429e3207816fb145b589ed273b448589.ppt
- Количество слайдов: 29
 Overview of the Pathway Tools Software and Pathway/Genome Databases
Overview of the Pathway Tools Software and Pathway/Genome Databases
 Introductions l BRG Staff l Peter Karp l Pallavi Kaipa l Mario Latendresse l Suzanne Paley l Markus Krummenacker l Ingrid Keseler l Ron Caspi l Alex Shearer l Carol Fulcher l Attendees l Where from, what genome? l What do you hope to get out of the tutorial? SRI International Bioinformatics
Introductions l BRG Staff l Peter Karp l Pallavi Kaipa l Mario Latendresse l Suzanne Paley l Markus Krummenacker l Ingrid Keseler l Ron Caspi l Alex Shearer l Carol Fulcher l Attendees l Where from, what genome? l What do you hope to get out of the tutorial? SRI International Bioinformatics
 SRI International l Private nonprofit research institute l No permanent funding sources l 1300 staff in Menlo Park – Founded in 1946 as Stanford Research Institute – Separated from Stanford University in 1970 – Name changed to SRI International in 1977 – David Sarnoff Research Center acquired in 1987 SRI International Bioinformatics
SRI International l Private nonprofit research institute l No permanent funding sources l 1300 staff in Menlo Park – Founded in 1946 as Stanford Research Institute – Separated from Stanford University in 1970 – Name changed to SRI International in 1977 – David Sarnoff Research Center acquired in 1987 SRI International Bioinformatics
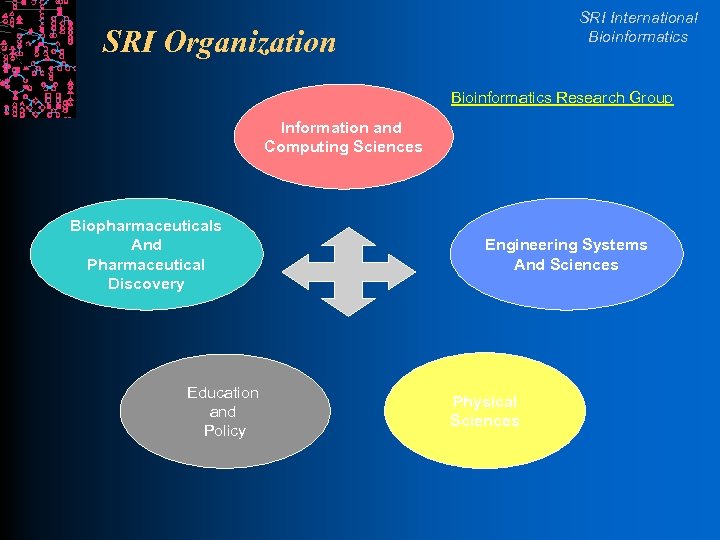 SRI International Bioinformatics SRI Organization Bioinformatics Research Group Information and Computing Sciences Biopharmaceuticals And Pharmaceutical Discovery Education and Policy Engineering Systems And Sciences Physical Sciences
SRI International Bioinformatics SRI Organization Bioinformatics Research Group Information and Computing Sciences Biopharmaceuticals And Pharmaceutical Discovery Education and Policy Engineering Systems And Sciences Physical Sciences
 Research in the SRI Bioinformatics Research Group l Eco. Cyc l Meta. Cyc l Pathway Tools l Pathway Holes l Bio. Warehouse l Enzyme Genomics SRI International Bioinformatics
Research in the SRI Bioinformatics Research Group l Eco. Cyc l Meta. Cyc l Pathway Tools l Pathway Holes l Bio. Warehouse l Enzyme Genomics SRI International Bioinformatics
 Outline for Tutorial l l SRI International Bioinformatics Monday l Introduction l Pathway/Genome Navigator l Patho. Logic tutorial and demo l Patho. Logic lab session – Make genome input files parsable Tuesday l Patho. Logic tutorial l Patho. Logic lab session – Build initial version of PGDB Wednesday l Pathway hole filler, operon predictor, transport inference parser Thursday l Editors l Feedback session
Outline for Tutorial l l SRI International Bioinformatics Monday l Introduction l Pathway/Genome Navigator l Patho. Logic tutorial and demo l Patho. Logic lab session – Make genome input files parsable Tuesday l Patho. Logic tutorial l Patho. Logic lab session – Build initial version of PGDB Wednesday l Pathway hole filler, operon predictor, transport inference parser Thursday l Editors l Feedback session
