c5aab5db837825271f69b39da72e00ef.ppt
- Количество слайдов: 47

Pathway structure inference and expansion from downstream perturbation effects Charles Vaske University of California Santa Cruz January 21, 2009 Stuart Lab Meeting

Glioblastoma p 53 Charles Vaske – UC Santa Cruz The Cancer Genome Atlas, Nature, 2008
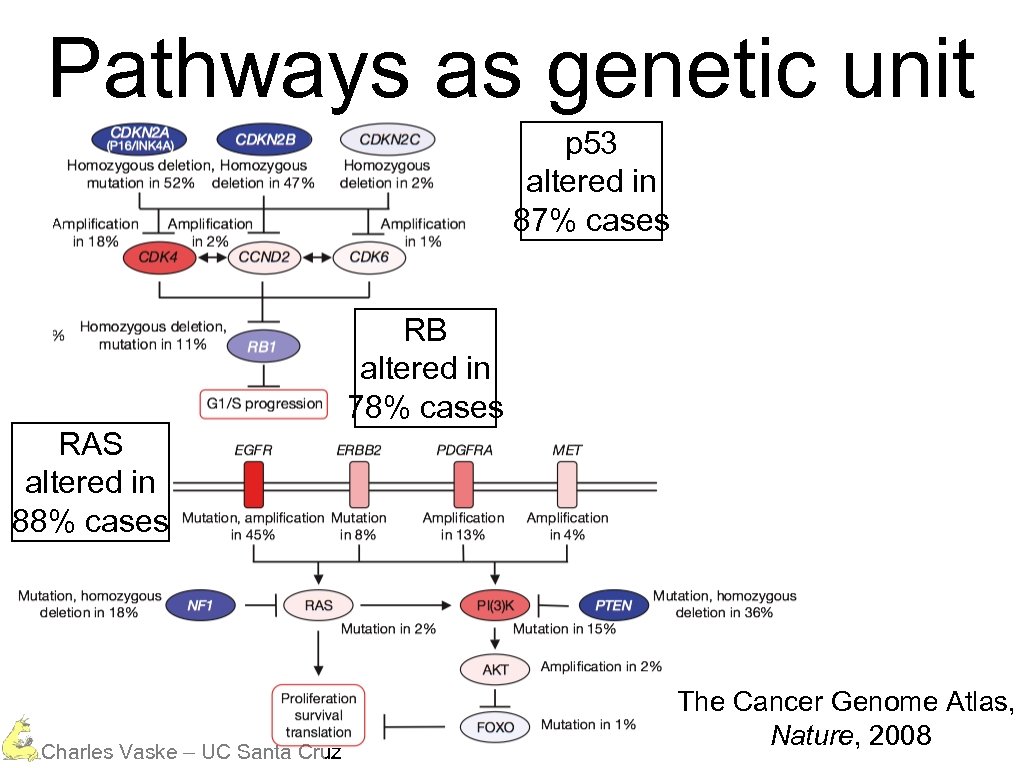
Pathways as genetic unit p 53 altered in 87% cases RB altered in 78% cases RAS altered in 88% cases Charles Vaske – UC Santa Cruz The Cancer Genome Atlas, Nature, 2008

Broad Goal: Learn Novel Pathways Many (most? ) pathways not yet understood 1. Find interactions (structure) between known genes using fast, available experimental techniques 2. Predict new members of the pathway 3. Validate, then iterate Charles Vaske – UC Santa Cruz
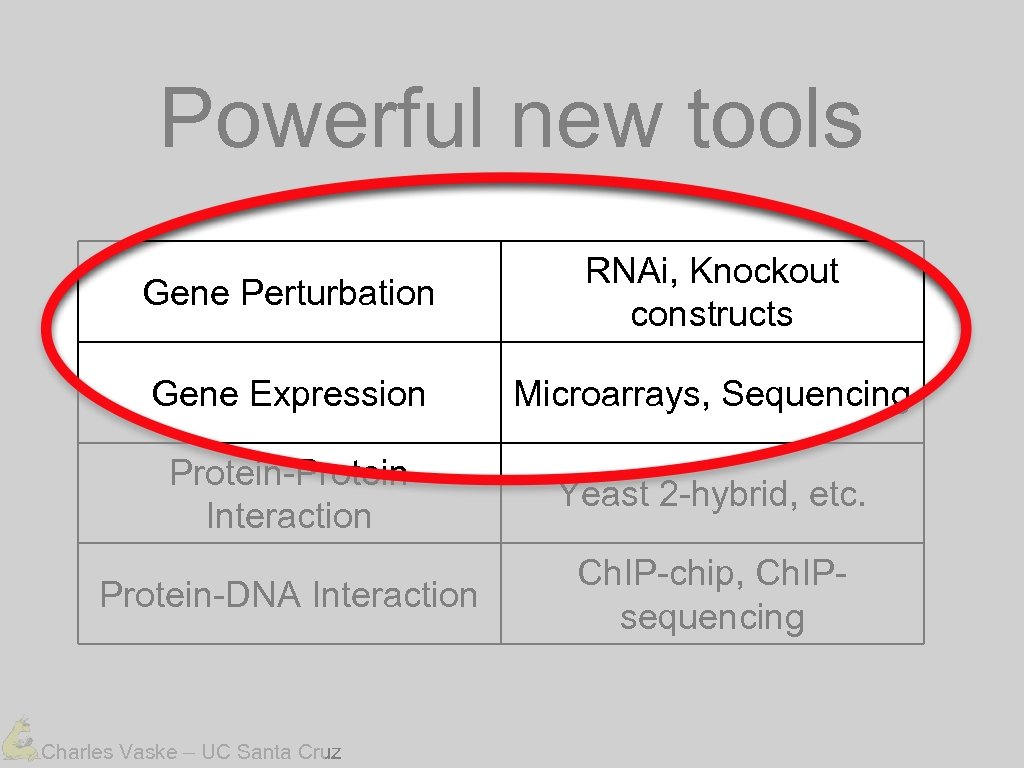
