8e5a44304dd7c948585b5d6267c479e9.ppt
- Количество слайдов: 12
 Paths to a Reference Architecture for an Open Bio Grid • Rick Stevens
Paths to a Reference Architecture for an Open Bio Grid • Rick Stevens
 Determining Requirements for the Open Bio. Grid • Model for Community Involvement • MPEG-7 process • Call for proposals • Technologies • Architectures • Interfaces and APIs • Requirements Collection • Input for an eventual RFP • Scope the components of a “Standard” • Related to existing Standards
Determining Requirements for the Open Bio. Grid • Model for Community Involvement • MPEG-7 process • Call for proposals • Technologies • Architectures • Interfaces and APIs • Requirements Collection • Input for an eventual RFP • Scope the components of a “Standard” • Related to existing Standards
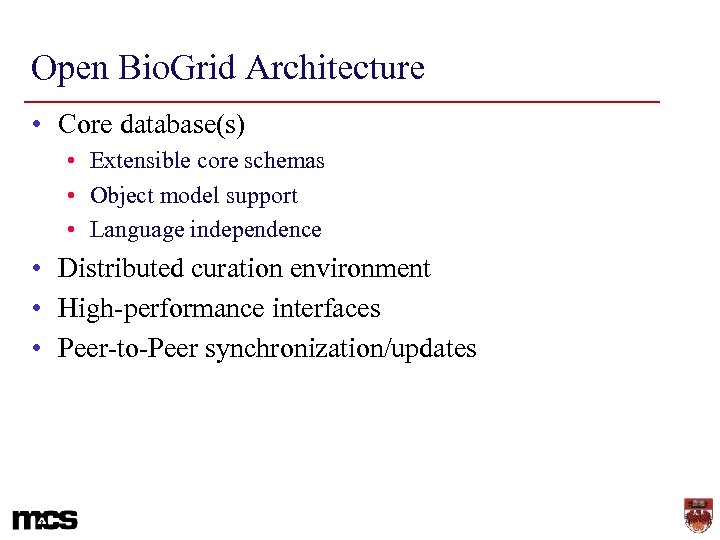 Open Bio. Grid Architecture • Core database(s) • Extensible core schemas • Object model support • Language independence • Distributed curation environment • High-performance interfaces • Peer-to-Peer synchronization/updates
Open Bio. Grid Architecture • Core database(s) • Extensible core schemas • Object model support • Language independence • Distributed curation environment • High-performance interfaces • Peer-to-Peer synchronization/updates
 Principal Partners and Stakeholders • Biology and Biomedical Communities • Computer Science Community • Industry • User community • Technology providers • Agencies (NIH, NSF, DOE, etc. ) • Standards Organizations • Professional Societies
Principal Partners and Stakeholders • Biology and Biomedical Communities • Computer Science Community • Industry • User community • Technology providers • Agencies (NIH, NSF, DOE, etc. ) • Standards Organizations • Professional Societies
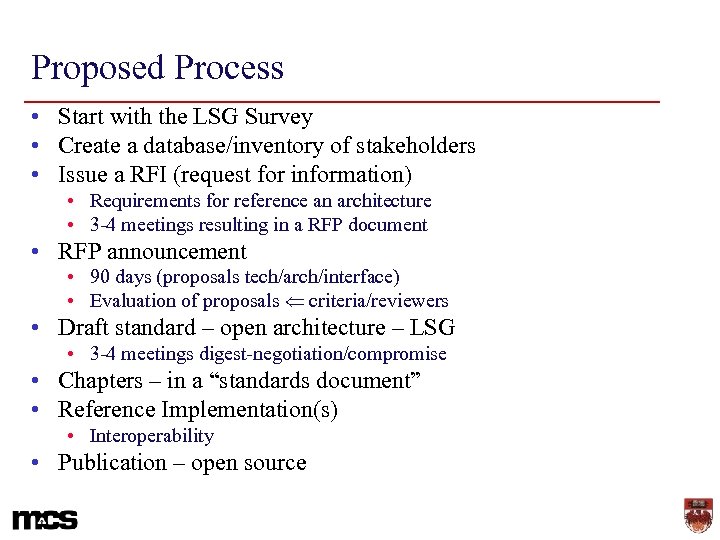 Proposed Process • Start with the LSG Survey • Create a database/inventory of stakeholders • Issue a RFI (request for information) • Requirements for reference an architecture • 3 -4 meetings resulting in a RFP document • RFP announcement • 90 days (proposals tech/arch/interface) • Evaluation of proposals criteria/reviewers • Draft standard – open architecture – LSG • 3 -4 meetings digest-negotiation/compromise • Chapters – in a “standards document” • Reference Implementation(s) • Interoperability • Publication – open source
