5bbc576feadb34ed4fdff59918699073.ppt
- Количество слайдов: 42

Pernille Haste Andersen, Ph. d. student Immunological Bioinformatics CBS, DTU CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU B cell epitopes and predictions
n n Antibodies are produced by B lymphocytes (B cells) Antibodies circulate in the blood They are referred to as “the first line of defense” against infection Antibodies play a central role in immunity by attaching to pathogens and recruiting effector systems that kill the invader CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU B cells and antibodies
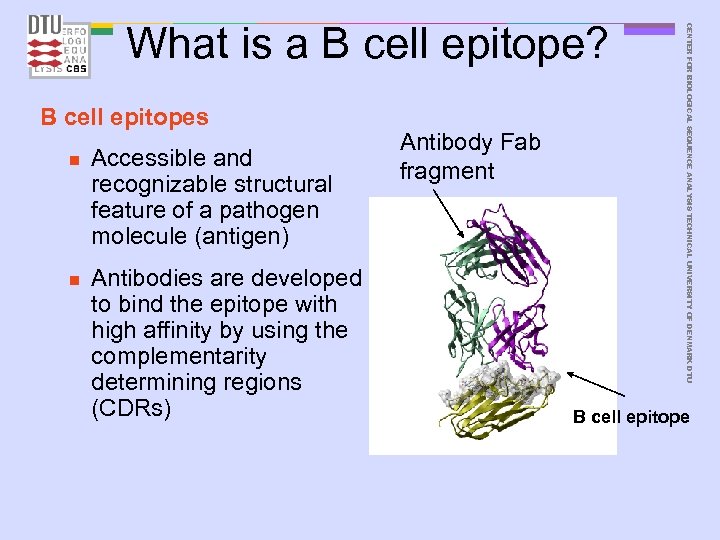
B cell epitopes n n Accessible and recognizable structural feature of a pathogen molecule (antigen) Antibodies are developed to bind the epitope with high affinity by using the complementarity determining regions (CDRs) Antibody Fab fragment CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU What is a B cell epitope? B cell epitope

§ Prediction of B cell epitopes can potentially guide experimental epitope mapping § Predictions of antigenicity in proteins can be used for selecting subunits in rational vaccine design § Predictions of B cell epitopes may also be valuable for interpretation of results from experiments based on antibody affinity binding such as ELISA, RIA CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU Motivations for prediction of B cell epitopes
>PATHOGEN PROTEIN KVFGRCELAAAMKRHGLDNYR GYSLGNWVCAAKFESNF Rational Vaccine Design CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU Computational Rational Vaccine Design
§ Classified into linear (~10%) and discontinuous epitopes (~90%) § Databases: Anti. Jen, IEDB, Bci. Pep, Los Alamos HIV database, Protein Data Bank § Large amount of data available for linear epitopes § Few data available for discontinuous epitopes § In general, B cell epitope prediction methods have relatively low performances CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU B cell epitopes, linear or discontinuous?