 Tutorial Goals SRI International Bioinformatics l General familiarity with Pathway Tools goals and functionality l Ability to create, edit, and navigate a new PGDB l Create new PGDB for genome(s) you brought with you l Familiarity with information resources available about Pathway Tools to continue your work
Tutorial Goals SRI International Bioinformatics l General familiarity with Pathway Tools goals and functionality l Ability to create, edit, and navigate a new PGDB l Create new PGDB for genome(s) you brought with you l Familiarity with information resources available about Pathway Tools to continue your work
 SRI’s Support for Pathway Tools SRI International Bioinformatics l NIH grant finances software development and user support l Additional grants finance other software development l Email us bug reports, suggestions, questions l Comprehensive bug reports are required for us to fix the problem you reported l Keep us posted regarding your progress
SRI’s Support for Pathway Tools SRI International Bioinformatics l NIH grant finances software development and user support l Additional grants finance other software development l Email us bug reports, suggestions, questions l Comprehensive bug reports are required for us to fix the problem you reported l Keep us posted regarding your progress
 Administrative Details l l l Please wear badges at all times Escort required outside this room/hallway Let us know when you are leaving l Use E-Bldg Entrance Phone numbers to call from entrance l Meals l Wednesday outing possible l Restrooms l SRI International Bioinformatics
Administrative Details l l l Please wear badges at all times Escort required outside this room/hallway Let us know when you are leaving l Use E-Bldg Entrance Phone numbers to call from entrance l Meals l Wednesday outing possible l Restrooms l SRI International Bioinformatics
 Tutorial Format l Questions SRI International Bioinformatics welcome during presentations l Lab sessions will take different amounts of time for different people l Refine your PGDB l Read Pathway Tools manuals l Buddy system for some computers l Computer logins l Internet connectivity
Tutorial Format l Questions SRI International Bioinformatics welcome during presentations l Lab sessions will take different amounts of time for different people l Refine your PGDB l Read Pathway Tools manuals l Buddy system for some computers l Computer logins l Internet connectivity
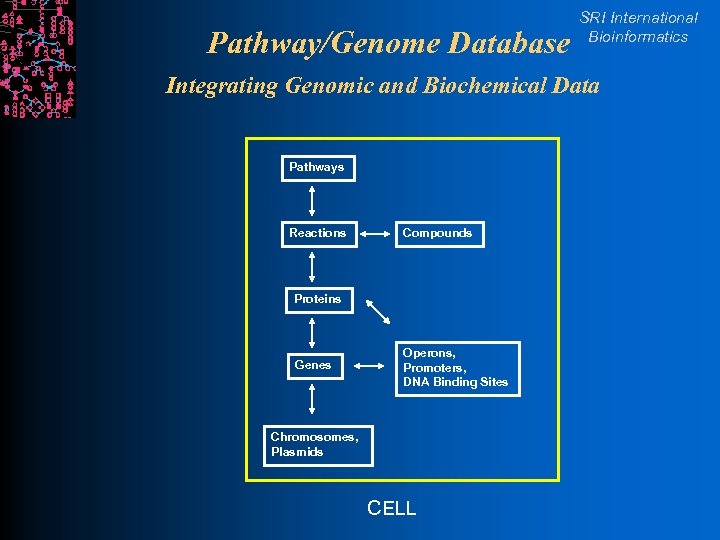 Pathway/Genome Database SRI International Bioinformatics Integrating Genomic and Biochemical Data Pathways Reactions Compounds Proteins Genes Operons, Promoters, DNA Binding Sites Chromosomes, Plasmids CELL
Pathway/Genome Database SRI International Bioinformatics Integrating Genomic and Biochemical Data Pathways Reactions Compounds Proteins Genes Operons, Promoters, DNA Binding Sites Chromosomes, Plasmids CELL
 Terminology l. Model Organism Database (MOD) – DB describing genome and other information about an organism l. Pathway/Genome Database (PGDB) – MOD that combines information about l Pathways, reactions, substrates l Enzymes, transporters l Genes, replicons l Transcription factors, promoters, operons, DNA binding sites l. Bio. Cyc – Collection of 205 PGDBs at Bio. Cyc. org l Eco. Cyc, Agro. Cyc, Human. Cyc SRI International Bioinformatics
