371ac7b44fdb9a4f66daf970eb44bf29.ppt
- Количество слайдов: 21
 Performance of CMAQ on a Mac OS X System Tracey Holloway, John Bachan, Scott Spak Center for Sustainability and the Global Environment University of Wisconsin-Madison A presentation to the 3 rd annual CMAS Models-3 conference October 19, 2004
Performance of CMAQ on a Mac OS X System Tracey Holloway, John Bachan, Scott Spak Center for Sustainability and the Global Environment University of Wisconsin-Madison A presentation to the 3 rd annual CMAS Models-3 conference October 19, 2004
 Thinking different. Motivation Methods Performance Hardware Release Ongoing Improvements
Thinking different. Motivation Methods Performance Hardware Release Ongoing Improvements
 Motivations. Simplified operation Easier development Easy clustering Improved performance
Motivations. Simplified operation Easier development Easy clustering Improved performance
 Motivation: Operation. Single platform for all research and academic computing User-friendly interface UNIX OS Open source software, hardware support Today’s cluster node = tomorrow’s desktop
Motivation: Operation. Single platform for all research and academic computing User-friendly interface UNIX OS Open source software, hardware support Today’s cluster node = tomorrow’s desktop
 Motivation: Development. Better Developer Tools Xcode (Interface Builder) CHUD performance & debugging suite Distribution Tools standardized profiles Package. Maker FAT binaries automated installation
Motivation: Development. Better Developer Tools Xcode (Interface Builder) CHUD performance & debugging suite Distribution Tools standardized profiles Package. Maker FAT binaries automated installation
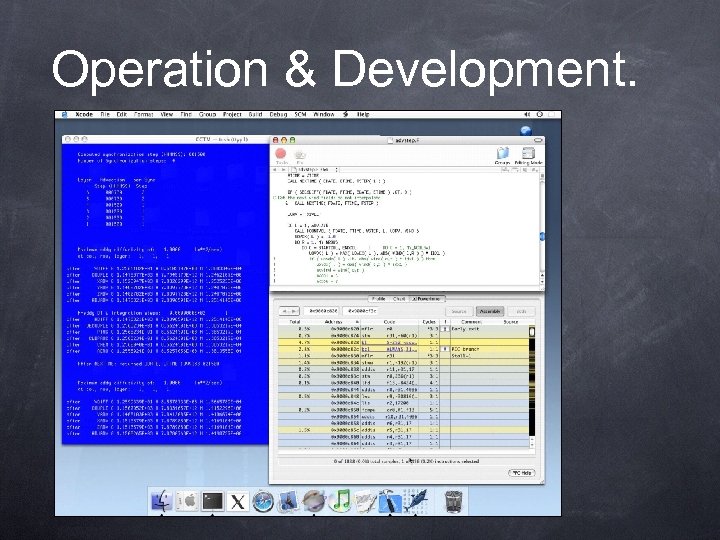 Operation & Development.
Operation & Development.
 Motivation: Performance. Unique Hardware Advantages powerful PPC 970 vector chip auto-vectorizing compilers 2000 NASA Langley report Populist Parallelization mix dedicated cluster nodes with free cycles on personal & lab machines off-the-shelf solutions simple GUI and command-line tools