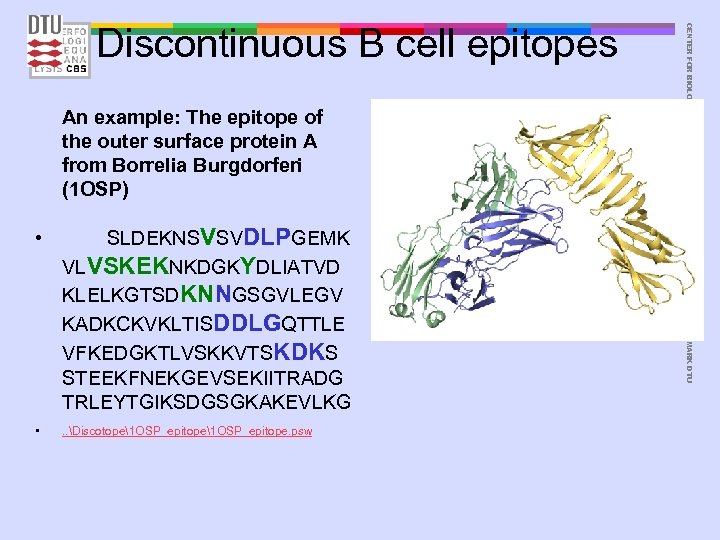
An example: The epitope of the outer surface protein A from Borrelia Burgdorferi (1 OSP) • SLDEKNSVSVDLPGEMK VLVSKEKNKDGKYDLIATVD KLELKGTSDKNNGSGVLEGV KADKCKVKLTISDDLGQTTLE VFKEDGKTLVSKKVTSKDKS STEEKFNEKGEVSEKIITRADG TRLEYTGIKSDGSGKAKEVLKG • . . Discotope1 OSP_epitope. psw CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU Discontinuous B cell epitopes
Binding strength § § Salt bridges Hydrogen bonds Hydrophobic interactions Van der Waals forces CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU The binding interactions
§ Many of the charged groups and hydrogen bonding partners are present on highly flexible amino acid side chains. § Most crystal structures of epitopes and antibodies in free and complexed forms have shown conformational rearrangements upon binding. § “Induced fit” model of interactions. CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU B-cell epitopes are dynamic
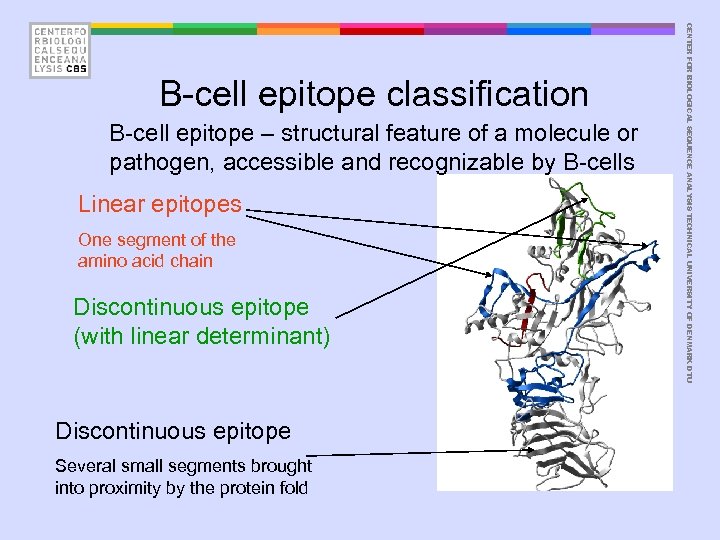
B-cell epitope – structural feature of a molecule or pathogen, accessible and recognizable by B-cells Linear epitopes One segment of the amino acid chain Discontinuous epitope (with linear determinant) Discontinuous epitope Several small segments brought into proximity by the protein fold CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU B-cell epitope classification
• Linear epitopes: – Chop sequence into small pieces and measure binding to antibody • Discontinuous epitopes: – Measure binding of whole protein to antibody • The best annotation method : X-ray crystal structure of the antibody-epitope complex CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU B-cell epitope annotation
• Databases: Anti. Jen, IEDB, Bci. Pep, Los Alamos HIV database, Protein Data Bank • Large amount of data available for linear epitopes • Few data available for discontinuous CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU B-cell epitope data bases

CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU B cell epitope prediction
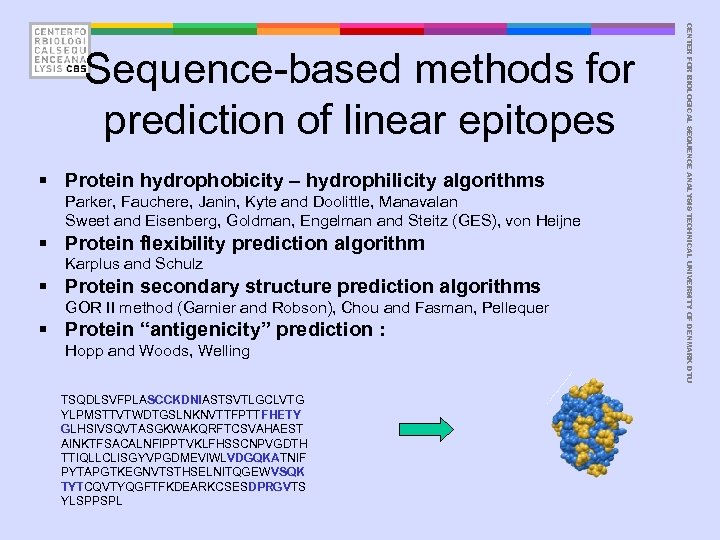
§ Protein hydrophobicity – hydrophilicity algorithms Parker, Fauchere, Janin, Kyte and Doolittle, Manavalan Sweet and Eisenberg, Goldman, Engelman and Steitz (GES), von Heijne § Protein flexibility prediction algorithm Karplus and Schulz § Protein secondary structure prediction algorithms GOR II method (Garnier and Robson), Chou and Fasman, Pellequer § Protein “antigenicity” prediction : Hopp and Woods, Welling TSQDLSVFPLASCCKDNIASTSVTLGCLVTG YLPMSTTVTWDTGSLNKNVTTFPTTFHETY GLHSIVSQVTASGKWAKQRFTCSVAHAEST AINKTFSACALNFIPPTVKLFHSSCNPVGDTH TTIQLLCLISGYVPGDMEVIWLVDGQKATNIF PYTAPGTKEGNVTSTHSELNITQGEWVSQK TYTCQVTYQGFTFKDEARKCSESDPRGVTS YLSPPSPL CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU Sequence-based methods for prediction of linear epitopes
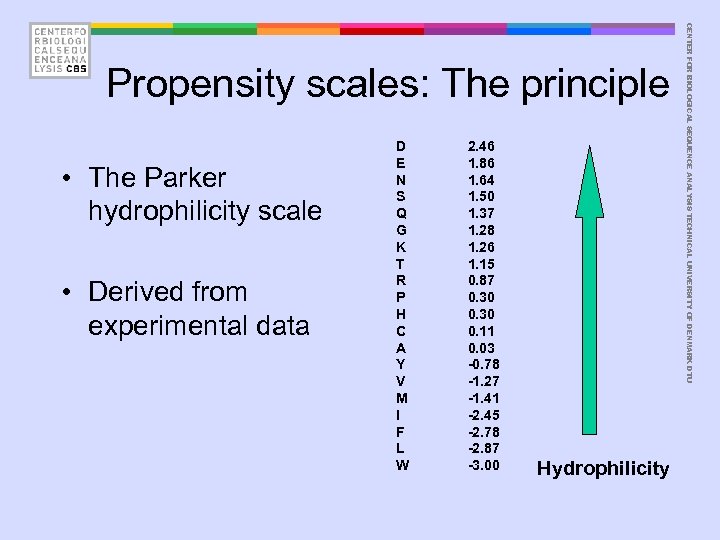
• The Parker hydrophilicity scale • Derived from experimental data D E N S Q G K T R P H C A Y V M I F L W 2. 46 1. 86 1. 64 1. 50 1. 37 1. 28 1. 26 1. 15 0. 87 0. 30 0. 11 0. 03 -0. 78 -1. 27 -1. 41 -2. 45 -2. 78 -2. 87 -3. 00 Hydrophilicity CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU Propensity scales: The principle
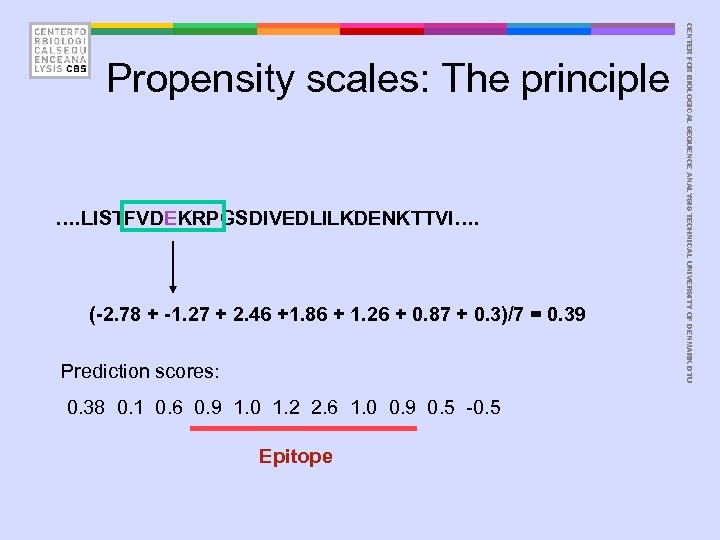
…. LISTFVDEKRPGSDIVEDLILKDENKTTVI…. (-2. 78 + -1. 27 + 2. 46 +1. 86 + 1. 26 + 0. 87 + 0. 3)/7 = 0. 39 Prediction scores: 0. 38 0. 1 0. 6 0. 9 1. 0 1. 2 2. 6 1. 0 0. 9 0. 5 -0. 5 Epitope CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU Propensity scales: The principle
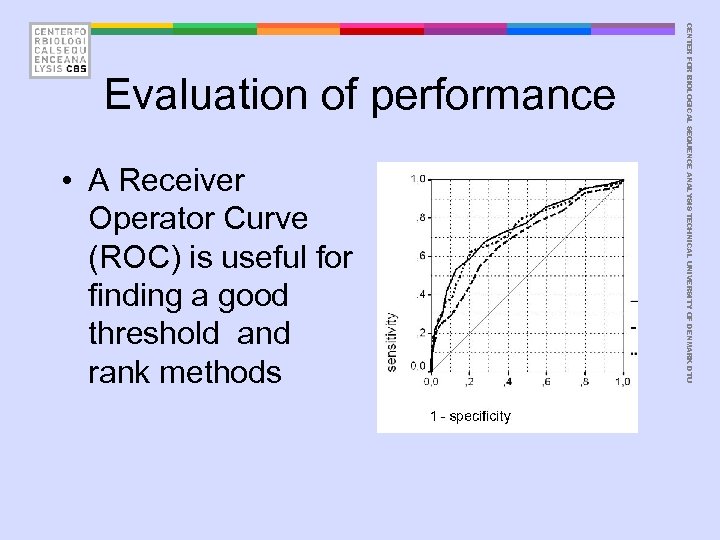
• A Receiver Operator Curve (ROC) is useful for finding a good threshold and rank methods CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU Evaluation of performance
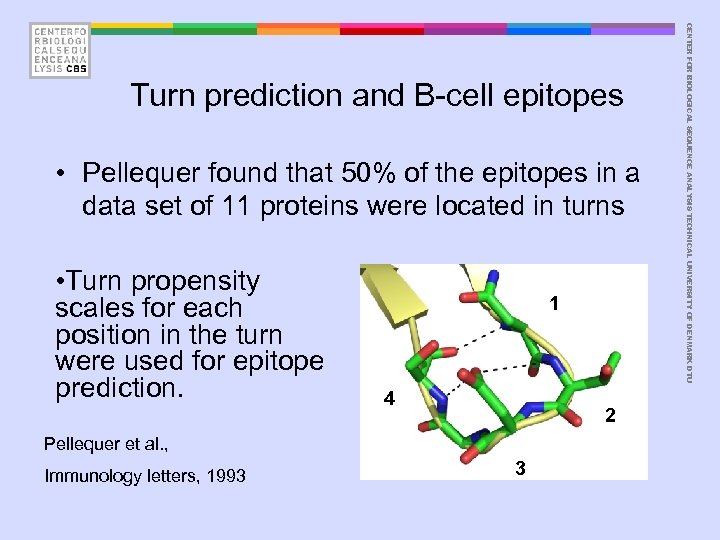