Powerful new tools Gene Perturbation RNAi, Knockout constructs Gene Expression Microarrays, Sequencing Protein-Protein Interaction Yeast 2 -hybrid, etc. Protein-DNA Interaction Ch. IP-chip, Ch. IPsequencing Charles Vaske – UC Santa Cruz
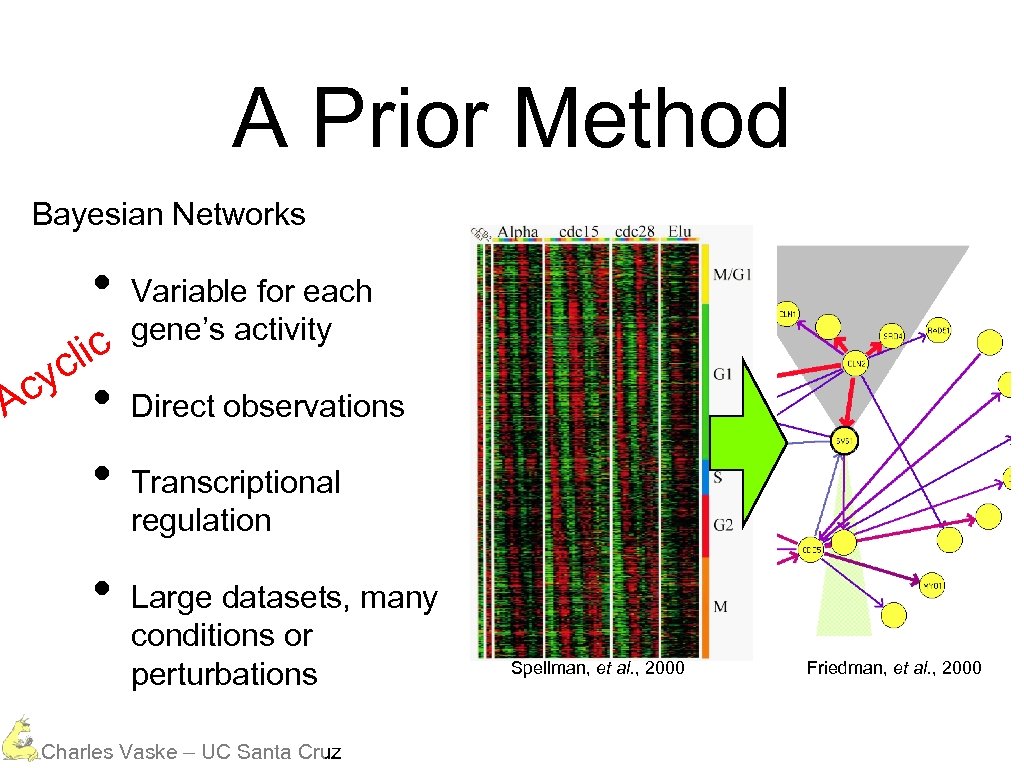
A Prior Method Bayesian Networks • lic c cy A • • • Variable for each gene’s activity Direct observations Transcriptional regulation Large datasets, many conditions or perturbations Charles Vaske – UC Santa Cruz Spellman, et al. , 2000 Friedman, et al. , 2000
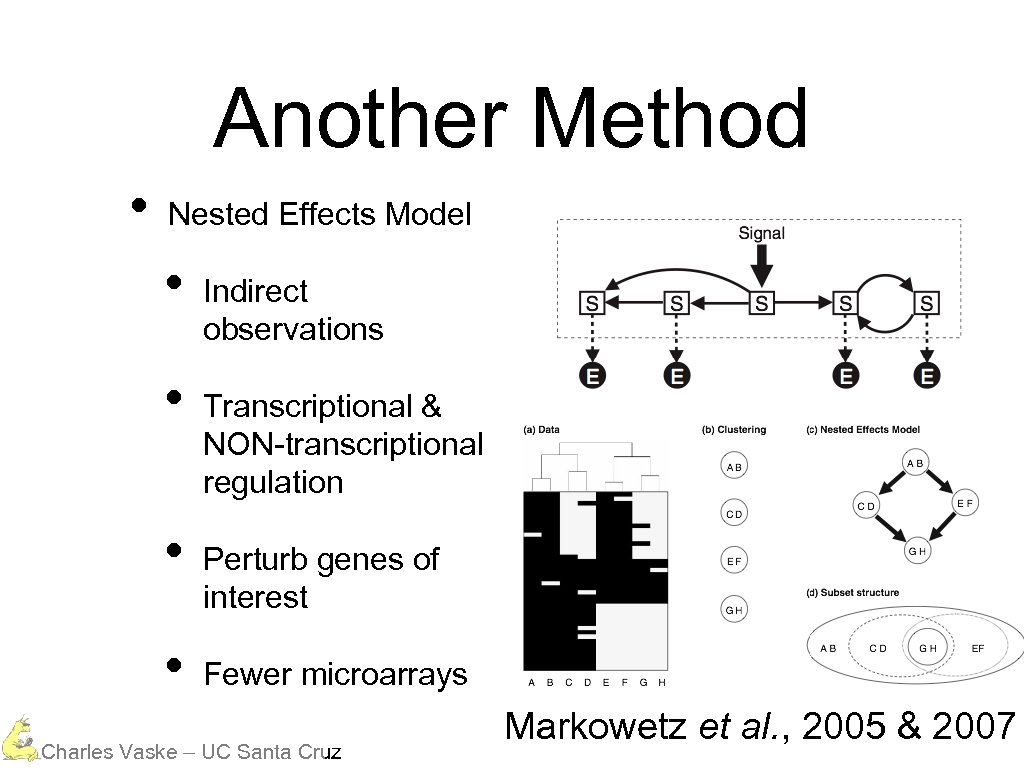
Another Method • Nested Effects Model • • Indirect observations Transcriptional & NON-transcriptional regulation Perturb genes of interest Fewer microarrays Charles Vaske – UC Santa Cruz Markowetz et al. , 2005 & 2007
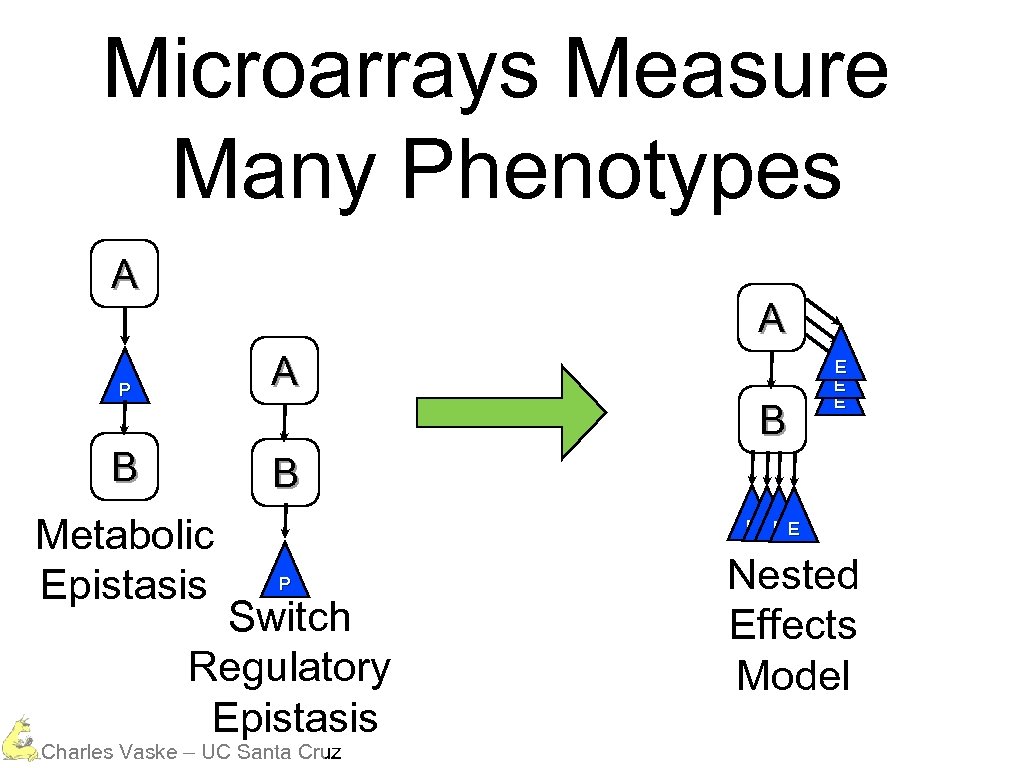
Microarrays Measure Many Phenotypes A A P A B B B Metabolic Epistasis E E EE E P Switch Regulatory Epistasis Charles Vaske – UC Santa Cruz Nested Effects Model
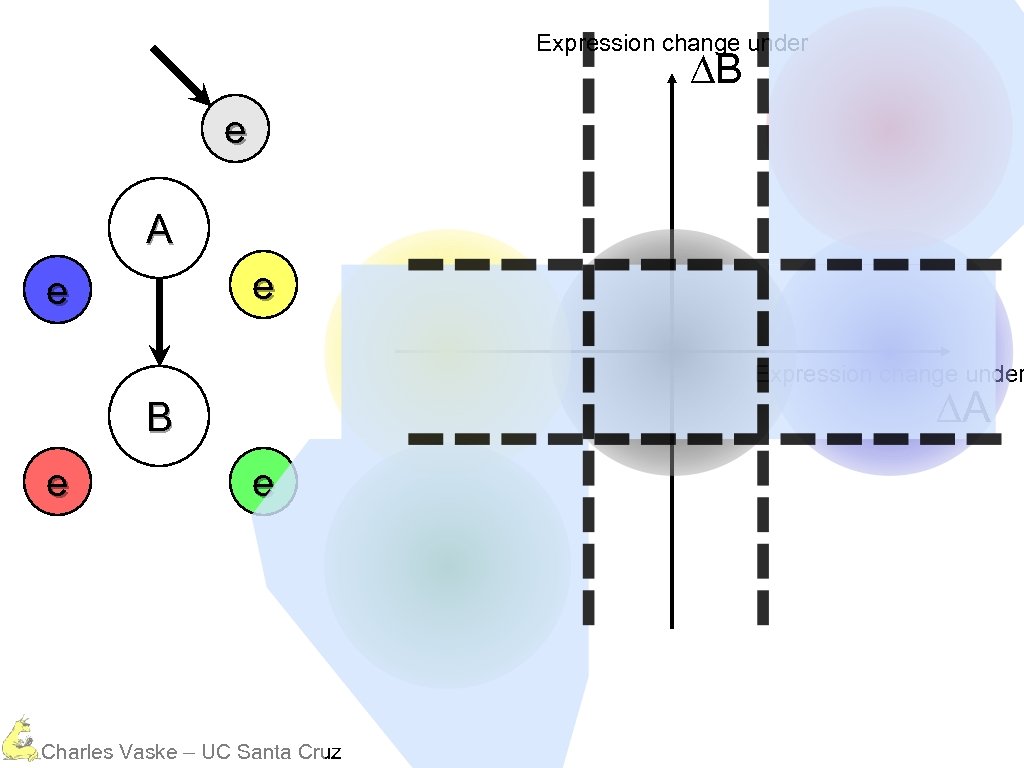