Motivation: Performance. Unique Hardware Advantages powerful PPC 970 vector chip auto-vectorizing compilers 2000 NASA Langley report Populist Parallelization mix dedicated cluster nodes with free cycles on personal & lab machines off-the-shelf solutions simple GUI and command-line tools
 Methods. IBM XL Fortan v 8. 1 compiler auto-vectorization equivalent to AIX Modifications flag conversion build settings array passing > 400 man-hours
Methods. IBM XL Fortan v 8. 1 compiler auto-vectorization equivalent to AIX Modifications flag conversion build settings array passing > 400 man-hours
 Performance. 2 Test Machines dual 2 GHz G 5, 5 GB RAM, 1 GHz bus stock dual 1 GHz G 4, 1. 5 GB RAM, 133 MHz bus Mac OS X 10. 3. 5 1 Test Run First day of CMAQ 4. 3 tutorial 1 day, 32 km x 32 km, 38 x 38, 6 layers default EBI CB 4 chemistry
Performance. 2 Test Machines dual 2 GHz G 5, 5 GB RAM, 1 GHz bus stock dual 1 GHz G 4, 1. 5 GB RAM, 133 MHz bus Mac OS X 10. 3. 5 1 Test Run First day of CMAQ 4. 3 tutorial 1 day, 32 km x 32 km, 38 x 38, 6 layers default EBI CB 4 chemistry
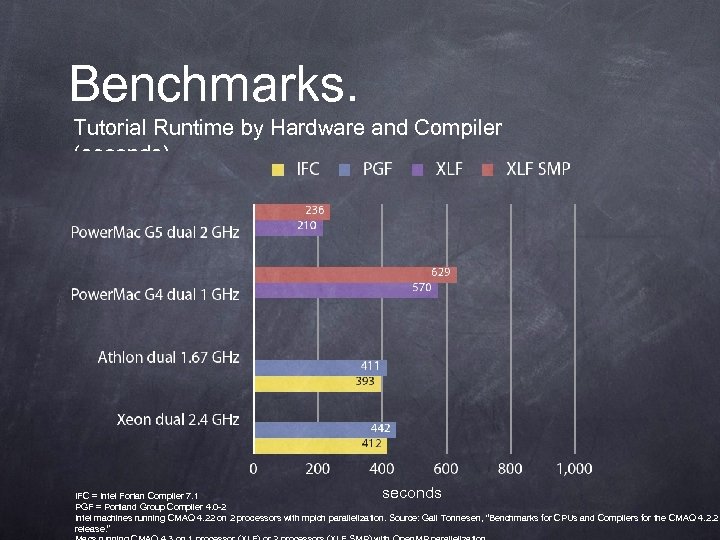 Benchmarks. Tutorial Runtime by Hardware and Compiler (seconds) seconds IFC = Intel Fortan Compiler 7. 1 PGF = Portland Group Compiler 4. 0 -2 Intel machines running CMAQ 4. 22 on 2 processors with mpich parallelization. Source: Gail Tonnesen, “Benchmarks for CPUs and Compilers for the CMAQ 4. 2. 2 release. ”
Benchmarks. Tutorial Runtime by Hardware and Compiler (seconds) seconds IFC = Intel Fortan Compiler 7. 1 PGF = Portland Group Compiler 4. 0 -2 Intel machines running CMAQ 4. 22 on 2 processors with mpich parallelization. Source: Gail Tonnesen, “Benchmarks for CPUs and Compilers for the CMAQ 4. 2. 2 release. ”
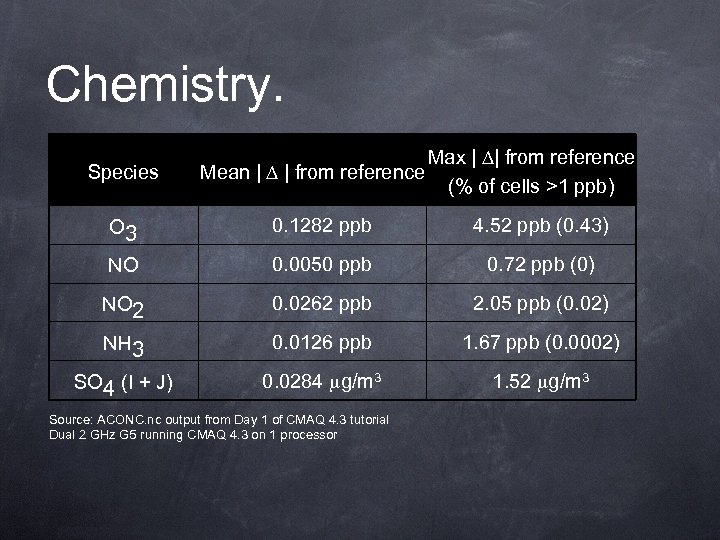 Chemistry. Species Mean | | from reference Max | | from reference (% of cells >1 ppb) O 3 0. 1282 ppb 4. 52 ppb (0. 43) NO 0. 0050 ppb 0. 72 ppb (0) NO 2 0. 0262 ppb 2. 05 ppb (0. 02) NH 3 0. 0126 ppb 1. 67 ppb (0. 0002) SO 4 (I + J) 0. 0284 g/m 3 1. 52 g/m 3 Source: ACONC. nc output from Day 1 of CMAQ 4. 3 tutorial Dual 2 GHz G 5 running CMAQ 4. 3 on 1 processor
