calculation was performed on the structure of the catalytic fragment Active site found prior to completion of the biochemical characterization Active site correctly located by THEMATICS
THEMATICS results Colicin E 3 n n [E 517, H 526, R 495, R 545, Y 519] Calculation was performed on the structure of the catalytic fragment Active site found prior to completion of the biochemical characterization Active site correctly located by THEMATICS
 HIV-1 protease n n n Acid protease Cathepsin D fold Active as a dimer D 25 and D 25’ are the catalytic groups THEMATICS – human observer found D 25 and D 25’ …
HIV-1 protease n n n Acid protease Cathepsin D fold Active as a dimer D 25 and D 25’ are the catalytic groups THEMATICS – human observer found D 25 and D 25’ …
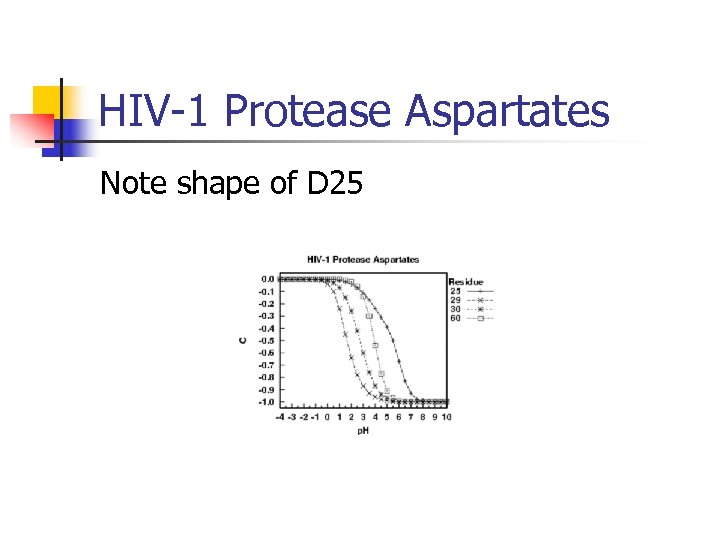 HIV-1 Protease Aspartates Note shape of D 25
HIV-1 Protease Aspartates Note shape of D 25
![THEMATICS on HIV-1 Protease n n n Human observer finds: [D 25, D 25’] THEMATICS on HIV-1 Protease n n n Human observer finds: [D 25, D 25’]](https://present5.com/presentation/79d474ae8e1cf3bbff9a8a02fca96309/image-34.jpg) THEMATICS on HIV-1 Protease n n n Human observer finds: [D 25, D 25’] Statistical analysis finds: [D 25, D 25’, R 87] R 8 and R 87 are believed to be involved in substrate recognition: Bardi, J. S. , I. Luque, E. Freire, Structure-based thermodynamic analysis of HIV-1 protease inhibitors. Biochemistry, 1997. 36: p. 6588 -6596
THEMATICS on HIV-1 Protease n n n Human observer finds: [D 25, D 25’] Statistical analysis finds: [D 25, D 25’, R 87] R 8 and R 87 are believed to be involved in substrate recognition: Bardi, J. S. , I. Luque, E. Freire, Structure-based thermodynamic analysis of HIV-1 protease inhibitors. Biochemistry, 1997. 36: p. 6588 -6596
 Conclusions n n n THEMATICS – simple, computationally fast, and reliable Simple connection with chemistry A cluster of two or more positive residues is predictive of active sites Has been automated Characterizes residues (reactivity) Positive clusters well conserved
Conclusions n n n THEMATICS – simple, computationally fast, and reliable Simple connection with chemistry A cluster of two or more positive residues is predictive of active sites Has been automated Characterizes residues (reactivity) Positive clusters well conserved
 Conclusions - continued n n Perturbed curves result from the polyprotic nature of proteins Working hypotheses about perturbed titration curves n n Afford catalytic advantage Afford advantage in reversible binding at recognition sites
Conclusions - continued n n Perturbed curves result from the polyprotic nature of proteins Working hypotheses about perturbed titration curves n n Afford catalytic advantage Afford advantage in reversible binding at recognition sites
 Thanks n n David Budil, Leo Murga, Terry Yang, Ying Wei, Alissa Bologna, Katie Boino, Wenxu Tong, Bob Futrelle, Ron Williams (Northeastern) Jaeju Ko (IUP) Ihsan Shehadi (UAEU) Dagmar Ringe & Jim Clifton (Brandeis)
Thanks n n David Budil, Leo Murga, Terry Yang, Ying Wei, Alissa Bologna, Katie Boino, Wenxu Tong, Bob Futrelle, Ron Williams (Northeastern) Jaeju Ko (IUP) Ihsan Shehadi (UAEU) Dagmar Ringe & Jim Clifton (Brandeis)
 Support Acknowledged National Science Foundation Institute for Complex Scientific Software (ICSS) - Northeastern
Support Acknowledged National Science Foundation Institute for Complex Scientific Software (ICSS) - Northeastern
 mjo@neu. edu M. J. Ondrechen, J. G. Clifton and D. Ringe, Proc. Natl. Acad Sci USA 98, 1247312478 (2001) I. A. Shehadi, H. Yang and M. J. Ondrechen, Mol. Biol. Rpts 29, 329335 (2002)
mjo@neu. edu M. J. Ondrechen, J. G. Clifton and D. Ringe, Proc. Natl. Acad Sci USA 98, 1247312478 (2001) I. A. Shehadi, H. Yang and M. J. Ondrechen, Mol. Biol. Rpts 29, 329335 (2002)