• Pellequer found that 50% of the epitopes in a data set of 11 proteins were located in turns • Turn propensity scales for each position in the turn were used for epitope prediction. 1 4 2 Pellequer et al. , Immunology letters, 1993 3 CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU Turn prediction and B-cell epitopes
• Extensive evaluation of propensity scales for epitope prediction • Conclusion: – Basically all the classical scales perform close to random! – Other methods must be used for epitope prediction CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU Blythe and Flower 2005
• Parker hydrophilicity scale • Hidden Markov model • Markov model based on linear epitopes extracted from the Anti. Jen database • Combination of the Parker prediction scores and Markov model leads to prediction score • Tested on the Pellequer dataset and epitopes in the HIV Los Alamos database CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU Bepi. Pred: CBS in-house tool
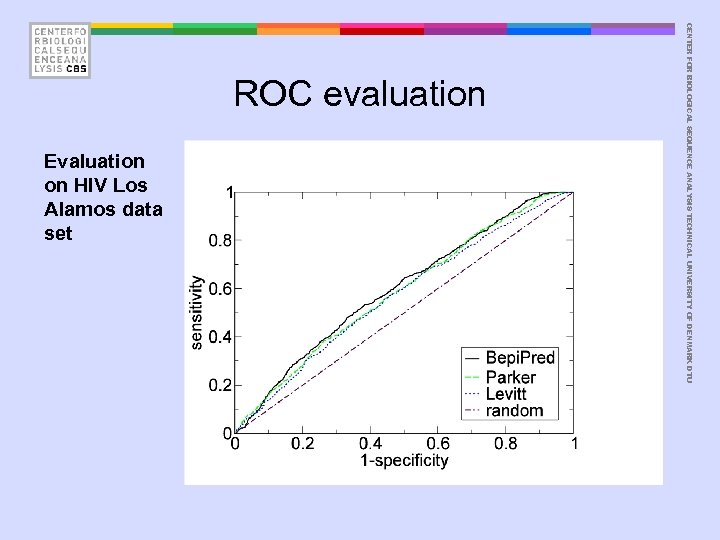
Evaluation on HIV Los Alamos data set CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU ROC evaluation
• Pellequer data set: – Levitt – Parker – Bepi. Pred AROC = 0. 66 AROC = 0. 65 AROC = 0. 68 • HIV Los Alamos data set – Levitt – Parker – Bepi. Pred AROC = 0. 57 AROC = 0. 59 AROC = 0. 60 CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU Bepi. Pred performance

• Bepi. Pred conclusion: – On both of the evaluation data sets, Bepipred was shown to perform better – Still the AROC value is low compared to Tcell epitope prediction tools! – Bepipred is available as a webserver: www. cbs. dtu. dk/services/Bepi. Pred CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU Bepi. Pred
Structural determination • X-ray crystallography • NMR spectroscopy Both methods are time consuming and not easily done in a larger scale Structure prediction • Homology modeling • Fold recognition Less time consuming, but there is a possibility of incorrect predictions, specially in loop regions CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU How can we get information about the three-dimensional structure?