Expression change under ∆B e A e e Expression change under ∆A B e e Charles Vaske – UC Santa Cruz
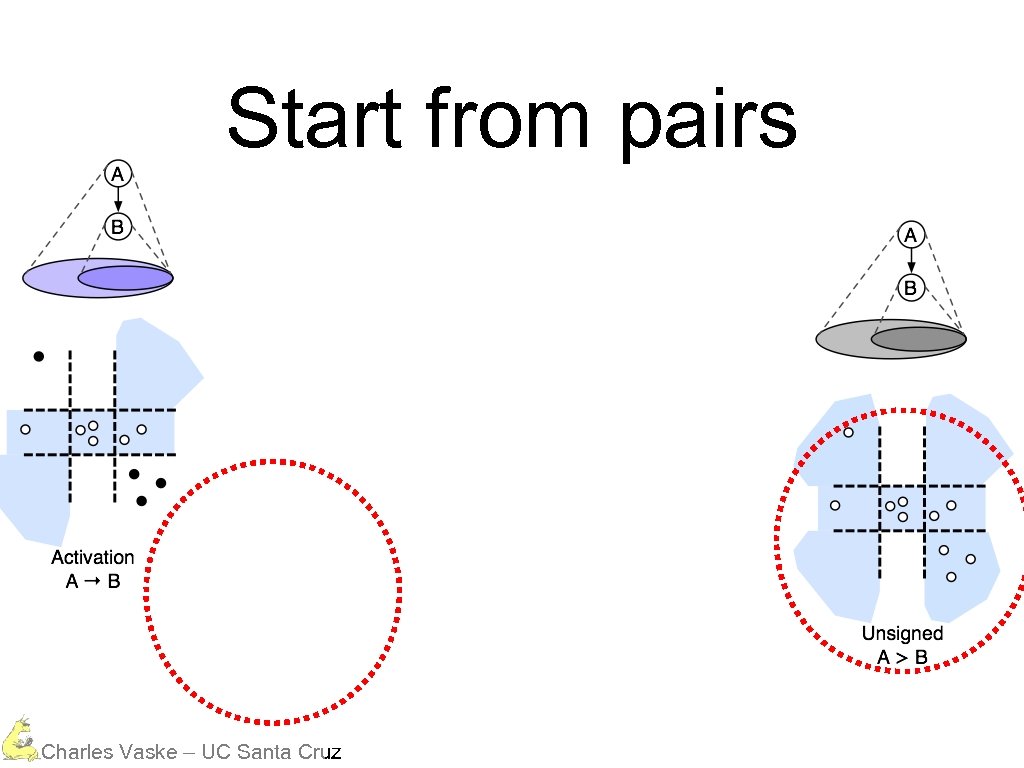
Start from pairs Charles Vaske – UC Santa Cruz
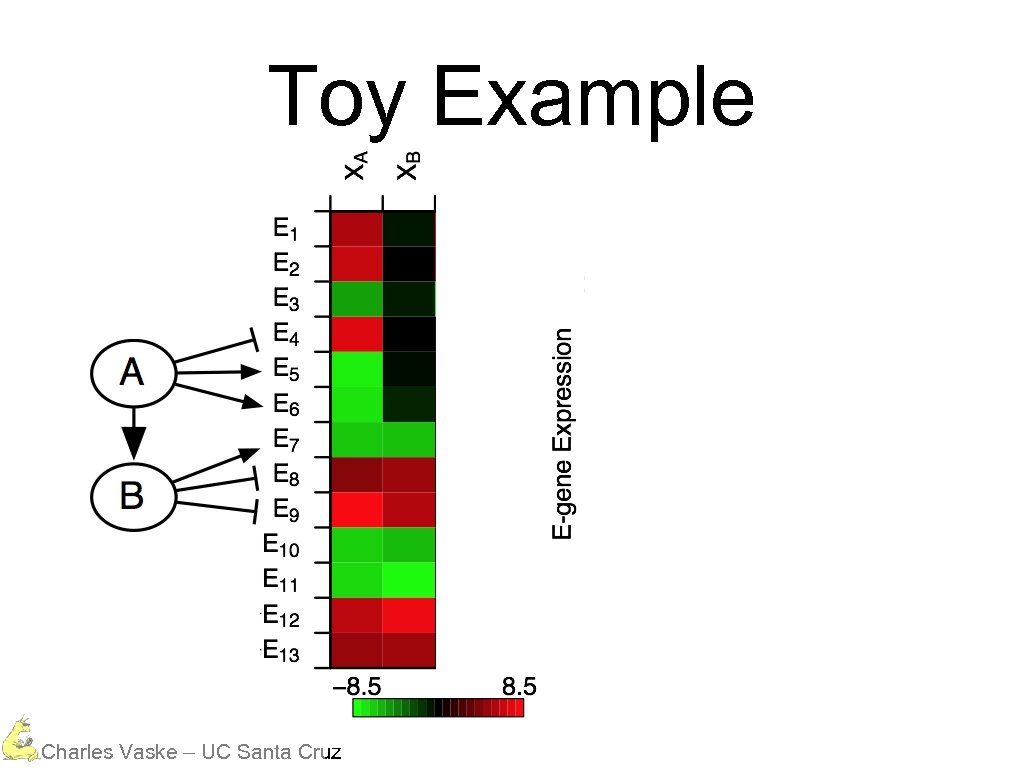
Toy Example Charles Vaske – UC Santa Cruz
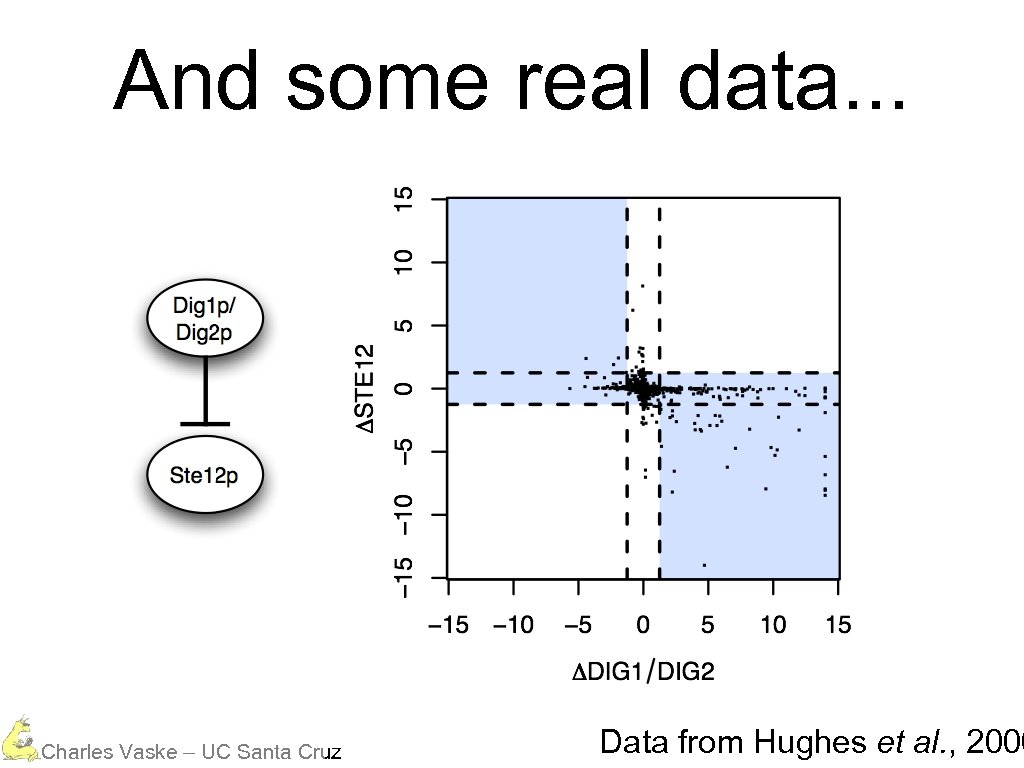
And some real data. . . Charles Vaske – UC Santa Cruz Data from Hughes et al. , 2000
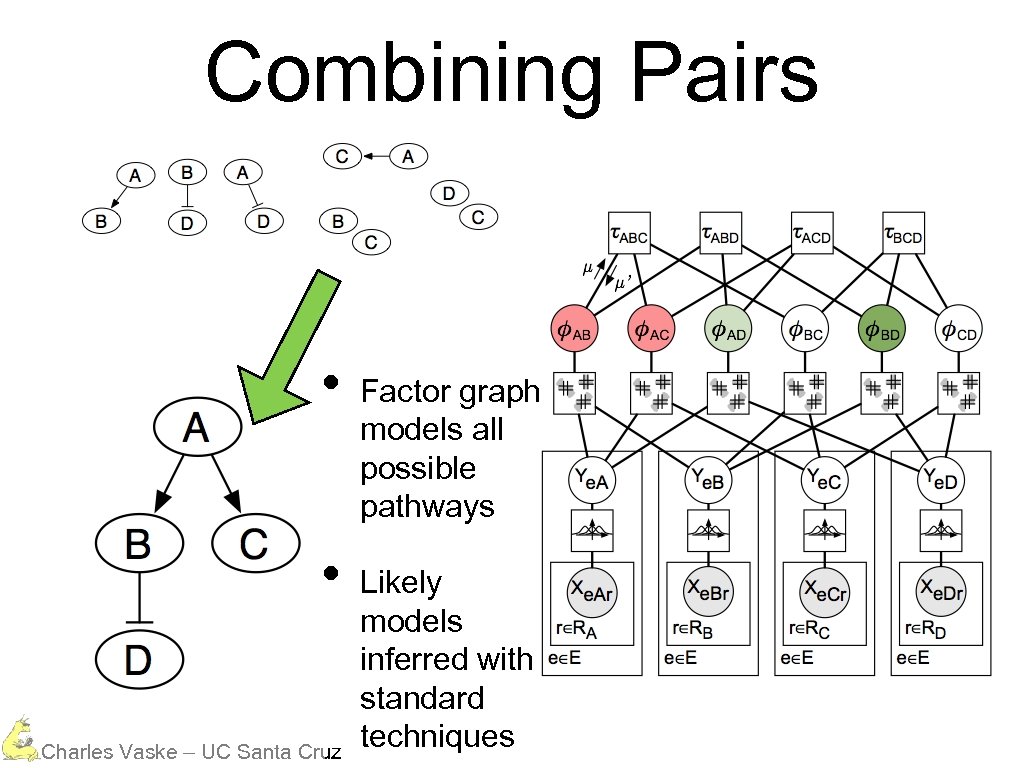
Combining Pairs • • Charles Vaske – UC Santa Cruz Factor graph models all possible pathways Likely models inferred with standard techniques
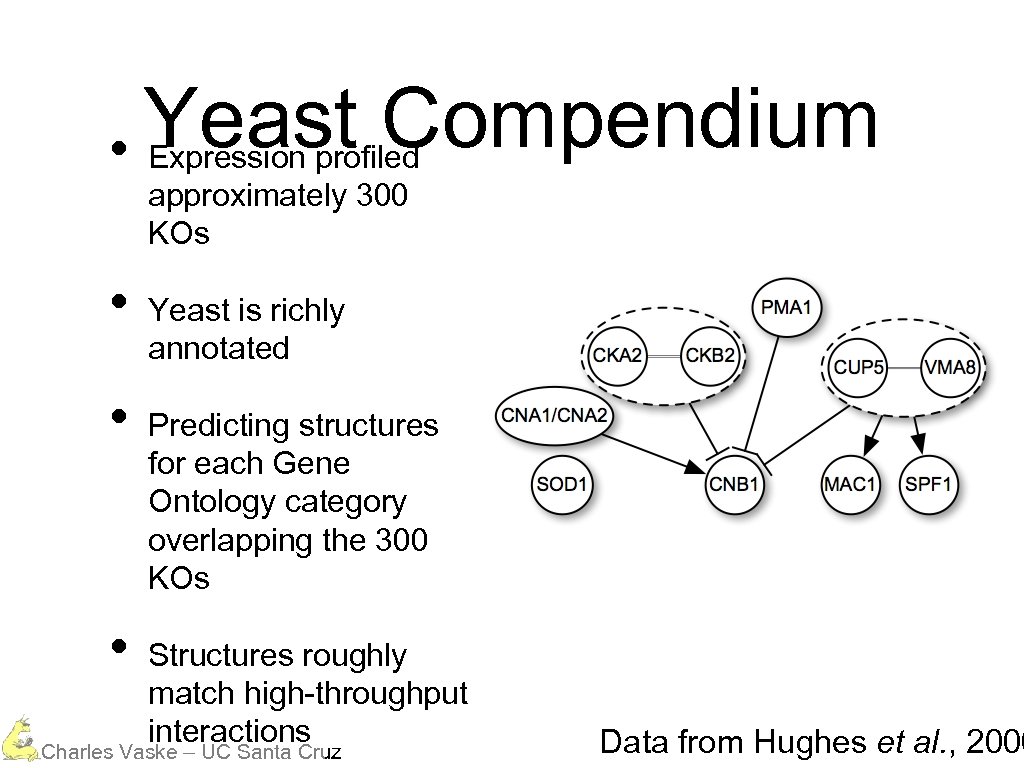
Yeast Compendium • Expression profiled approximately 300 KOs • • • Yeast is richly annotated Predicting structures for each Gene Ontology category overlapping the 300 KOs Structures roughly match high-throughput interactions Charles Vaske – UC Santa Cruz Data from Hughes et al. , 2000
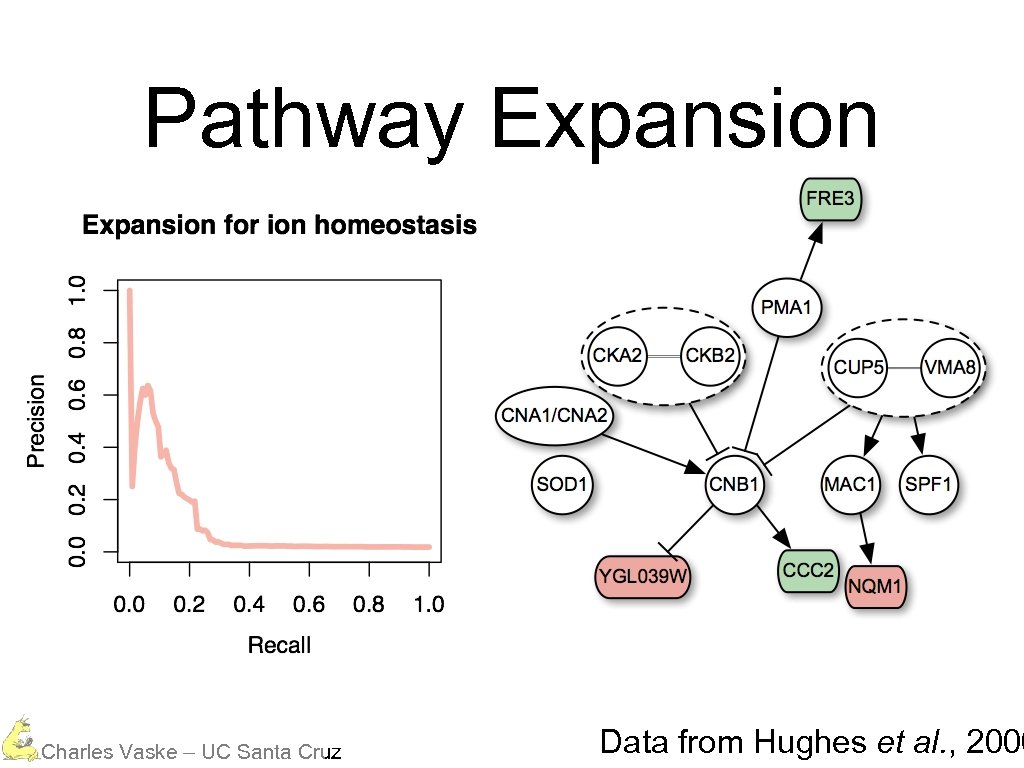