Terminology l. Model Organism Database (MOD) – DB describing genome and other information about an organism l. Pathway/Genome Database (PGDB) – MOD that combines information about l Pathways, reactions, substrates l Enzymes, transporters l Genes, replicons l Transcription factors, promoters, operons, DNA binding sites l. Bio. Cyc – Collection of 205 PGDBs at Bio. Cyc. org l Eco. Cyc, Agro. Cyc, Human. Cyc SRI International Bioinformatics
 Bio. Cyc Collection of Pathway/Genome Databases l. Pathway/Genome Database (PGDB) – combines information about l Pathways, reactions, substrates l Enzymes, transporters l Genes, replicons l Transcription factors/sites, promoters, operons l. Tier 1: Literature-Derived PGDBs l Meta. Cyc l Eco. Cyc -- Escherichia coli K-12 l Bio. Cyc Open Chemical Database l. Tier 2: Computationally-derived DBs, Some Curation -- 12 PGDBs l Human. Cyc l Mycobacterium tuberculosis l. Tier 3: Computationally-derived DBs, No Curation -- 191 DBs SRI International Bioinformatics
Bio. Cyc Collection of Pathway/Genome Databases l. Pathway/Genome Database (PGDB) – combines information about l Pathways, reactions, substrates l Enzymes, transporters l Genes, replicons l Transcription factors/sites, promoters, operons l. Tier 1: Literature-Derived PGDBs l Meta. Cyc l Eco. Cyc -- Escherichia coli K-12 l Bio. Cyc Open Chemical Database l. Tier 2: Computationally-derived DBs, Some Curation -- 12 PGDBs l Human. Cyc l Mycobacterium tuberculosis l. Tier 3: Computationally-derived DBs, No Curation -- 191 DBs SRI International Bioinformatics
 Terminology – Pathway Tools Software SRI International Bioinformatics l Patho. Logic l Predicts operons, metabolic network, pathway hole fillers, from genome l Computational creation of new Pathway/Genome Databases l Pathway/Genome Editors l Distributed curation of PGDBs l Distributed object database system, interactive editing tools l Pathway/Genome Navigator l WWW publishing of PGDBs l Querying, visualization of pathways, chromosomes, operons l Analysis operations u u Pathway visualization of gene-expression data Global comparisons of metabolic networks Bioinformatics 18: S 225 2002
Terminology – Pathway Tools Software SRI International Bioinformatics l Patho. Logic l Predicts operons, metabolic network, pathway hole fillers, from genome l Computational creation of new Pathway/Genome Databases l Pathway/Genome Editors l Distributed curation of PGDBs l Distributed object database system, interactive editing tools l Pathway/Genome Navigator l WWW publishing of PGDBs l Querying, visualization of pathways, chromosomes, operons l Analysis operations u u Pathway visualization of gene-expression data Global comparisons of metabolic networks Bioinformatics 18: S 225 2002