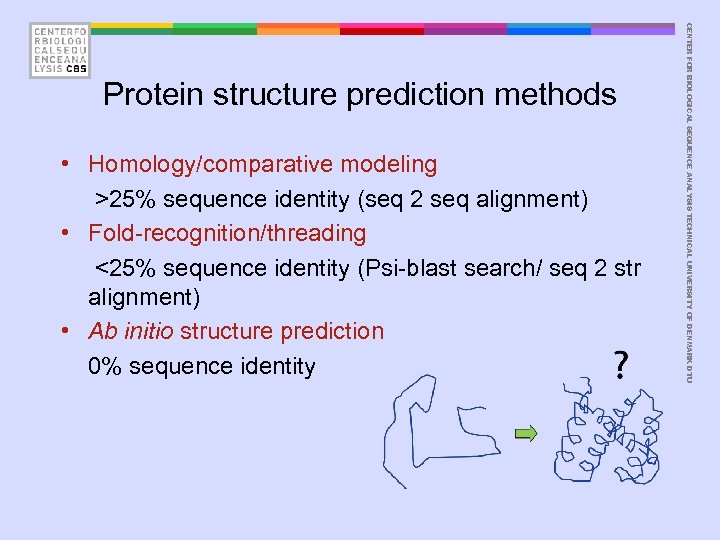
• Homology/comparative modeling >25% sequence identity (seq 2 seq alignment) • Fold-recognition/threading <25% sequence identity (Psi-blast search/ seq 2 str alignment) • Ab initio structure prediction 0% sequence identity CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU Protein structure prediction methods
§ A data set of 75 discontinuous epitopes was compiled from structures of antibodies/protein antigen complexes in the PDB § The data set has been used for developing a method for predictions of discontinuous B cell epitopes § Since about 30 of the PDB entries represented Lysozyme, I have used homology grouping (25 groups of non-homologous antigens) and 5 fold cross -validation for training of the method § Performance was measured using ROC curves on a per antigen basis, and by weighted averaging of AUC values CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU A data set of 3 D discontinuous epitopes
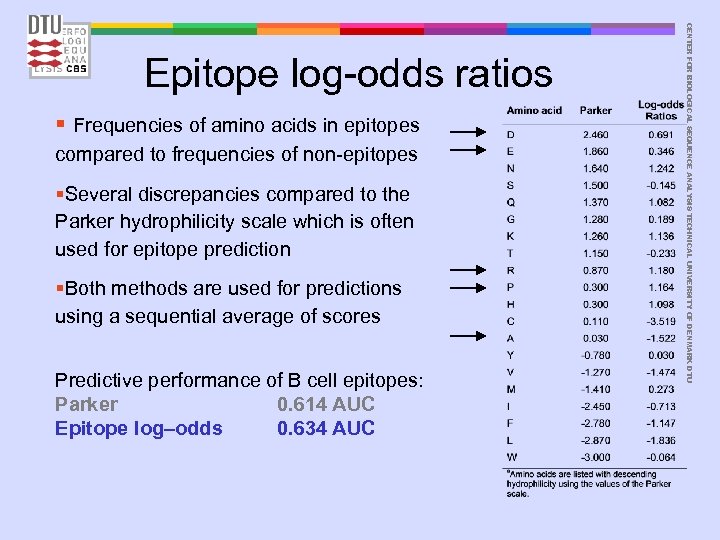
§ Frequencies of amino acids in epitopes compared to frequencies of non-epitopes §Several discrepancies compared to the Parker hydrophilicity scale which is often used for epitope prediction §Both methods are used for predictions using a sequential average of scores Predictive performance of B cell epitopes: Parker 0. 614 AUC Epitope log–odds 0. 634 AUC CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU Epitope log-odds ratios
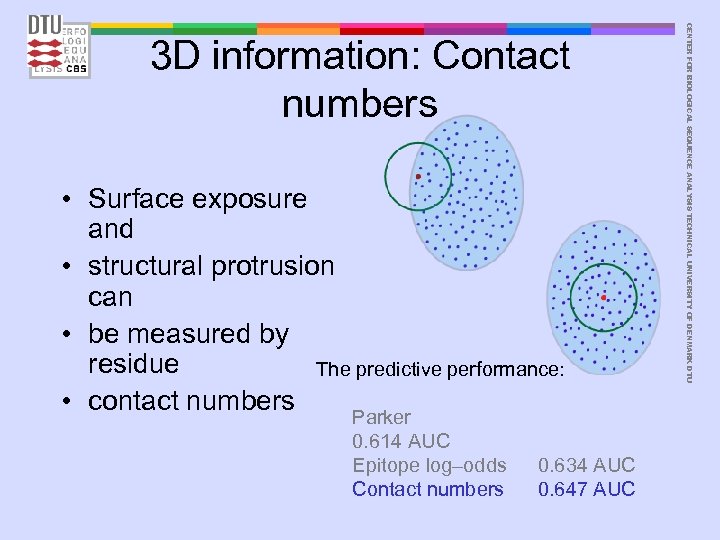
