7cab1ed43107b8fa8e3e2646dbab206d.ppt
- Количество слайдов: 17
 Overview of the Encyclopedia of Life (EOL) Project
Overview of the Encyclopedia of Life (EOL) Project
 Background • Biology has become a data driven science • We have the blueprint (genomes) of over 800 organisms • This number will increase rapidly to the point in 5 -10 years where your blueprint becomes a tool in your medical diagnosis • First we must understand the buildings (proteins) that control life’s processes • EOL strives to be the 21 st century “Britannica” that everyone will turn to
Background • Biology has become a data driven science • We have the blueprint (genomes) of over 800 organisms • This number will increase rapidly to the point in 5 -10 years where your blueprint becomes a tool in your medical diagnosis • First we must understand the buildings (proteins) that control life’s processes • EOL strives to be the 21 st century “Britannica” that everyone will turn to
 EOL Project Description • The Encyclopedia of Life is a joint development of the San Diego Supercomputer Center (SDSC) and scientists and biological resources worldwide • EOL involves SDSC staff from HPC, DAKS, Grids and clusters and visualization • EOL has three parts: 1. Putative functional and 3 -D structure assignment through the largest computation ever attempted 2. True API level integration with key biological resources 3. A focus for future collaborative developments via the EOL Notebook
EOL Project Description • The Encyclopedia of Life is a joint development of the San Diego Supercomputer Center (SDSC) and scientists and biological resources worldwide • EOL involves SDSC staff from HPC, DAKS, Grids and clusters and visualization • EOL has three parts: 1. Putative functional and 3 -D structure assignment through the largest computation ever attempted 2. True API level integration with key biological resources 3. A focus for future collaborative developments via the EOL Notebook
 Type of Questions to be Addressed by EOL • If a knockout gene in arabidopsis leads to an average phenotypic response of 10% increased growth, will the same likely happen in rice? • Is protein X found in anthrax? • Is protein X a drug target, that is, does it exist predominantly in pathogenic bacteria of is it found in eukaryotes also? • Has caspase-1, a protein involved in cell death and aging been identified in any plants, if so what species and do the proposed protein structures look similar? • Give me all available information on caspase-1