 Pathway/Genome DBs Created by External Users SRI International Bioinformatics l 600+ licensees -- 50 groups applying software to 100+ organisms l. Software freely available to academics; Each PGDB owned by its creator l. Saccharomyces cerevisiae, SGD project, Stanford University l pathway. yeastgenome. org/biocyc/ l. TAIR, Carnegie Institution of Washington Arabidopsis. org: 1555 ldicty. Base, Northwestern University l. Gramene. DB, Cold Spring Harbor Laboratory l. Planned: l CGD (Candida albicans), Stanford University l MGD (Mouse), Jackson Laboratory l RGD (Rat), Medical College of Wisconsin l Worm. Base (C. elegans), Caltech l. Large l l scale users: C. Medigue, Genoscope, 67 PGDBs G. Burger, U Montreal, 20 PGDBs DOE GTL contractors: l G. Church, Harvard, Prochlorococcus marinus MED 4 l Larimer/Uberbacher, ORNL, Shewanella onedensis l J. Keasling, UC Berkeley, Desulfovibrio vulgaris l. Fiona Brinkman, Simon Fraser Univ, Pseudomonas aeruginosa l
Pathway/Genome DBs Created by External Users SRI International Bioinformatics l 600+ licensees -- 50 groups applying software to 100+ organisms l. Software freely available to academics; Each PGDB owned by its creator l. Saccharomyces cerevisiae, SGD project, Stanford University l pathway. yeastgenome. org/biocyc/ l. TAIR, Carnegie Institution of Washington Arabidopsis. org: 1555 ldicty. Base, Northwestern University l. Gramene. DB, Cold Spring Harbor Laboratory l. Planned: l CGD (Candida albicans), Stanford University l MGD (Mouse), Jackson Laboratory l RGD (Rat), Medical College of Wisconsin l Worm. Base (C. elegans), Caltech l. Large l l scale users: C. Medigue, Genoscope, 67 PGDBs G. Burger, U Montreal, 20 PGDBs DOE GTL contractors: l G. Church, Harvard, Prochlorococcus marinus MED 4 l Larimer/Uberbacher, ORNL, Shewanella onedensis l J. Keasling, UC Berkeley, Desulfovibrio vulgaris l. Fiona Brinkman, Simon Fraser Univ, Pseudomonas aeruginosa l
 Terminology l “Database” SRI International Bioinformatics = “DB” = “Knowledge Base” = “KB” = “Pathway/Genome Database” = “PGDB”
Terminology l “Database” SRI International Bioinformatics = “DB” = “Knowledge Base” = “KB” = “Pathway/Genome Database” = “PGDB”
 Why Create PGDBs? SRI International Bioinformatics l Extract more information from your genome l Create an up-to-date computable information repository about an organism l Perform analyses on the genome and pathway complement of the organism, e. g. , analyses of omics data l Perform comparative analyses with other organisms l Generate a genome poster and metabolic wall chart
Why Create PGDBs? SRI International Bioinformatics l Extract more information from your genome l Create an up-to-date computable information repository about an organism l Perform analyses on the genome and pathway complement of the organism, e. g. , analyses of omics data l Perform comparative analyses with other organisms l Generate a genome poster and metabolic wall chart
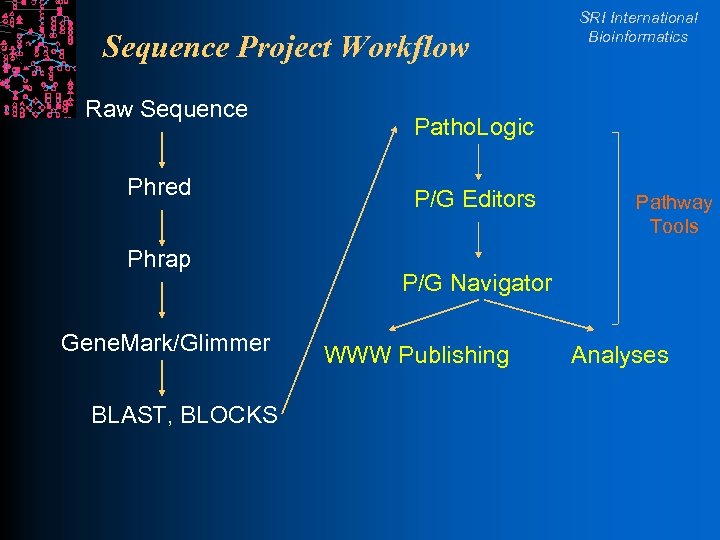 Sequence Project Workflow Raw Sequence Phred Phrap Gene. Mark/Glimmer BLAST, BLOCKS SRI International Bioinformatics Patho. Logic P/G Editors Pathway Tools P/G Navigator WWW Publishing Analyses
Sequence Project Workflow Raw Sequence Phred Phrap Gene. Mark/Glimmer BLAST, BLOCKS SRI International Bioinformatics Patho. Logic P/G Editors Pathway Tools P/G Navigator WWW Publishing Analyses