Pathway Expansion Charles Vaske – UC Santa Cruz Data from Hughes et al. , 2000

Gene Ontology Category Improved Pathway Expansion Charles Vaske – UC Santa Cruz With Priors No Priors Unsigned Correlation Method Data from Hughes et al. , 2000
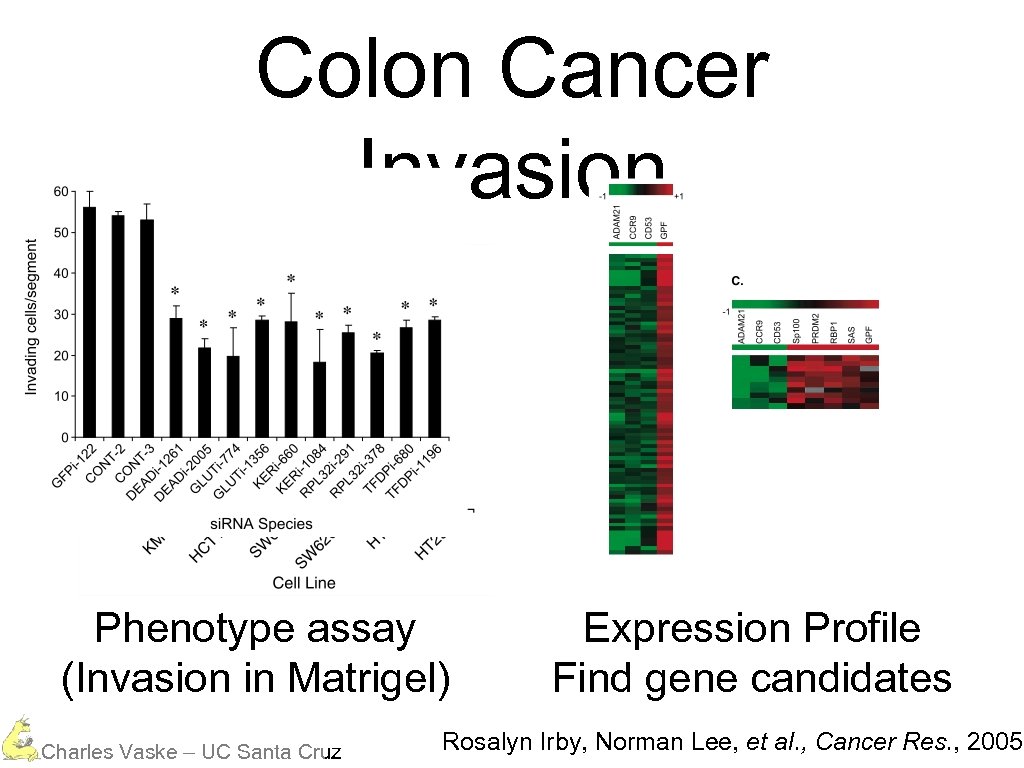
Colon Cancer Invasion Phenotype assay (Invasion in Matrigel) Charles Vaske – UC Santa Cruz Expression Profile Find gene candidates Rosalyn Irby, Norman Lee, et al. , Cancer Res. , 2005
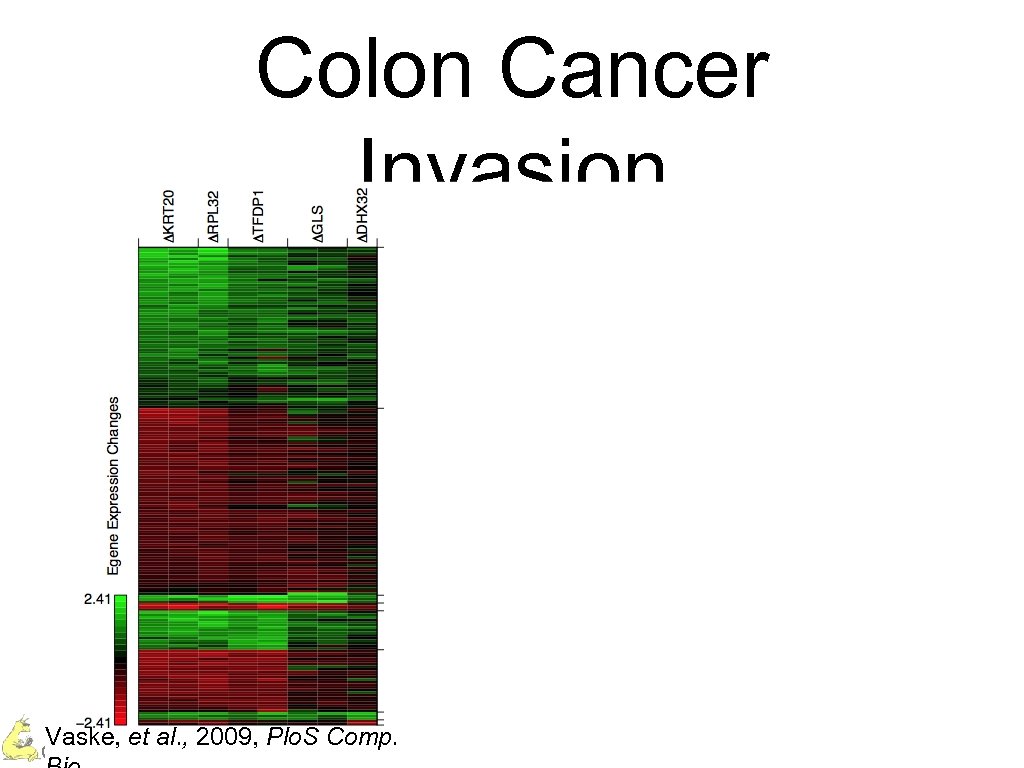
Colon Cancer Invasion Vaske, et al. , 2009, Plo. S Comp. Charles Vaske – UC Santa Cruz
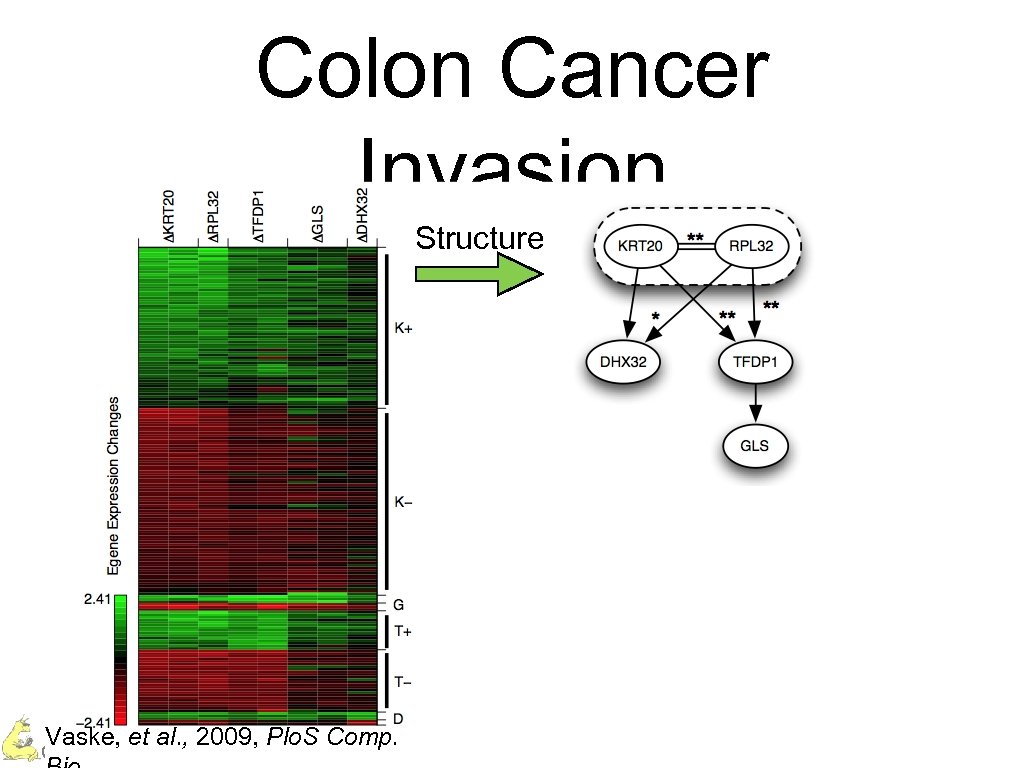
Colon Cancer Invasion Structure Vaske, et al. , 2009, Plo. S Comp. Charles Vaske – UC Santa Cruz
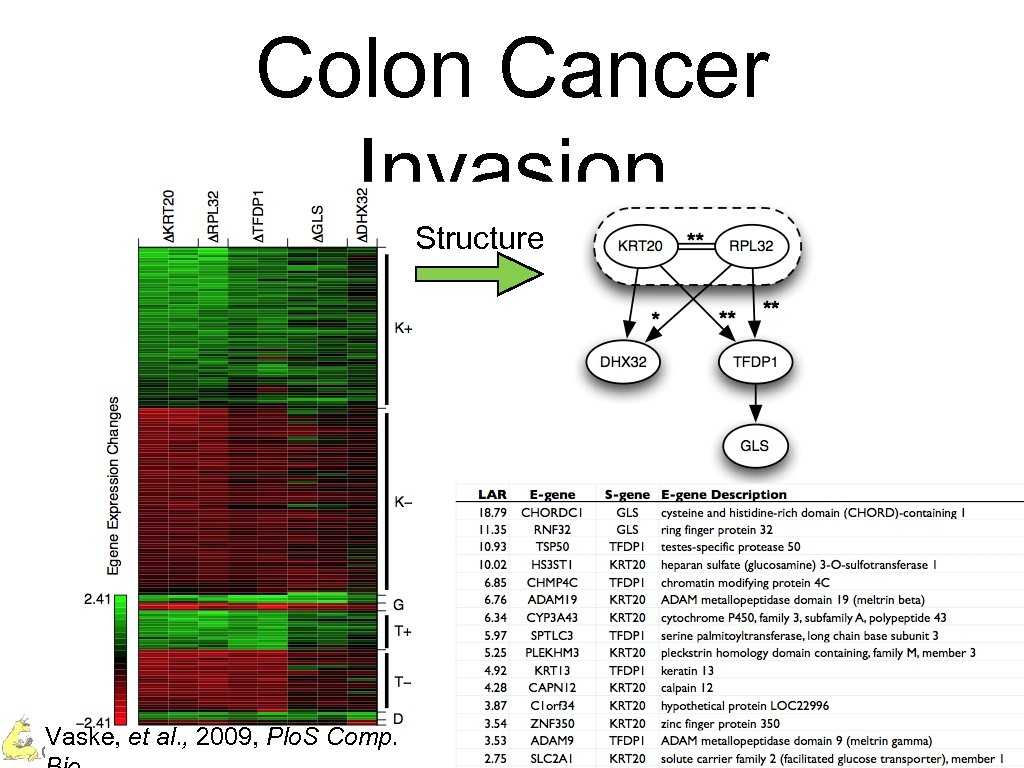
Colon Cancer Invasion Structure Vaske, et al. , 2009, Plo. S Comp. Charles Vaske – UC Santa Cruz
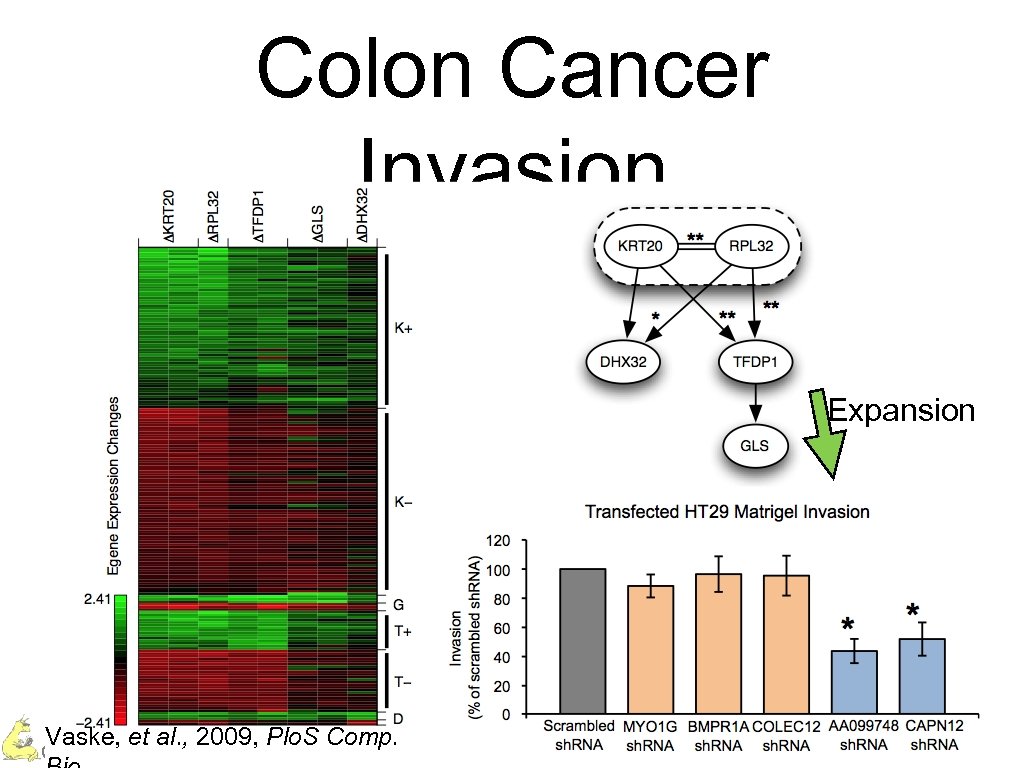