Type of Questions to be Addressed by EOL • If a knockout gene in arabidopsis leads to an average phenotypic response of 10% increased growth, will the same likely happen in rice? • Is protein X found in anthrax? • Is protein X a drug target, that is, does it exist predominantly in pathogenic bacteria of is it found in eukaryotes also? • Has caspase-1, a protein involved in cell death and aging been identified in any plants, if so what species and do the proposed protein structures look similar? • Give me all available information on caspase-1
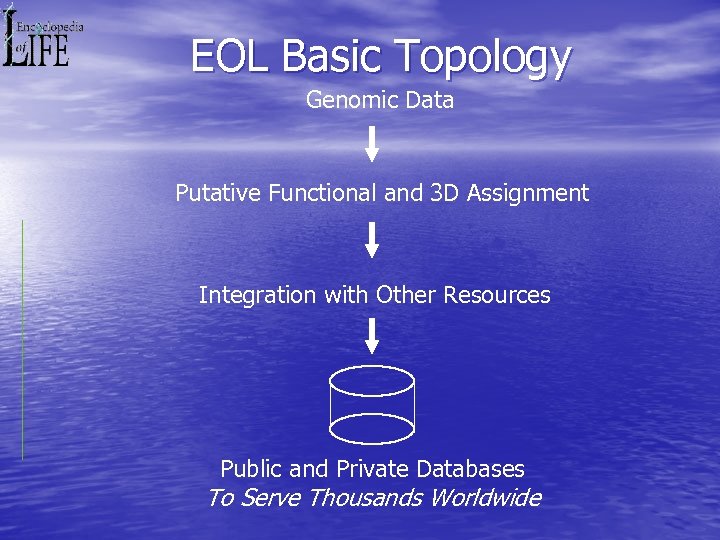 EOL Basic Topology Genomic Data Putative Functional and 3 D Assignment Integration with Other Resources Public and Private Databases To Serve Thousands Worldwide
EOL Basic Topology Genomic Data Putative Functional and 3 D Assignment Integration with Other Resources Public and Private Databases To Serve Thousands Worldwide
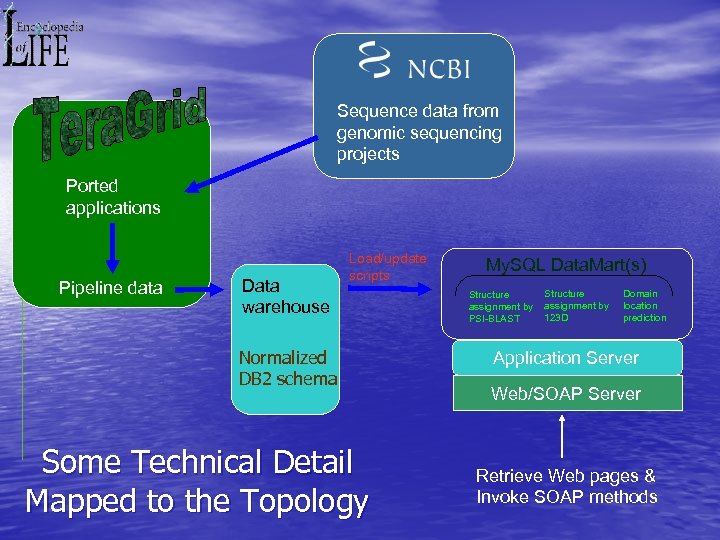 Sequence data from genomic sequencing projects Ported applications Pipeline data Data warehouse Load/update scripts Normalized DB 2 schema Some Technical Detail Mapped to the Topology My. SQL Data. Mart(s) Structure assignment by PSI-BLAST Structure assignment by 123 D Domain location prediction Application Server Web/SOAP Server Retrieve Web pages & Invoke SOAP methods
Sequence data from genomic sequencing projects Ported applications Pipeline data Data warehouse Load/update scripts Normalized DB 2 schema Some Technical Detail Mapped to the Topology My. SQL Data. Mart(s) Structure assignment by PSI-BLAST Structure assignment by 123 D Domain location prediction Application Server Web/SOAP Server Retrieve Web pages & Invoke SOAP methods
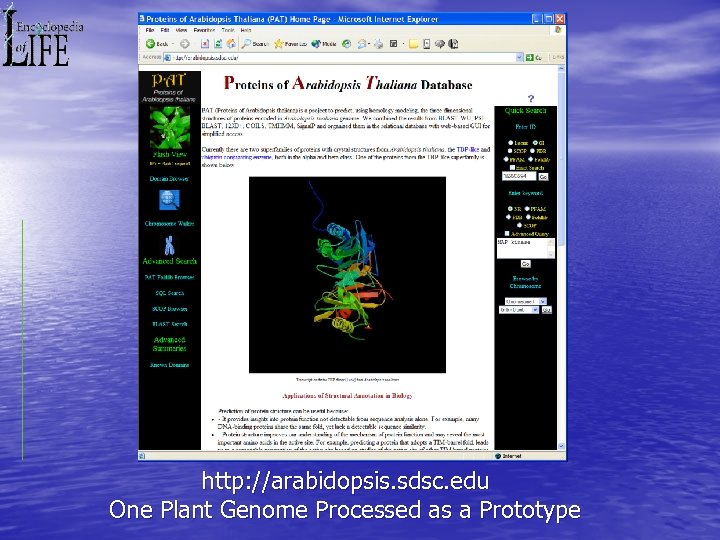 http: //arabidopsis. sdsc. edu One Plant Genome Processed as a Prototype
http: //arabidopsis. sdsc. edu One Plant Genome Processed as a Prototype