 SRI International Bioinformatics Meta. Cyc: Metabolic Encyclopedia l Nonredundant metabolic pathway database l Describe a representative sample of every experimentally determined metabolic pathway l Literature-based DB with extensive references and commentary l Pathways, reactions, enzymes, substrates l Jointly developed by SRI and Carnegie Institution Nucleic Acids Research 34: D 511 -D 516 2006
SRI International Bioinformatics Meta. Cyc: Metabolic Encyclopedia l Nonredundant metabolic pathway database l Describe a representative sample of every experimentally determined metabolic pathway l Literature-based DB with extensive references and commentary l Pathways, reactions, enzymes, substrates l Jointly developed by SRI and Carnegie Institution Nucleic Acids Research 34: D 511 -D 516 2006
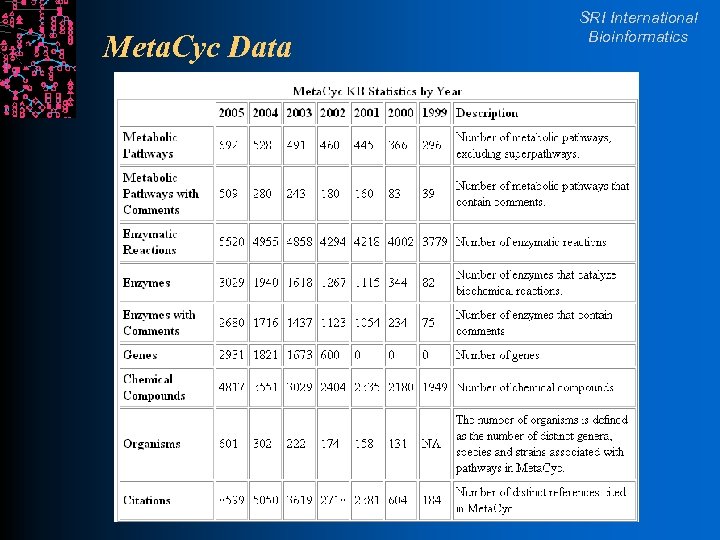 Meta. Cyc Data SRI International Bioinformatics
Meta. Cyc Data SRI International Bioinformatics
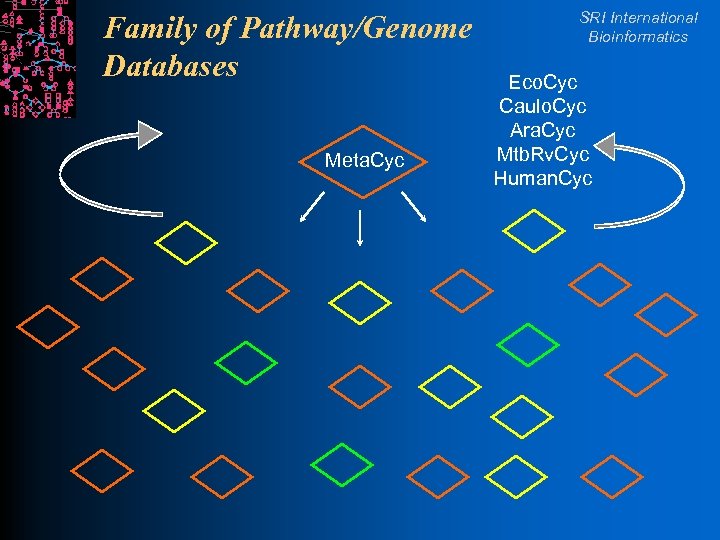 Family of Pathway/Genome Databases Meta. Cyc SRI International Bioinformatics Eco. Cyc Caulo. Cyc Ara. Cyc Mtb. Rv. Cyc Human. Cyc
Family of Pathway/Genome Databases Meta. Cyc SRI International Bioinformatics Eco. Cyc Caulo. Cyc Ara. Cyc Mtb. Rv. Cyc Human. Cyc
 SRI International Bioinformatics Omics Viewer l Import gene expression, proteomics, metabolomics data l Obtain pathway based visualizations of omics data l Numerical spectrum of expression values mapped to a color spectrum l Steps of overview painted with color corresponding to expression level(s) of genes that encode enzyme(s) for that step
SRI International Bioinformatics Omics Viewer l Import gene expression, proteomics, metabolomics data l Obtain pathway based visualizations of omics data l Numerical spectrum of expression values mapped to a color spectrum l Steps of overview painted with color corresponding to expression level(s) of genes that encode enzyme(s) for that step
 SRI International Bioinformatics Environment for Computational Exploration of Genomes l Powerful ontology opens many facets of the biology to computational exploration l Global characterization of metabolic network l Analysis of interface between transport and metabolism l Nutrient analysis of metabolic network
SRI International Bioinformatics Environment for Computational Exploration of Genomes l Powerful ontology opens many facets of the biology to computational exploration l Global characterization of metabolic network l Analysis of interface between transport and metabolism l Nutrient analysis of metabolic network
 SRI International Bioinformatics Pathway Tools Implementation Details l Allegro Common Lisp l Sun, Linux, Windows platforms l Ocelot object database l 300, 000+ lines of code l Lisp-based WWW server at Bio. Cyc. org l Manages 205 PGDBs
SRI International Bioinformatics Pathway Tools Implementation Details l Allegro Common Lisp l Sun, Linux, Windows platforms l Ocelot object database l 300, 000+ lines of code l Lisp-based WWW server at Bio. Cyc. org l Manages 205 PGDBs
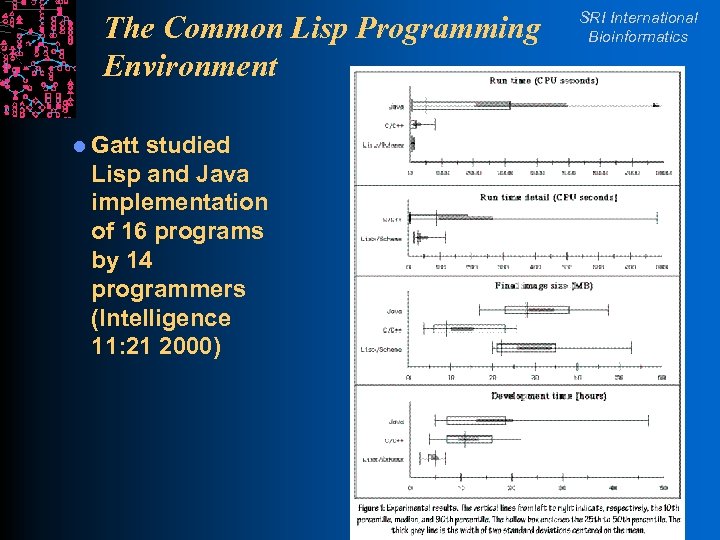 The Common Lisp Programming Environment l Gatt studied Lisp and Java implementation of 16 programs by 14 programmers (Intelligence 11: 21 2000) SRI International Bioinformatics