Colon Cancer Invasion Expansion Vaske, et al. , 2009, Plo. S Comp. Charles Vaske – UC Santa Cruz
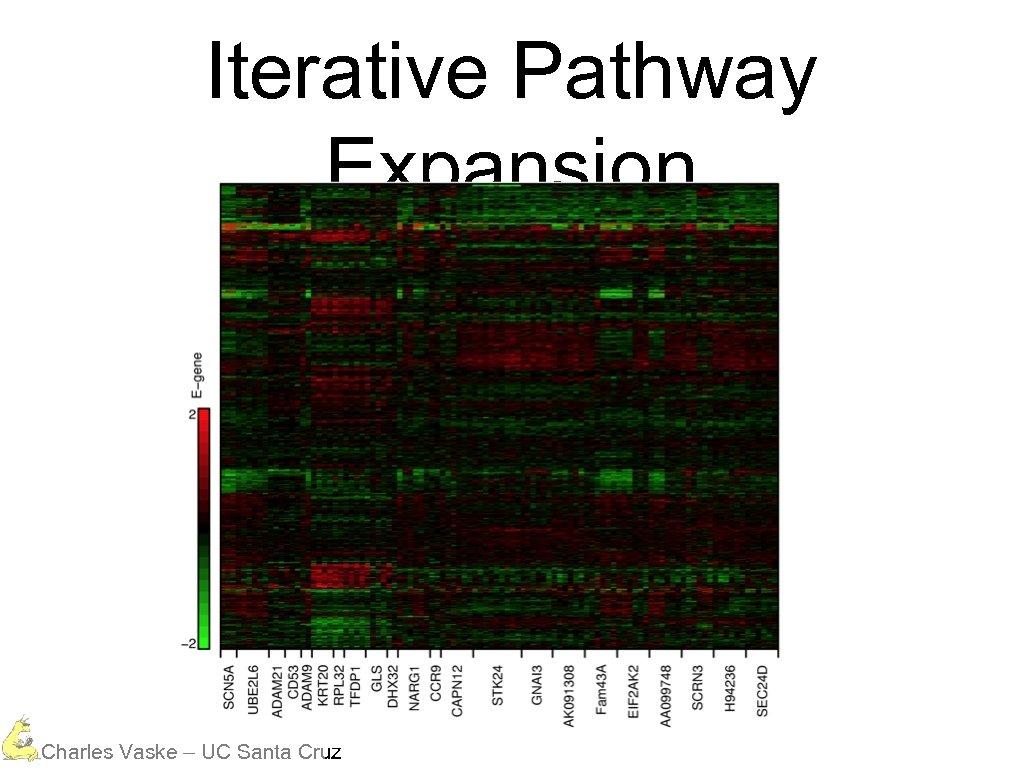
Iterative Pathway Expansion Charles Vaske – UC Santa Cruz
Three Different Tiers Mostly everything is in the same data range Charles Vaske – UC Santa Cruz
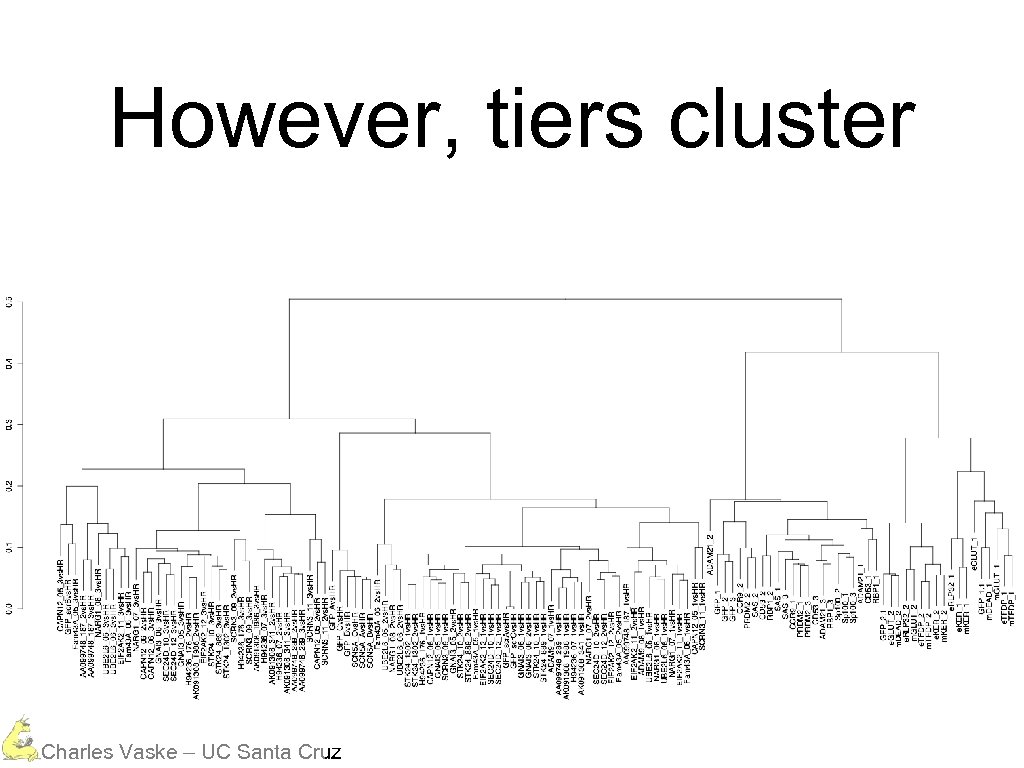
However, tiers cluster Charles Vaske – UC Santa Cruz
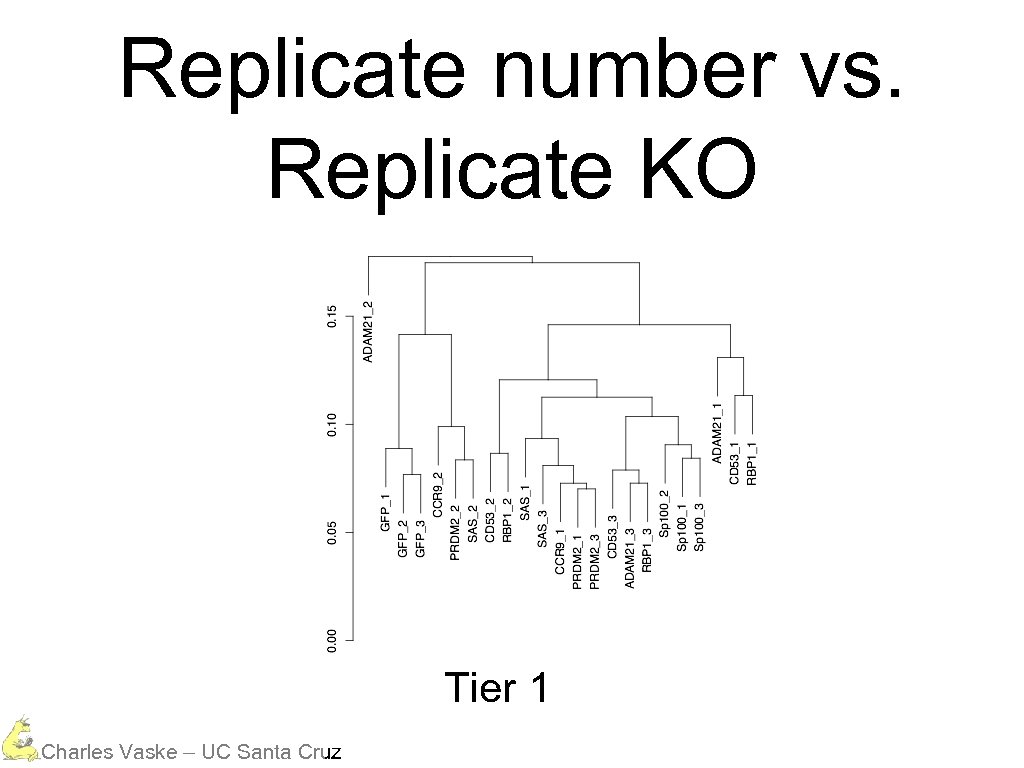
Replicate number vs. Replicate KO Tier 1 Charles Vaske – UC Santa Cruz
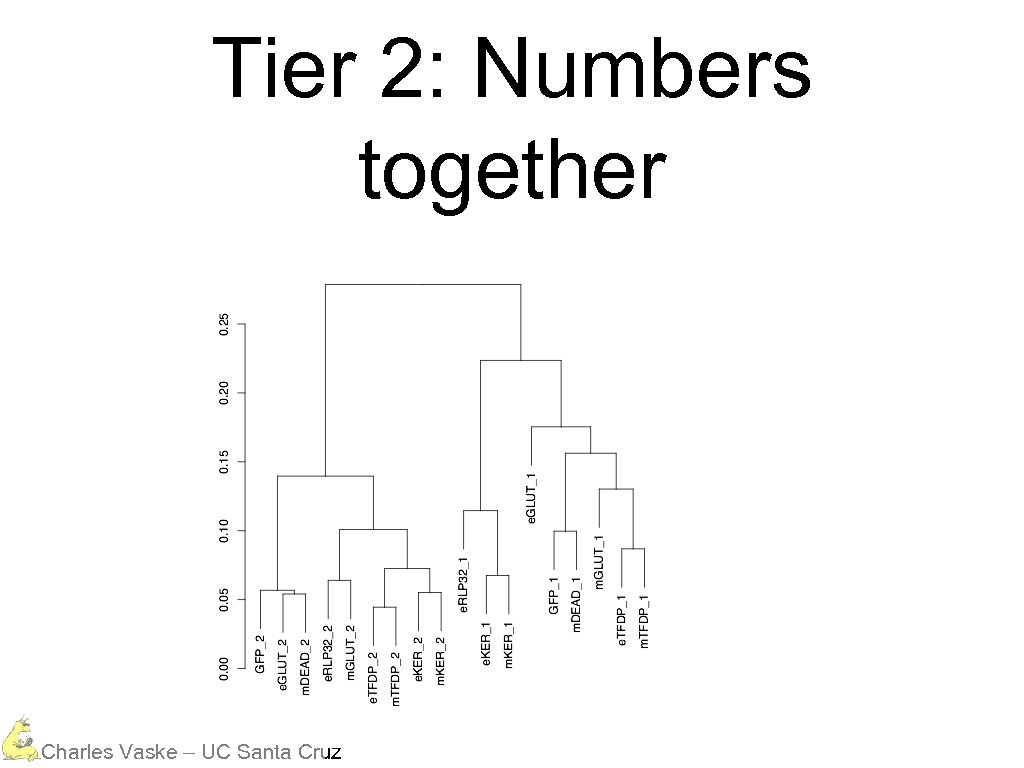
Tier 2: Numbers together Charles Vaske – UC Santa Cruz
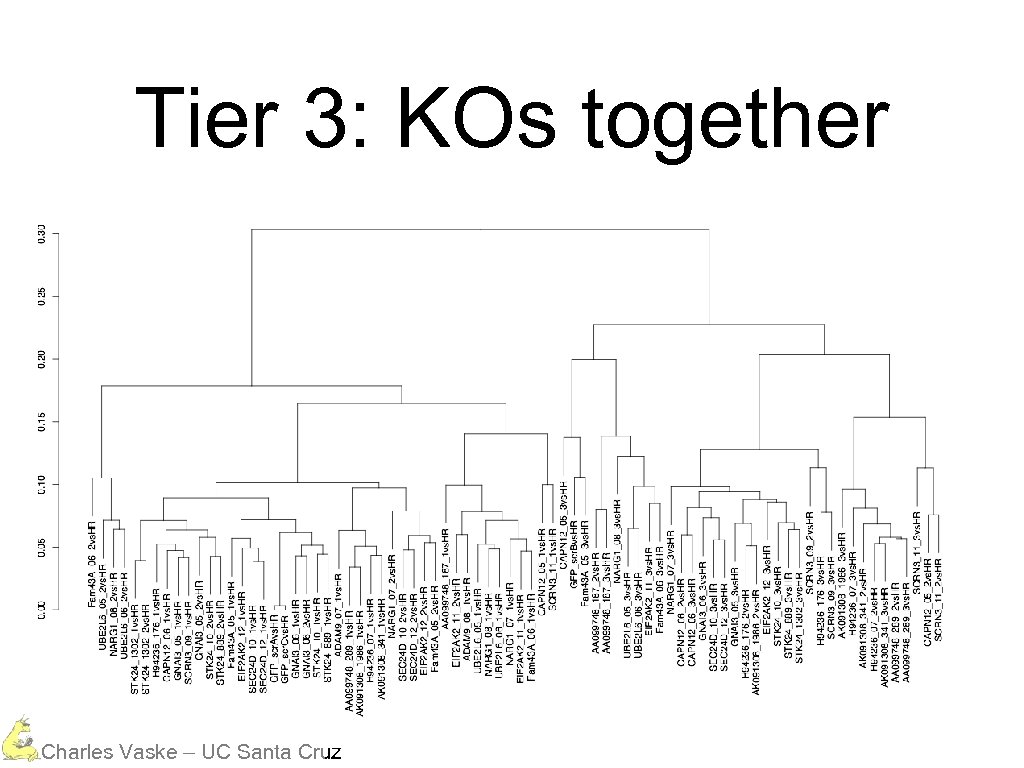
Tier 3: KOs together Charles Vaske – UC Santa Cruz
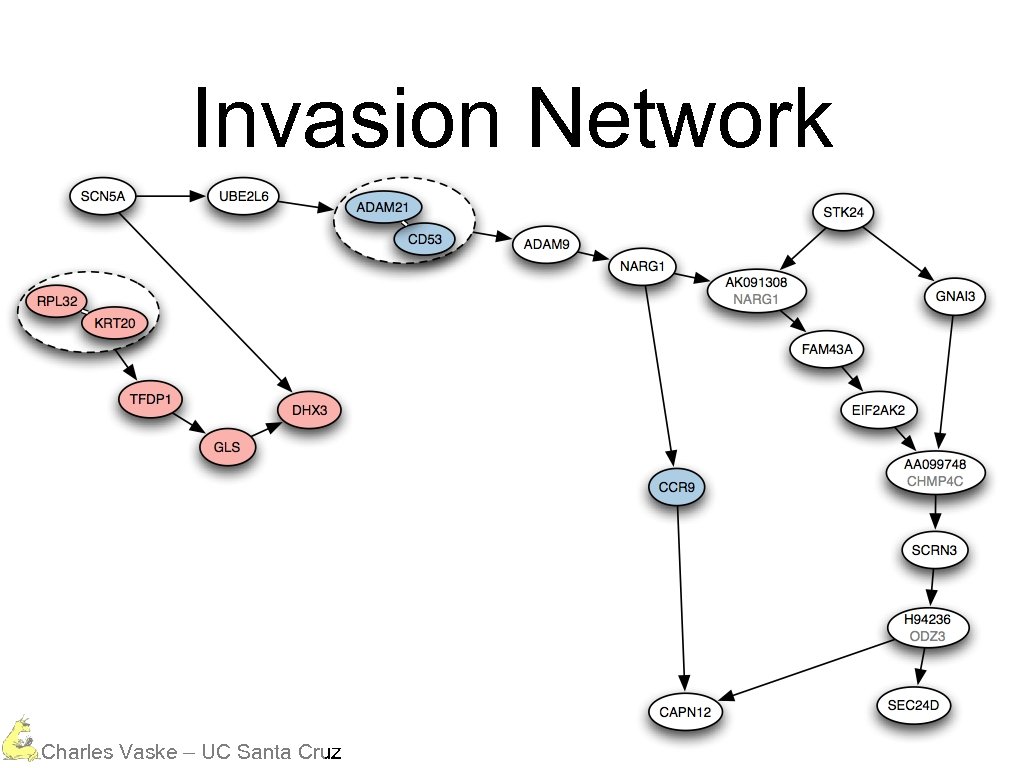
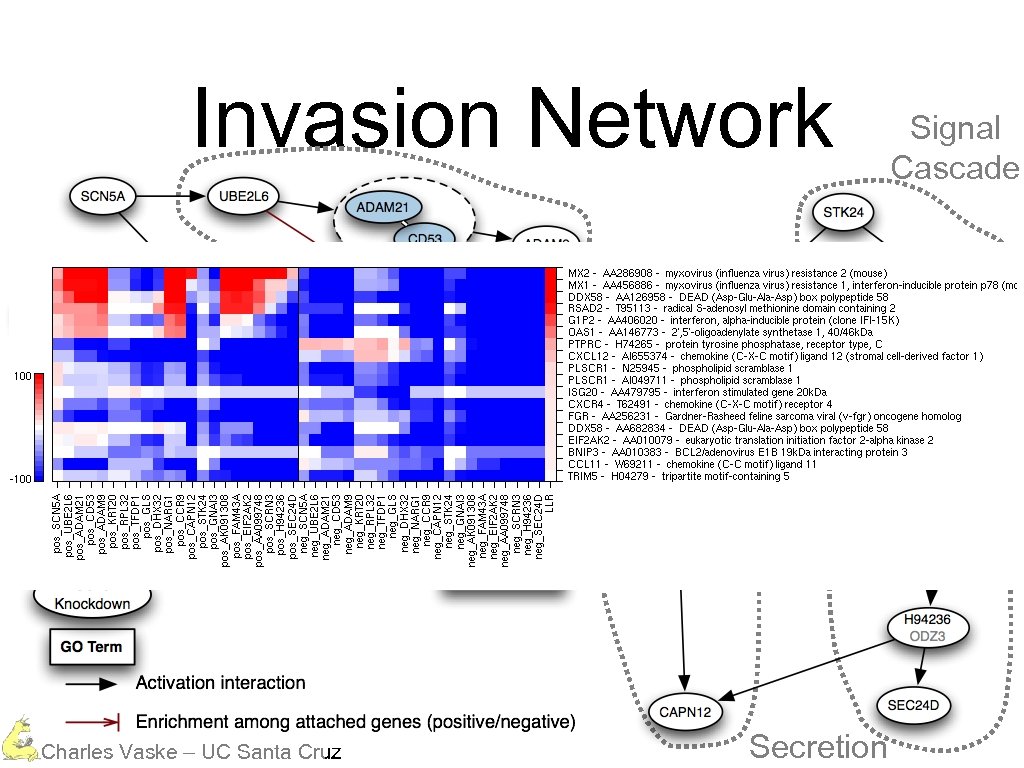
Invasion Network Charles Vaske – UC Santa Cruz
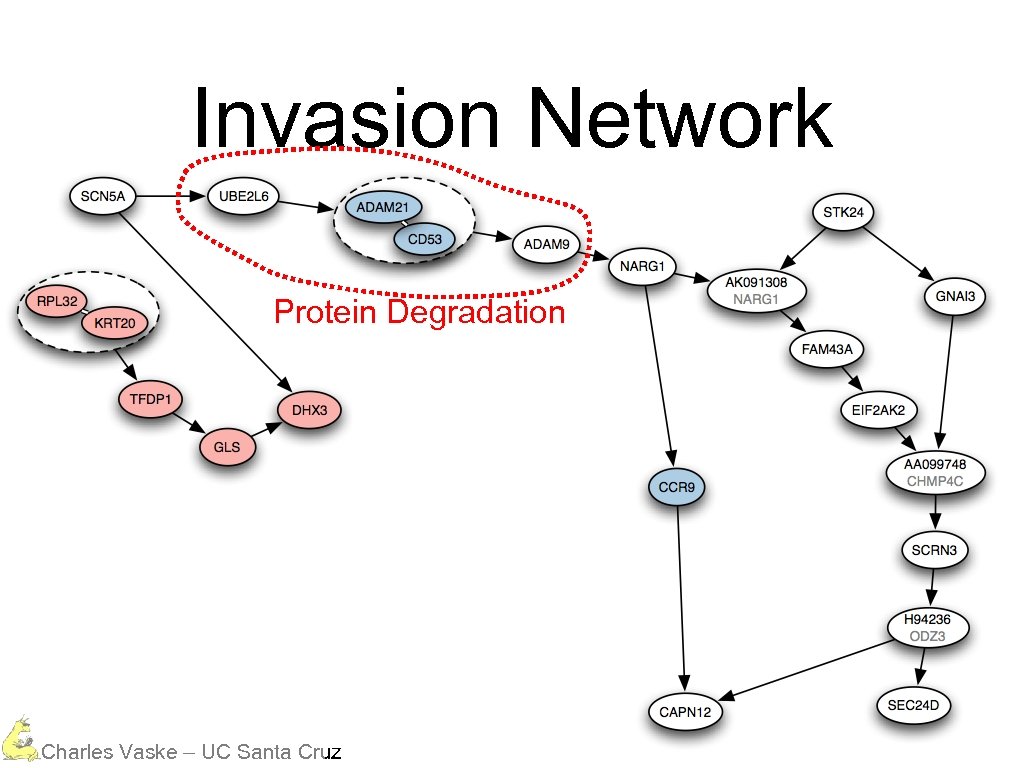
Invasion Network Protein Degradation Charles Vaske – UC Santa Cruz
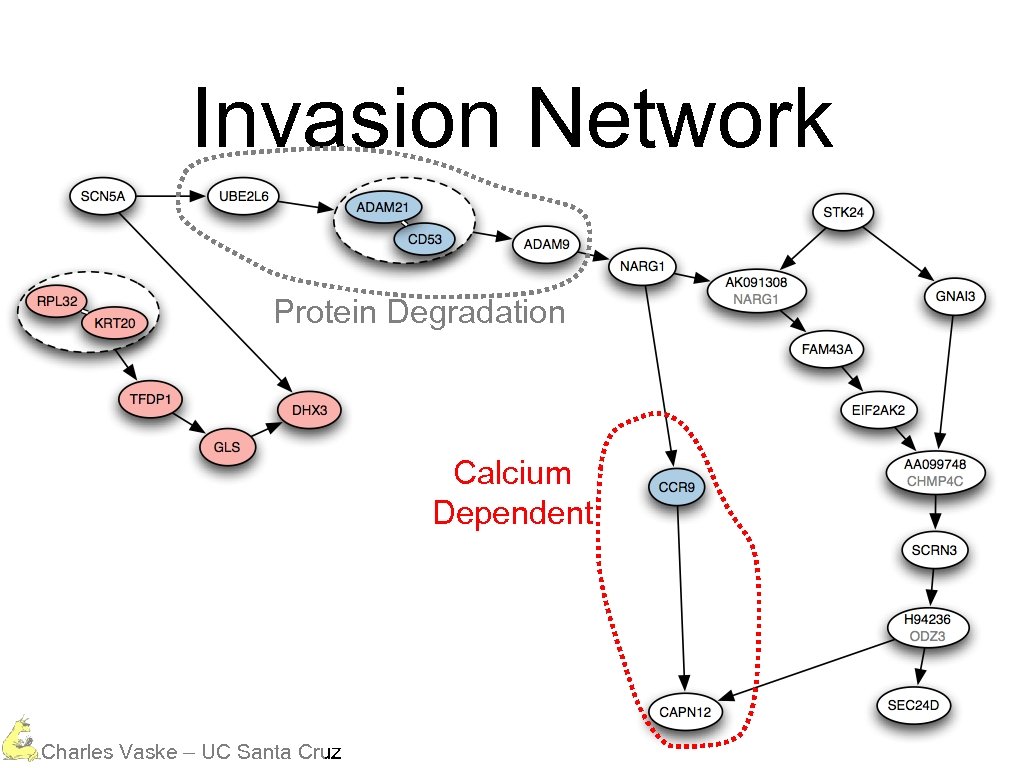