Chemistry. Species Mean | | from reference Max | | from reference (% of cells >1 ppb) O 3 0. 1282 ppb 4. 52 ppb (0. 43) NO 0. 0050 ppb 0. 72 ppb (0) NO 2 0. 0262 ppb 2. 05 ppb (0. 02) NH 3 0. 0126 ppb 1. 67 ppb (0. 0002) SO 4 (I + J) 0. 0284 g/m 3 1. 52 g/m 3 Source: ACONC. nc output from Day 1 of CMAQ 4. 3 tutorial Dual 2 GHz G 5 running CMAQ 4. 3 on 1 processor
 Good Chemistry. Small difference from reference set greater than difference among Intel machines and compilers Noise, floating point calculations, initialization greatest at surface level, early in run ambient concentrations only random distribution no bias does not propagate in time or space not correlated to high or low concentrations Consistent G 4/G 5 chemistry modules compiler flags
Good Chemistry. Small difference from reference set greater than difference among Intel machines and compilers Noise, floating point calculations, initialization greatest at surface level, early in run ambient concentrations only random distribution no bias does not propagate in time or space not correlated to high or low concentrations Consistent G 4/G 5 chemistry modules compiler flags
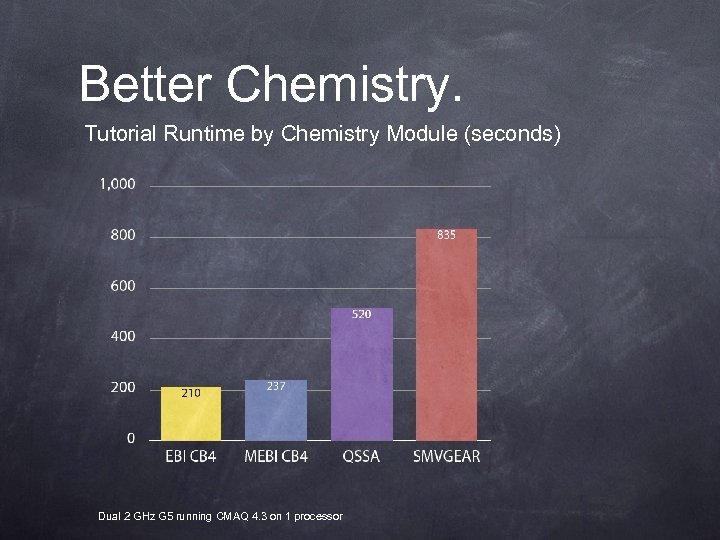 Better Chemistry. Tutorial Runtime by Chemistry Module (seconds) Dual 2 GHz G 5 running CMAQ 4. 3 on 1 processor
Better Chemistry. Tutorial Runtime by Chemistry Module (seconds) Dual 2 GHz G 5 running CMAQ 4. 3 on 1 processor
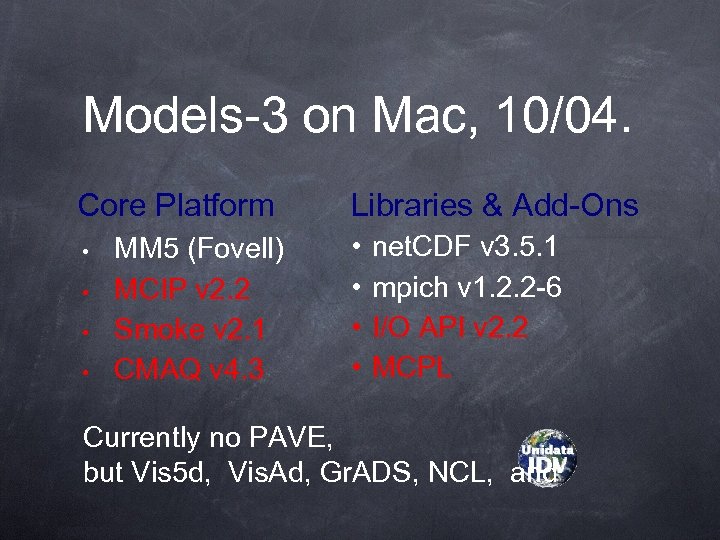 Models-3 on Mac, 10/04. Core Platform • • MM 5 (Fovell) MCIP v 2. 2 Smoke v 2. 1 CMAQ v 4. 3 Libraries & Add-Ons • • net. CDF v 3. 5. 1 mpich v 1. 2. 2 -6 I/O API v 2. 2 MCPL Currently no PAVE, but Vis 5 d, Vis. Ad, Gr. ADS, NCL, and
Models-3 on Mac, 10/04. Core Platform • • MM 5 (Fovell) MCIP v 2. 2 Smoke v 2. 1 CMAQ v 4. 3 Libraries & Add-Ons • • net. CDF v 3. 5. 1 mpich v 1. 2. 2 -6 I/O API v 2. 2 MCPL Currently no PAVE, but Vis 5 d, Vis. Ad, Gr. ADS, NCL, and
 Hardware.