The Common Lisp Programming Environment l Gatt studied Lisp and Java implementation of 16 programs by 14 programmers (Intelligence 11: 21 2000) SRI International Bioinformatics
 Survey l Please SRI International Bioinformatics complete survey at end of each day
Survey l Please SRI International Bioinformatics complete survey at end of each day
 PGDB(s) That You Build l Before SRI International Bioinformatics you leave l Tar up your PGDB directory and FTP it home, email it home, or copy it to flash disk l We will create a backup copy of your PGDB directory if the directory is still there at the end of the tutorial l Delete the PGDB directory if you don’t want us to back it up l We will not give the backed up data to anyone else
PGDB(s) That You Build l Before SRI International Bioinformatics you leave l Tar up your PGDB directory and FTP it home, email it home, or copy it to flash disk l We will create a backup copy of your PGDB directory if the directory is still there at the end of the tutorial l Delete the PGDB directory if you don’t want us to back it up l We will not give the backed up data to anyone else
 Summary SRI International Bioinformatics l Pathway Tools and Pathway/Genome Databases l Not just for pathways! l Computational inferences u Operons, metabolic pathways, pathway hole fillers Editing tools l Analysis tools: Omics data on pathways l Web publishing of PGDBs l l Main classes of users: l Develop PGDB to extract more information from genome for genome paper l Develop a model-organism DB for the organism that is updated regularly and published on the web
Summary SRI International Bioinformatics l Pathway Tools and Pathway/Genome Databases l Not just for pathways! l Computational inferences u Operons, metabolic pathways, pathway hole fillers Editing tools l Analysis tools: Omics data on pathways l Web publishing of PGDBs l l Main classes of users: l Develop PGDB to extract more information from genome for genome paper l Develop a model-organism DB for the organism that is updated regularly and published on the web
 Information Sources l Pathway Tools User’s Guide l /root/aicexport/ecocyc/genopath/released/doc/manuals/userguide 1. p df u u l l Pathway/Genome Navigator Appendix A: Guide to the Pathway Tools Schema /toot/aicexport/ecocyc/genopath/released/doc/manuals/userguide 2. p df u l SRI International Bioinformatics Patho. Logic, Editing Tools NOTE: Location of the aic-export directory can vary across different computers Pathway Tools Web Site l http: //bioinformatics. ai. sri. com/ptools/ l Publications, programming examples, etc.
Information Sources l Pathway Tools User’s Guide l /root/aicexport/ecocyc/genopath/released/doc/manuals/userguide 1. p df u u l l Pathway/Genome Navigator Appendix A: Guide to the Pathway Tools Schema /toot/aicexport/ecocyc/genopath/released/doc/manuals/userguide 2. p df u l SRI International Bioinformatics Patho. Logic, Editing Tools NOTE: Location of the aic-export directory can vary across different computers Pathway Tools Web Site l http: //bioinformatics. ai. sri. com/ptools/ l Publications, programming examples, etc.