• Surface exposure and • structural protrusion can • be measured by residue The predictive performance: • contact numbers Parker 0. 614 AUC Epitope log–odds Contact numbers 0. 634 AUC 0. 647 AUC CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU 3 D information: Contact numbers
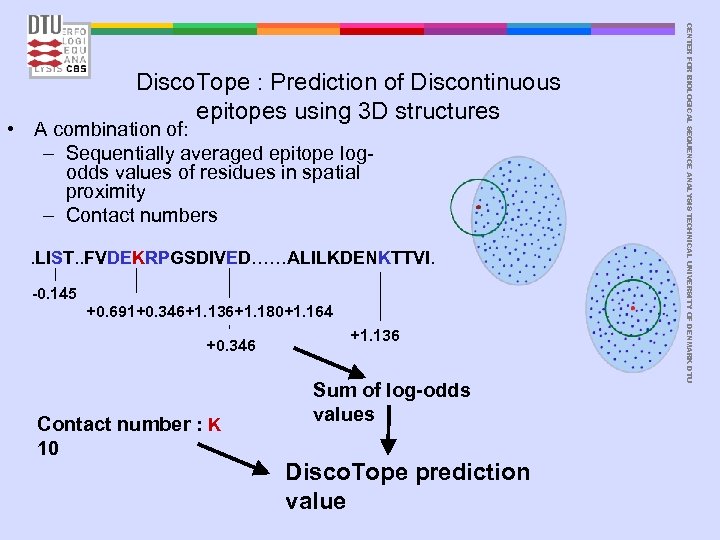
• A combination of: – Sequentially averaged epitope logodds values of residues in spatial proximity – Contact numbers . LIST. . FVDEKRPGSDIVED……ALILKDENKTTVI. -0. 145 +0. 691+0. 346+1. 136+1. 180+1. 164 +0. 346 Contact number : K 10 +1. 136 Sum of log-odds values Disco. Tope prediction value CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU Disco. Tope : Prediction of Discontinuous epitopes using 3 D structures

§ Improved prediction of residues in discontinuous B cell epitopes in the data set § The predictive performance on B cell epitopes: Parker 0. 614 AUC Epitope log–odds 0. 634 AUC Contact numbers 0. 647 AUC Disco. Tope 0. 711 AUC CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU Disco. Tope : Prediction of Discontinuous epi. Topes
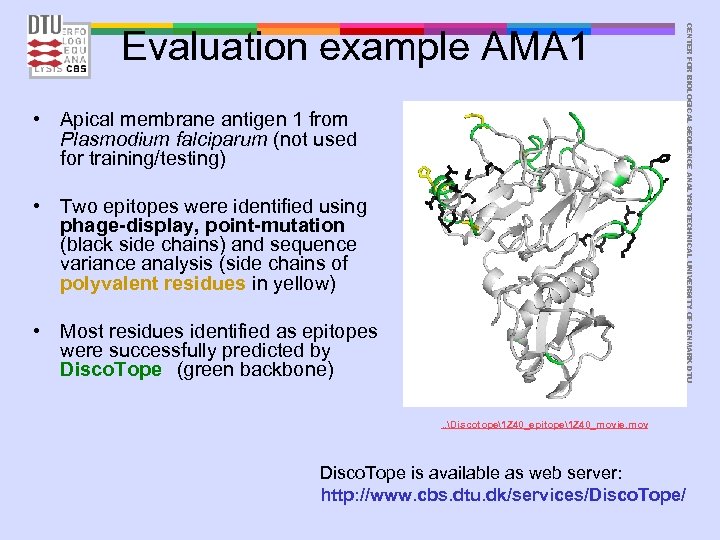
• Apical membrane antigen 1 from Plasmodium falciparum (not used for training/testing) • Two epitopes were identified using phage-display, point-mutation (black side chains) and sequence variance analysis (side chains of polyvalent residues in yellow) • Most residues identified as epitopes were successfully predicted by Disco. Tope (green backbone) CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU Evaluation example AMA 1 . . Discotope1 Z 40_epitope1 Z 40_movie. mov Disco. Tope is available as web server: http: //www. cbs. dtu. dk/services/Disco. Tope/