Invasion Network Protein Degradation Calcium Dependent Charles Vaske – UC Santa Cruz
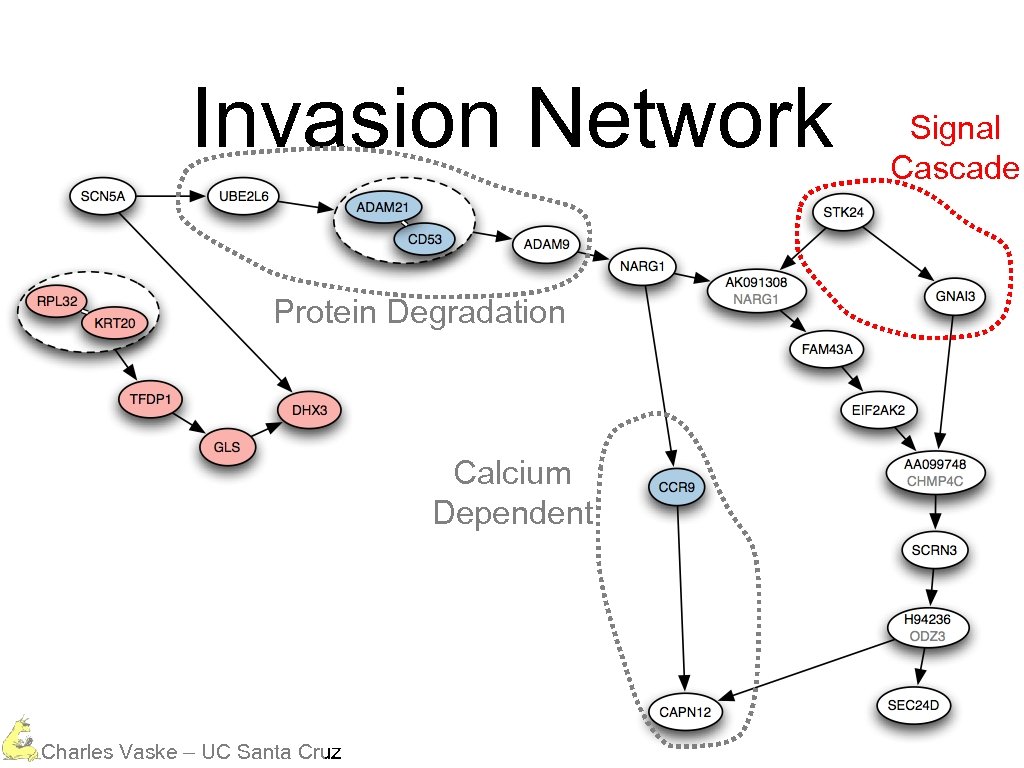
Invasion Network Protein Degradation Calcium Dependent Charles Vaske – UC Santa Cruz Signal Cascade
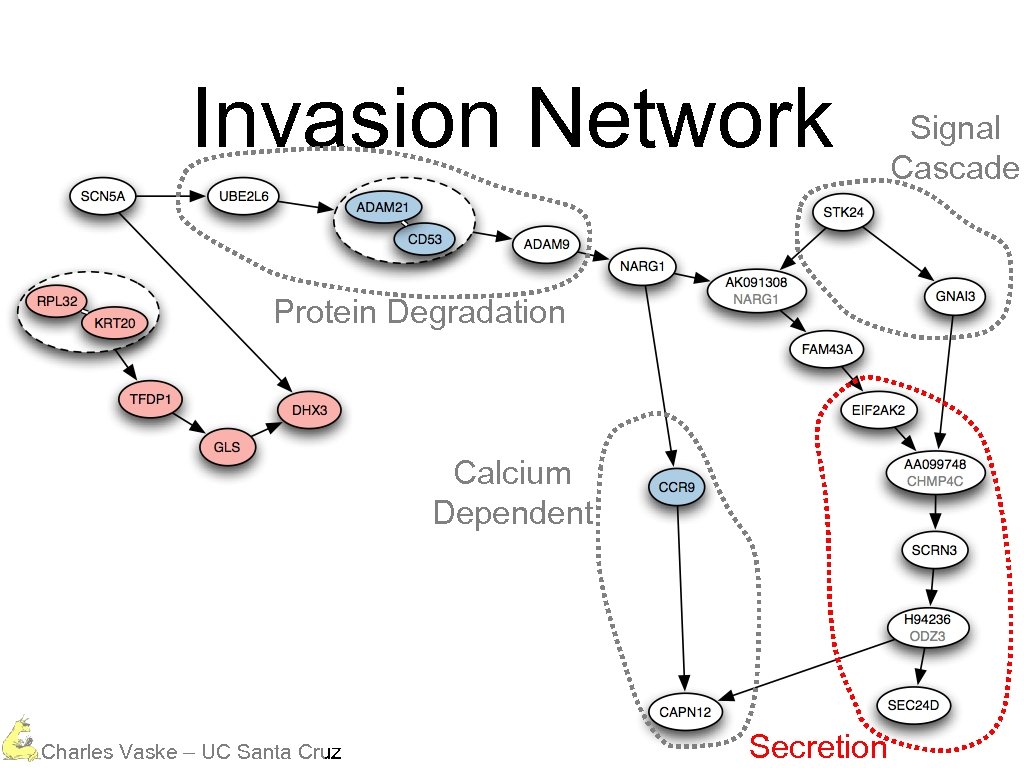
Invasion Network Protein Degradation Calcium Dependent Charles Vaske – UC Santa Cruz Secretion Signal Cascade
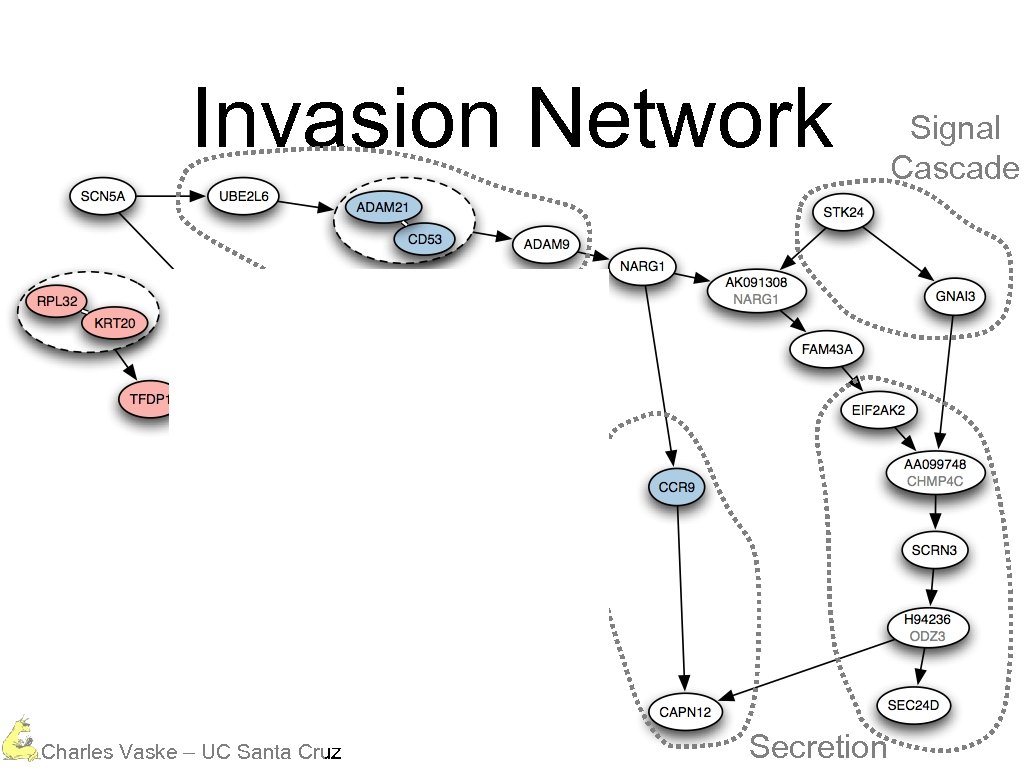
Invasion Network Protein Degradation Calcium Dependent Charles Vaske – UC Santa Cruz Secretion Signal Cascade
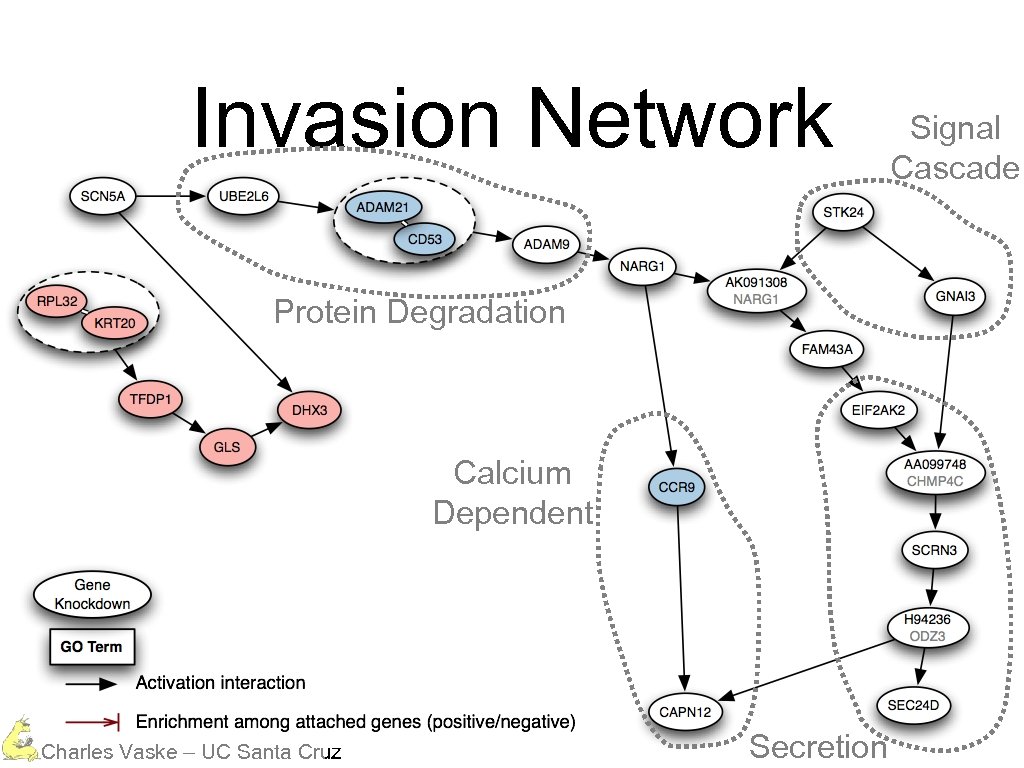
Invasion Network Protein Degradation Calcium Dependent Charles Vaske – UC Santa Cruz Secretion Signal Cascade
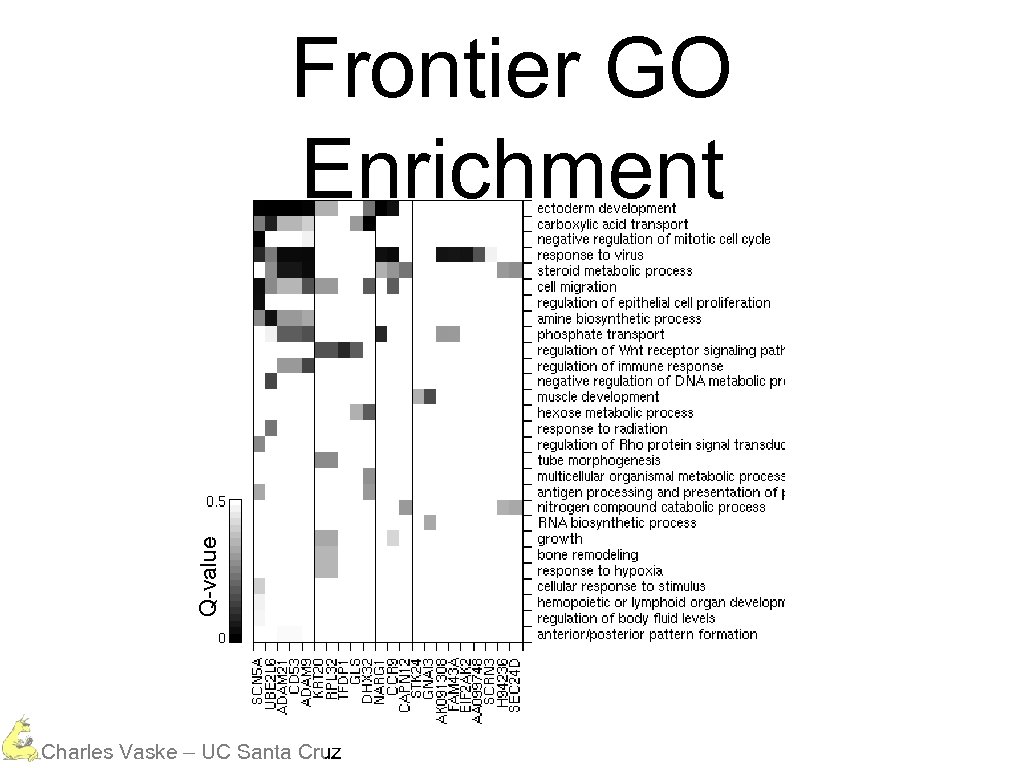
Q-value Frontier GO Enrichment Charles Vaske – UC Santa Cruz
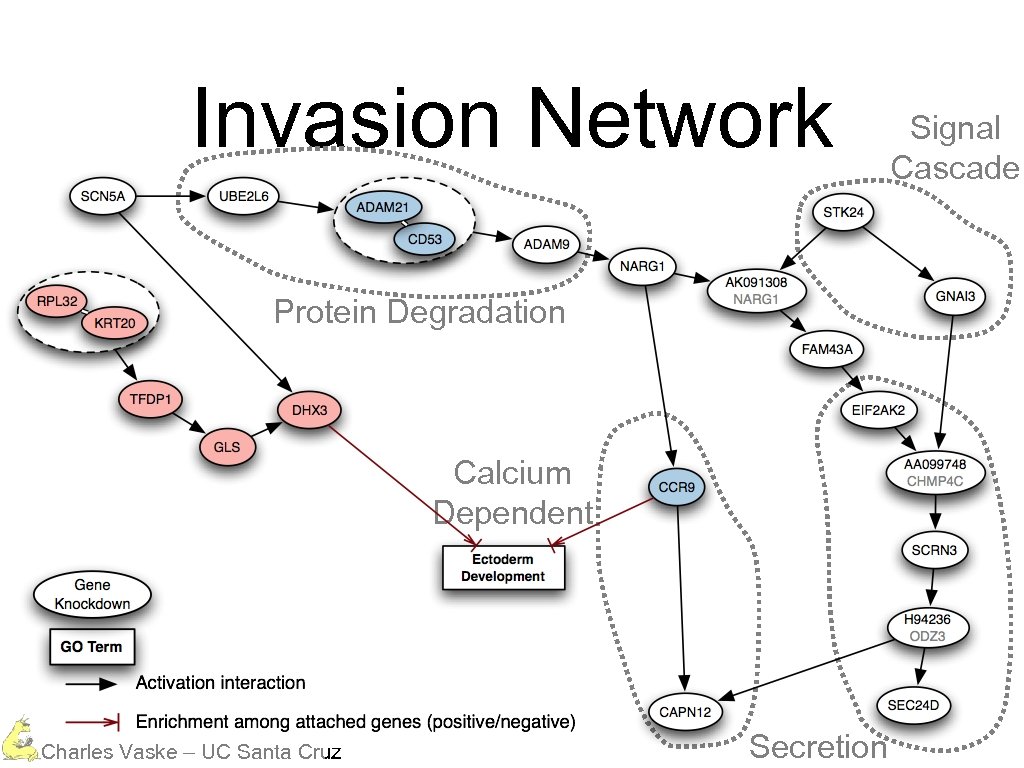
Invasion Network Protein Degradation Calcium Dependent Charles Vaske – UC Santa Cruz Secretion Signal Cascade
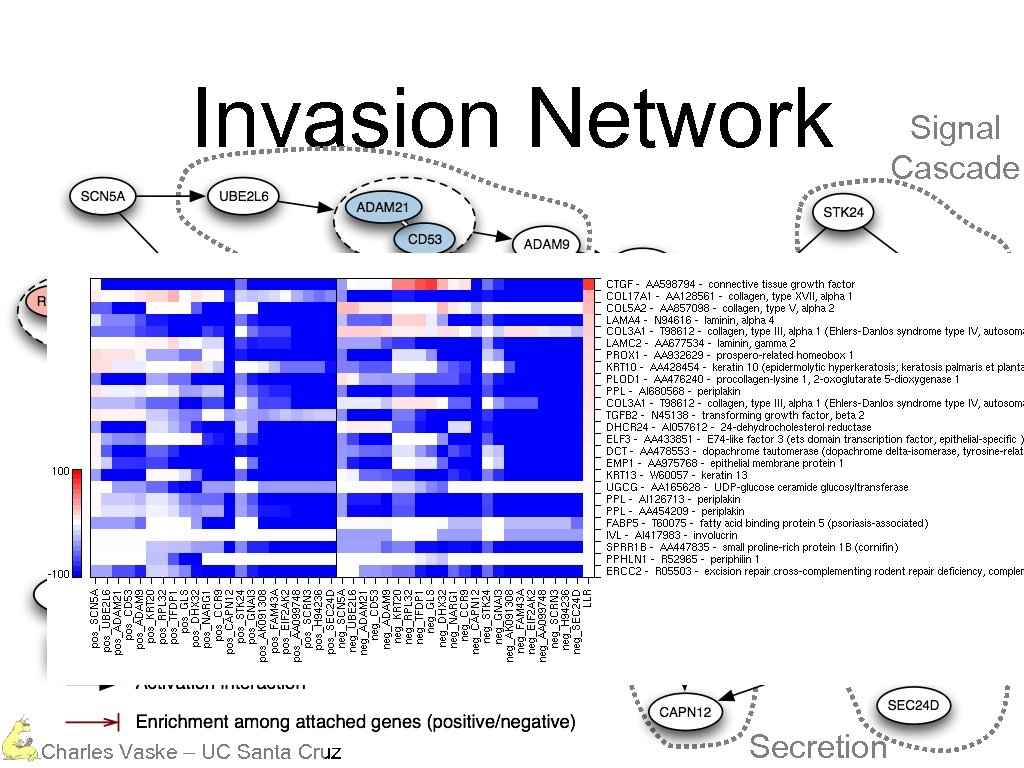