Proposed Process • Start with the LSG Survey • Create a database/inventory of stakeholders • Issue a RFI (request for information) • Requirements for reference an architecture • 3 -4 meetings resulting in a RFP document • RFP announcement • 90 days (proposals tech/arch/interface) • Evaluation of proposals criteria/reviewers • Draft standard – open architecture – LSG • 3 -4 meetings digest-negotiation/compromise • Chapters – in a “standards document” • Reference Implementation(s) • Interoperability • Publication – open source
 Open Issues • • Determining scope of “The Standard” Core team Fast track (meeting every 6 weeks (2 -3 days)) Buy-in from stakeholders Sponsorship Open Source (license issues) Time Frame for completion
Open Issues • • Determining scope of “The Standard” Core team Fast track (meeting every 6 weeks (2 -3 days)) Buy-in from stakeholders Sponsorship Open Source (license issues) Time Frame for completion
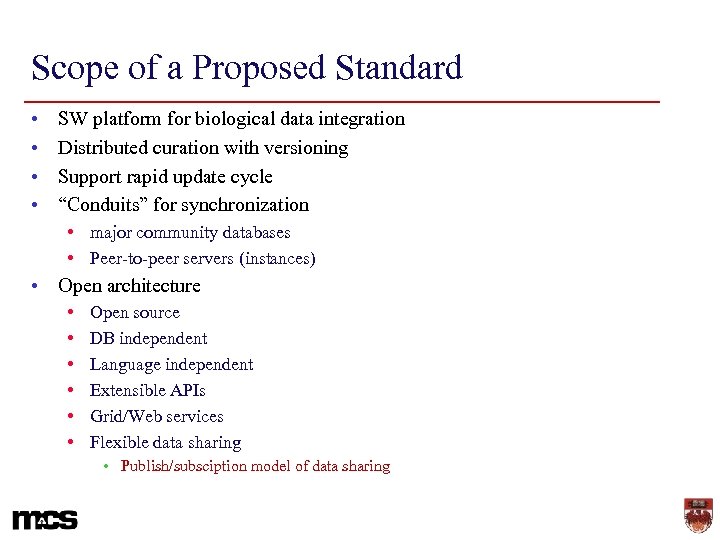 Scope of a Proposed Standard • • SW platform for biological data integration Distributed curation with versioning Support rapid update cycle “Conduits” for synchronization • major community databases • Peer-to-peer servers (instances) • Open architecture • • • Open source DB independent Language independent Extensible APIs Grid/Web services Flexible data sharing • Publish/subsciption model of data sharing
Scope of a Proposed Standard • • SW platform for biological data integration Distributed curation with versioning Support rapid update cycle “Conduits” for synchronization • major community databases • Peer-to-peer servers (instances) • Open architecture • • • Open source DB independent Language independent Extensible APIs Grid/Web services Flexible data sharing • Publish/subsciption model of data sharing
 Scope II • Supports multiple views and proprietary data • Private data integrated with public data • Public data • Interfaces • • Transactions High-throughput data paths, bulk transfers Simulation/DB connections Import/export APIs • Scalability • Security • Portability