§ Add epitope predictions for protein-protein complexes § Visualization of epitopes integrated in web server § Testing a score for sequence variability fx based on entropy of positions in the antigens § Combination with glycosylation site predictions § Combination with predictions of trans-membrane regions § Assembling predicted residues into whole epitopes CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU Future improvements

CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU Presentation of the web server
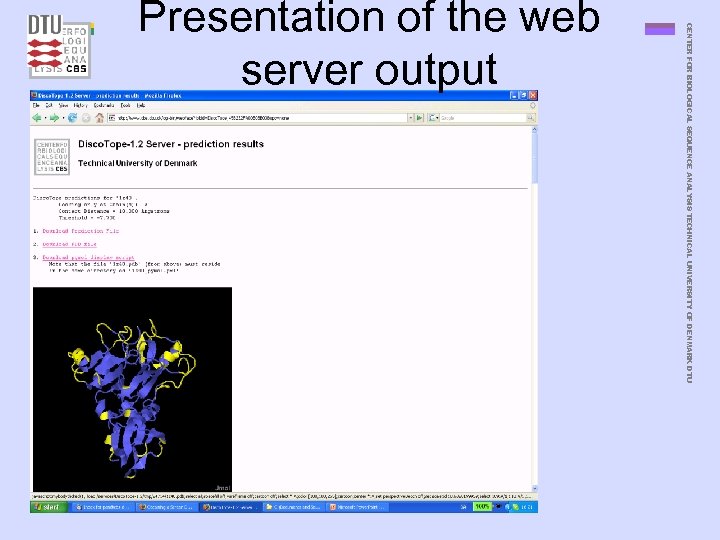
CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU Presentation of the web server output
• Conformational epitope server http: //202. 41. 70. 74: 8080/cgi-bin/cep. pl • Uses protein structure as input • Finds stretches in sequences which are surface exposed CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU Use the CEP server

• CBS server for prediction of discontinuous epitopes • Uses protein structure as input • Combines propensity scale values of amino acids in discontinuous epitopes with surface exposure • www. cbs. dtu. dk/services/Disco. Tope CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU Use the Disco. Tope server
>PATHOGEN PROTEIN KVFGRCELAAAMKRHGLDNYR GYSLGNWVCAAKFESNF Rational Vaccine Design CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU Rational vaccine design
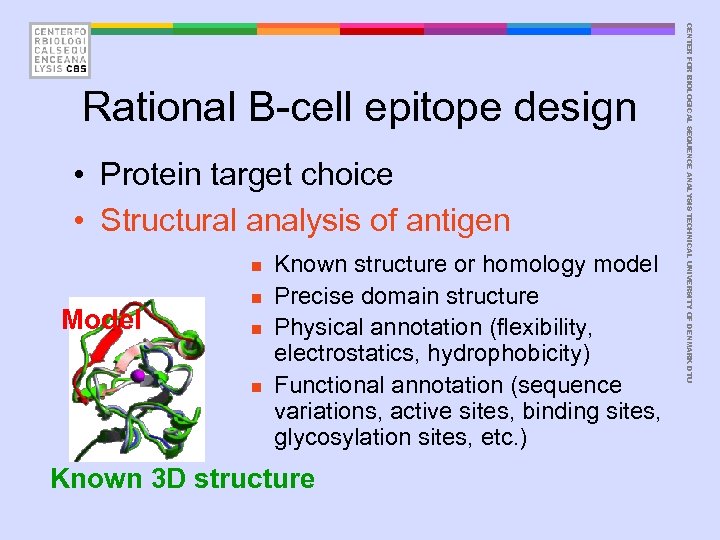
• Protein target choice • Structural analysis of antigen n Model n n n Known structure or homology model Precise domain structure Physical annotation (flexibility, electrostatics, hydrophobicity) Functional annotation (sequence variations, active sites, binding sites, glycosylation sites, etc. ) Known 3 D structure CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU Rational B-cell epitope design