Hardware.
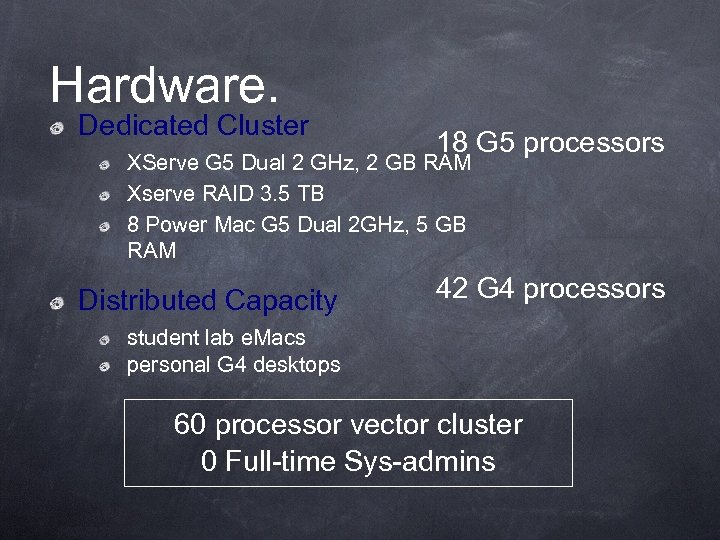 Hardware. Dedicated Cluster 18 G 5 processors XServe G 5 Dual 2 GHz, 2 GB RAM Xserve RAID 3. 5 TB 8 Power Mac G 5 Dual 2 GHz, 5 GB RAM Distributed Capacity 42 G 4 processors student lab e. Macs personal G 4 desktops 60 processor vector cluster 0 Full-time Sys-admins
Hardware. Dedicated Cluster 18 G 5 processors XServe G 5 Dual 2 GHz, 2 GB RAM Xserve RAID 3. 5 TB 8 Power Mac G 5 Dual 2 GHz, 5 GB RAM Distributed Capacity 42 G 4 processors student lab e. Macs personal G 4 desktops 60 processor vector cluster 0 Full-time Sys-admins
 Cost Competitive. Apple Xserve Dual G 5 2 GHz < $3500 RAID storage at $3 per GB G 5 Desktop $2000 - 4000 Compare to Dell Power. Vault RAID at $5 per GB Dell Precision dual Xeon 2. 8 GHz, $1200 4200 Sysadmin costs
Cost Competitive. Apple Xserve Dual G 5 2 GHz < $3500 RAID storage at $3 per GB G 5 Desktop $2000 - 4000 Compare to Dell Power. Vault RAID at $5 per GB Dell Precision dual Xeon 2. 8 GHz, $1200 4200 Sysadmin costs
 JOHN SCOTT
JOHN SCOTT
 Release. Following input from the CMAS Center alpha code to CMAS by November, 2004 CMAS testing potential support Following CMAS Testing, preliminary code, scripts, binaries, instructions available for download at www. sage. wisc. edu/cmaq Scott Spak will answer questions for early users: snspak@wisc. edu
Release. Following input from the CMAS Center alpha code to CMAS by November, 2004 CMAS testing potential support Following CMAS Testing, preliminary code, scripts, binaries, instructions available for download at www. sage. wisc. edu/cmaq Scott Spak will answer questions for early users: snspak@wisc. edu
 Ongoing improvements. Our planned activities g 95 - GNU compilation parallel implementations Condor Xgrid Pooch/Appleseed further optimization Dual 2. 5 GHz benchmarks CMAQ MADRID A community effort? CMAQ Unified MIMS PAVE
Ongoing improvements. Our planned activities g 95 - GNU compilation parallel implementations Condor Xgrid Pooch/Appleseed further optimization Dual 2. 5 GHz benchmarks CMAQ MADRID A community effort? CMAQ Unified MIMS PAVE
 Acknowledgements. Mary Sternitzky, UW Seth Price, UW Hans Vahlenkamp and NOAA GFDL Zac Adelman and the CMAS Help Desk Dr. Gail Tonnesen and Glen Kaukola, UCR Models-3 Listserv All funding provided by the University of Wisconsin. Madison.
Acknowledgements. Mary Sternitzky, UW Seth Price, UW Hans Vahlenkamp and NOAA GFDL Zac Adelman and the CMAS Help Desk Dr. Gail Tonnesen and Glen Kaukola, UCR Models-3 Listserv All funding provided by the University of Wisconsin. Madison.