Invasion Network Protein Degradation Calcium Dependent Charles Vaske – UC Santa Cruz Secretion Signal Cascade
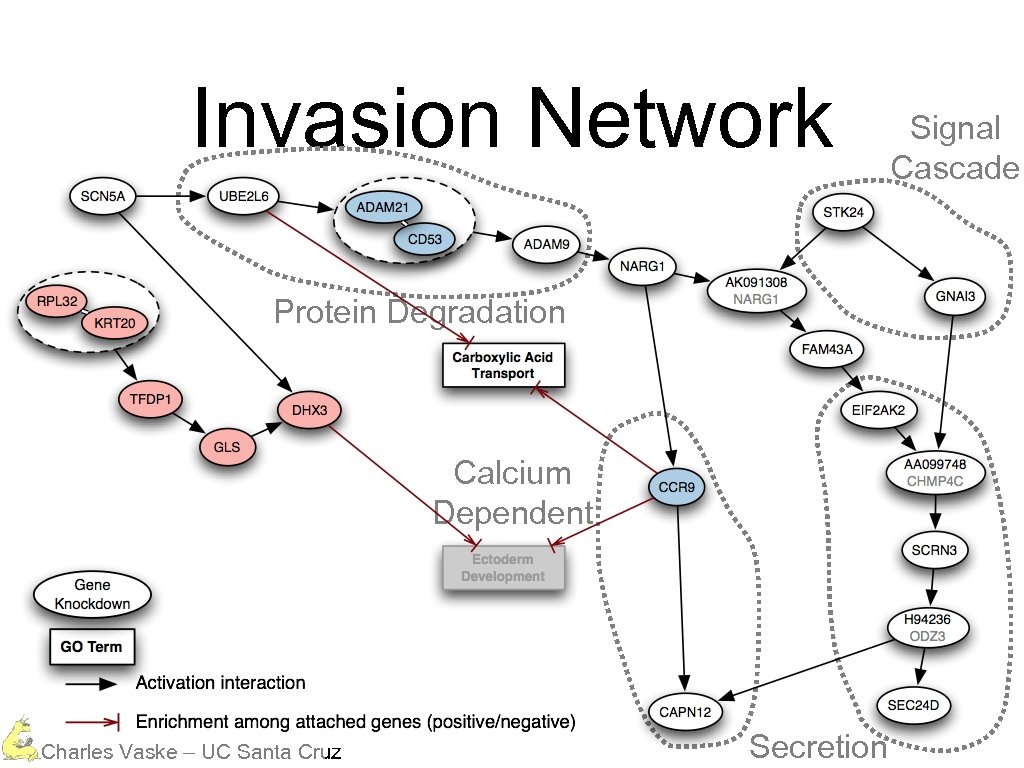
Invasion Network Protein Degradation Calcium Dependent Charles Vaske – UC Santa Cruz Secretion Signal Cascade
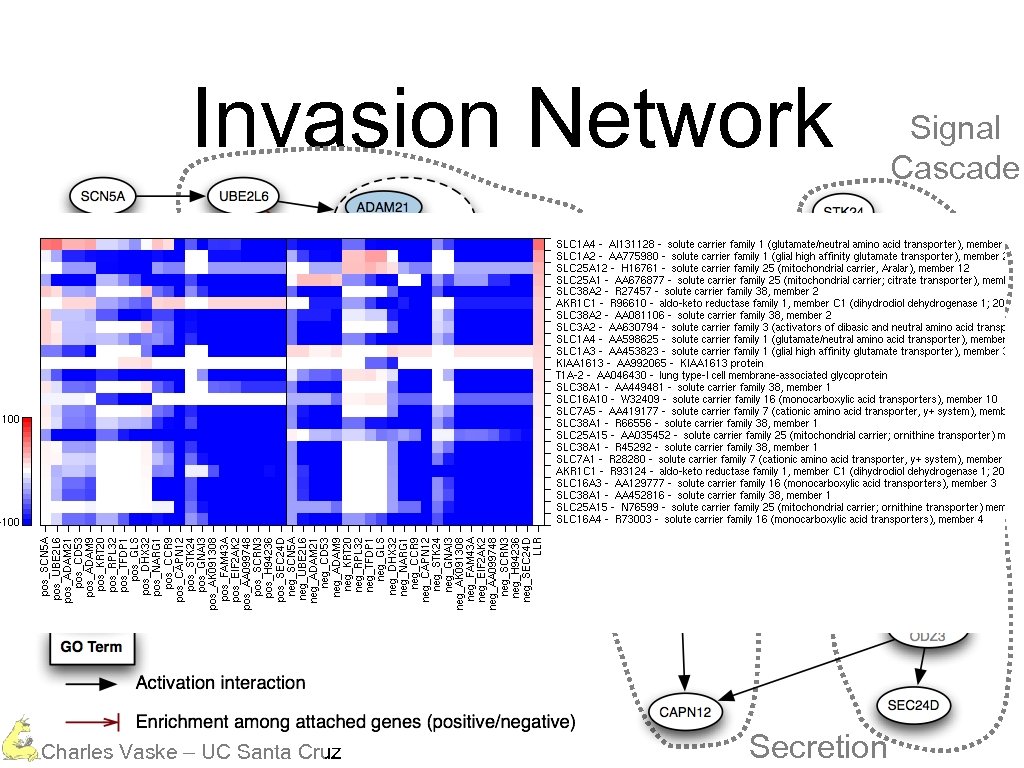
Invasion Network Protein Degradation Calcium Dependent Charles Vaske – UC Santa Cruz Secretion Signal Cascade
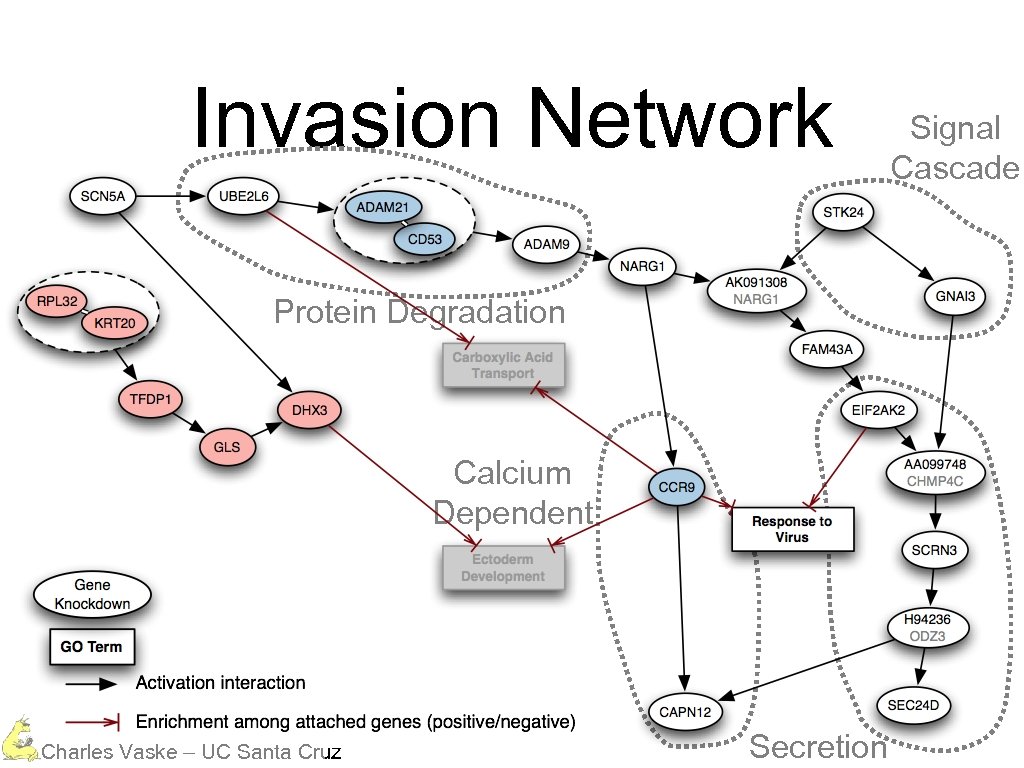
Invasion Network Protein Degradation Calcium Dependent Charles Vaske – UC Santa Cruz Secretion Signal Cascade
Invasion Network Protein Degradation Calcium Dependent Charles Vaske – UC Santa Cruz Secretion Signal Cascade
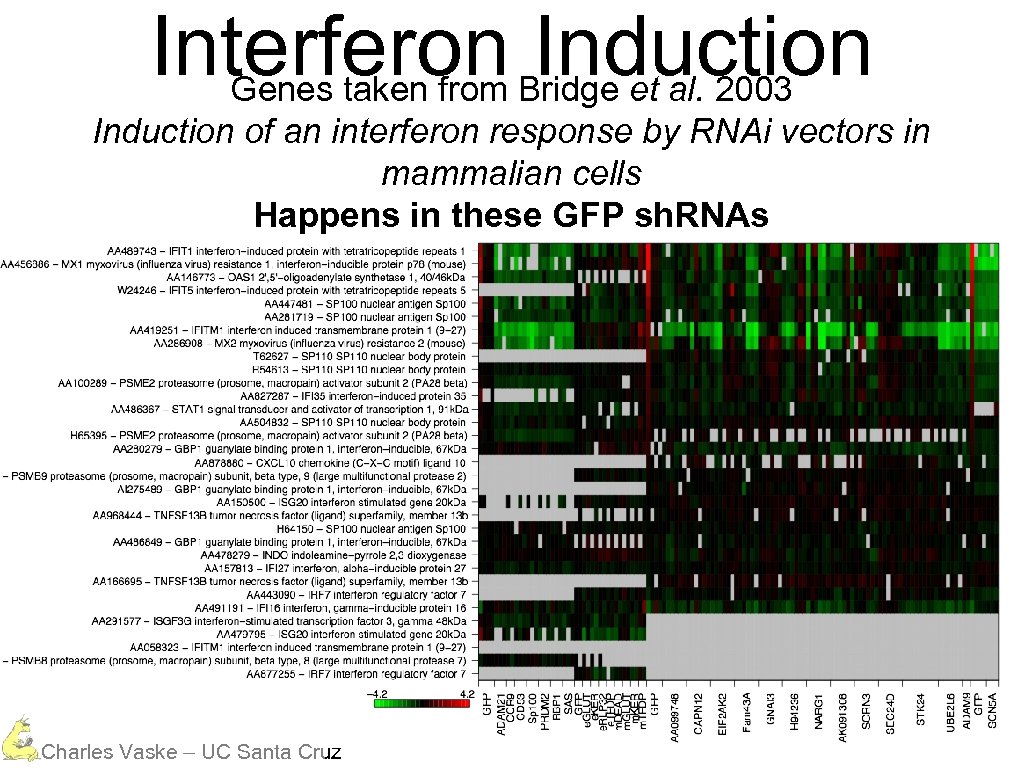
Interferon Bridge et al. 2003 Induction Genes taken from Induction of an interferon response by RNAi vectors in mammalian cells Happens in these GFP sh. RNAs Charles Vaske – UC Santa Cruz