Scope II • Supports multiple views and proprietary data • Private data integrated with public data • Public data • Interfaces • • Transactions High-throughput data paths, bulk transfers Simulation/DB connections Import/export APIs • Scalability • Security • Portability
 Scalability Goals • Millions of genes and gene products • GBs-TBs of annotation per gene • 100, 000 s of genomes • Many close variants • Millions of “phenomes” • Instances of “k” • Thousands of cooperating sites • Update Channels (pub/sub) • Thousands (some private, some open)
Scalability Goals • Millions of genes and gene products • GBs-TBs of annotation per gene • 100, 000 s of genomes • Many close variants • Millions of “phenomes” • Instances of “k” • Thousands of cooperating sites • Update Channels (pub/sub) • Thousands (some private, some open)
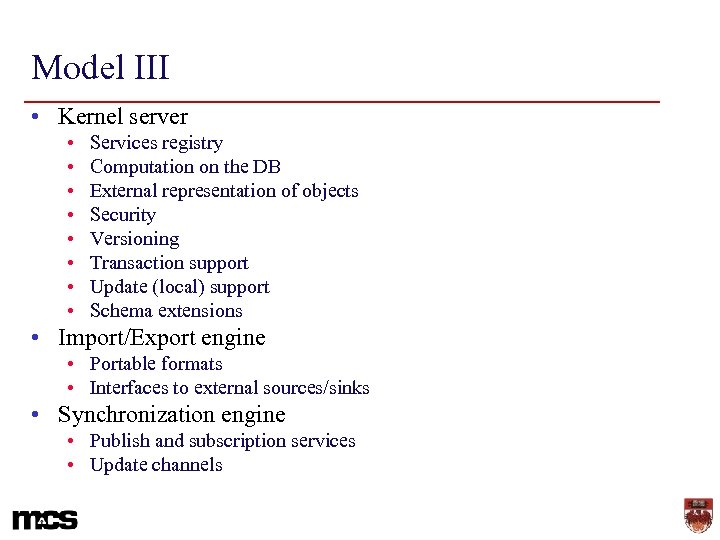 Model III • Kernel server • • Services registry Computation on the DB External representation of objects Security Versioning Transaction support Update (local) support Schema extensions • Import/Export engine • Portable formats • Interfaces to external sources/sinks • Synchronization engine • Publish and subscription services • Update channels
Model III • Kernel server • • Services registry Computation on the DB External representation of objects Security Versioning Transaction support Update (local) support Schema extensions • Import/Export engine • Portable formats • Interfaces to external sources/sinks • Synchronization engine • Publish and subscription services • Update channels
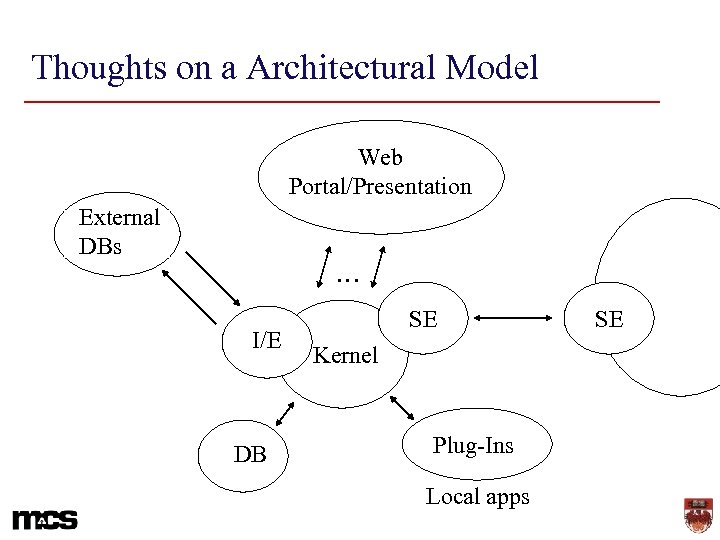 Thoughts on a Architectural Model Web Portal/Presentation External DBs … I/E DB SE Kernel Plug-Ins Local apps SE
Thoughts on a Architectural Model Web Portal/Presentation External DBs … I/E DB SE Kernel Plug-Ins Local apps SE
 Model II • OLSG Services • • • Directory services Namespace/ontology services Brokering Channel Services Computing services • Grid service • Security • Transport • Etc.
Model II • OLSG Services • • • Directory services Namespace/ontology services Brokering Channel Services Computing services • Grid service • Security • Transport • Etc.