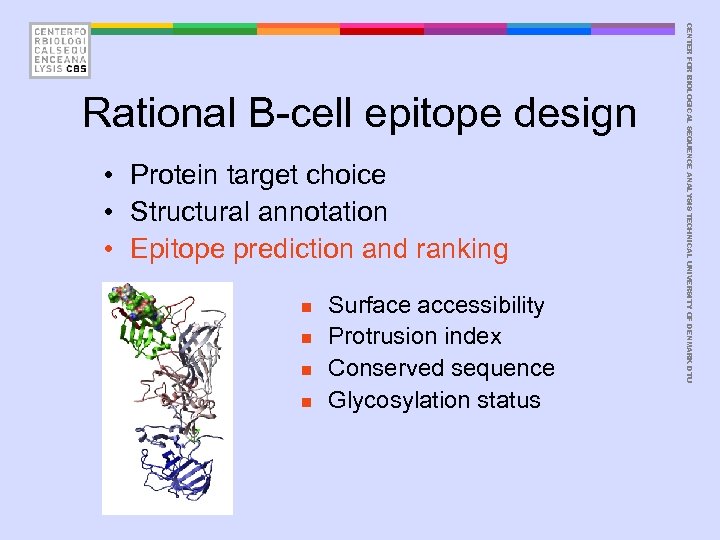
• Protein target choice • Structural annotation • Epitope prediction and ranking n n Surface accessibility Protrusion index Conserved sequence Glycosylation status CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU Rational B-cell epitope design
• • Protein target choice Structural annotation Epitope prediction and ranking Optimal Epitope presentation n n Fold minimization, or Design of structural mimics Choice of carrier (conjugates, DNA plasmids, virus like particles) Multiple chain protein engineering CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU Rational B-cell epitope design
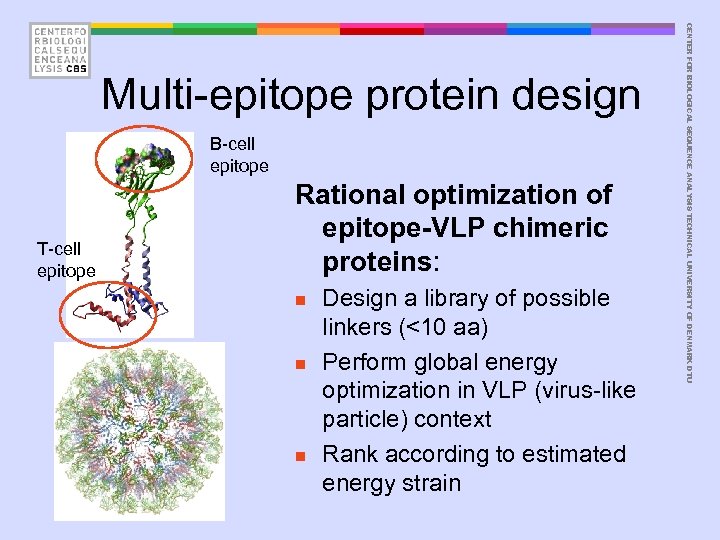
B-cell epitope T-cell epitope Rational optimization of epitope-VLP chimeric proteins: n n n Design a library of possible linkers (<10 aa) Perform global energy optimization in VLP (virus-like particle) context Rank according to estimated energy strain CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU Multi-epitope protein design

• Selection of protective B-cell epitopes involves structural, functional and immunogenic analysis of the pathogenic proteins • When you can: Use protein structure for prediction • Structural modeling tools are helpful in prediction of epitopes, design of epitope mimics and optimal epitope presentation CENTER FOR BIOLOGICAL SEQUENCE ANALYSIS TECHNICAL UNIVERSITY OF DENMARK DTU Conclusions
5bbc576feadb34ed4fdff59918699073.ppt