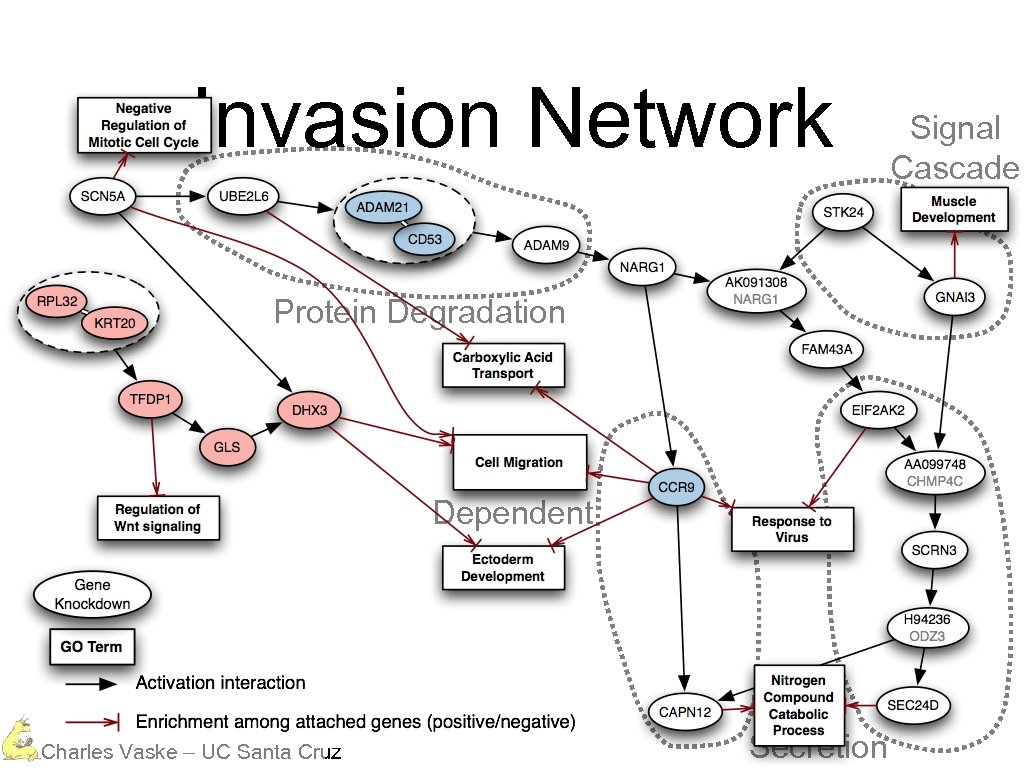
Invasion Network Protein Degradation Calcium Dependent Charles Vaske – UC Santa Cruz Secretion Signal Cascade
Summary • Pathways can be learned from a small number of microarrays • Pathway analysis is more predictive than correlation methods for association • A pathway structure is better at guiding hypotheses than gene-based analysis Charles Vaske – UC Santa Cruz
Future Work • Multiple perturbations/epistasis (in PSB 2009 proceedings) • Transfer to population studies (CNV, expression, genotype, etc. ) • Use whole pathways as basal genetic units • Incorporate further genetic models (Quantitative Trait Pathways? ) Charles Vaske – UC Santa Cruz

Acknowledgements UCSC Biomolecular Engineering Josh Stuart Pinal Kanabar Matt Weirauch • • • UCSC Environmental Toxicology Fitnat Yildiz Sinem Beyhan • • Academica Sinica Chen-Hsiang Yeang • George Washington University Norm Lee •

c5aab5db837825271f69b39da72e00ef.ppt