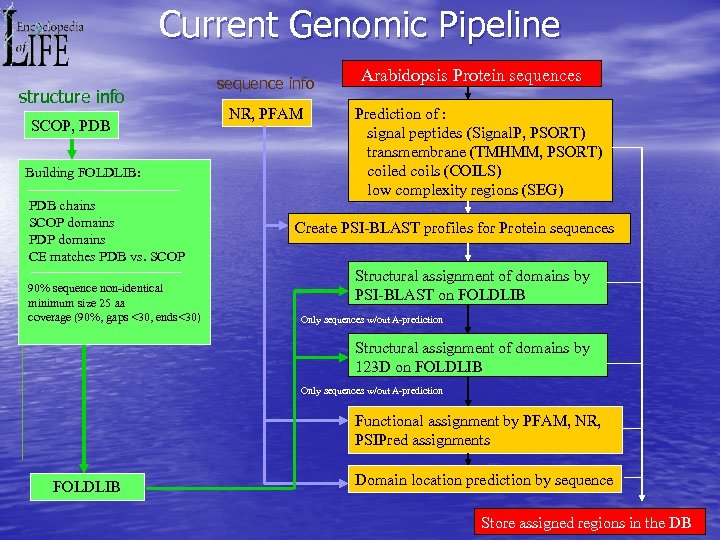 Current Genomic Pipeline structure info SCOP, PDB Building FOLDLIB: PDB chains SCOP domains PDP domains CE matches PDB vs. SCOP 90% sequence non-identical minimum size 25 aa coverage (90%, gaps <30, ends<30) sequence info NR, PFAM Arabidopsis Protein sequences Prediction of : signal peptides (Signal. P, PSORT) transmembrane (TMHMM, PSORT) coiled coils (COILS) low complexity regions (SEG) Create PSI-BLAST profiles for Protein sequences Structural assignment of domains by PSI-BLAST on FOLDLIB Only sequences w/out A-prediction Structural assignment of domains by 123 D on FOLDLIB Only sequences w/out A-prediction Functional assignment by PFAM, NR, PSIPred assignments FOLDLIB Domain location prediction by sequence Store assigned regions in the DB
Current Genomic Pipeline structure info SCOP, PDB Building FOLDLIB: PDB chains SCOP domains PDP domains CE matches PDB vs. SCOP 90% sequence non-identical minimum size 25 aa coverage (90%, gaps <30, ends<30) sequence info NR, PFAM Arabidopsis Protein sequences Prediction of : signal peptides (Signal. P, PSORT) transmembrane (TMHMM, PSORT) coiled coils (COILS) low complexity regions (SEG) Create PSI-BLAST profiles for Protein sequences Structural assignment of domains by PSI-BLAST on FOLDLIB Only sequences w/out A-prediction Structural assignment of domains by 123 D on FOLDLIB Only sequences w/out A-prediction Functional assignment by PFAM, NR, PSIPred assignments FOLDLIB Domain location prediction by sequence Store assigned regions in the DB
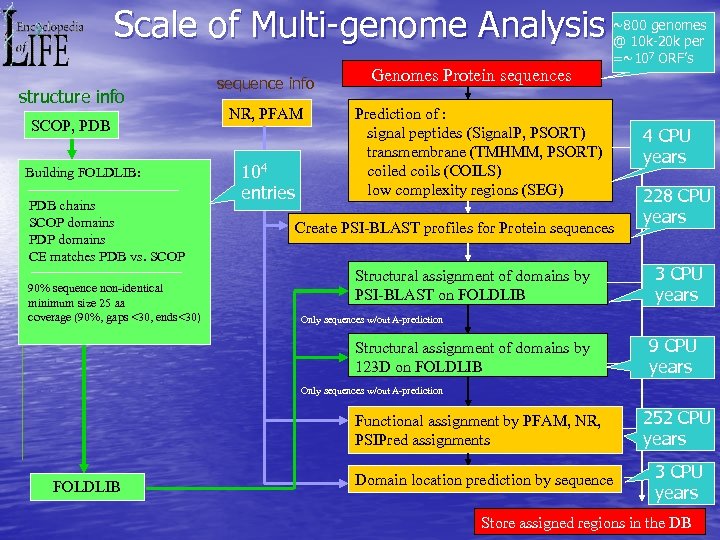 Scale of Multi-genome Analysis structure info SCOP, PDB Building FOLDLIB: PDB chains SCOP domains PDP domains CE matches PDB vs. SCOP 90% sequence non-identical minimum size 25 aa coverage (90%, gaps <30, ends<30) sequence info NR, PFAM 104 entries Genomes Protein sequences ~800 genomes @ 10 k-20 k per =~107 ORF’s Prediction of : signal peptides (Signal. P, PSORT) transmembrane (TMHMM, PSORT) coiled coils (COILS) low complexity regions (SEG) Create PSI-BLAST profiles for Protein sequences Structural assignment of domains by PSI-BLAST on FOLDLIB 4 CPU years 228 CPU years 3 CPU years Only sequences w/out A-prediction Structural assignment of domains by 123 D on FOLDLIB 9 CPU years Only sequences w/out A-prediction Functional assignment by PFAM, NR, PSIPred assignments FOLDLIB Domain location prediction by sequence 252 CPU years 3 CPU years Store assigned regions in the DB
Scale of Multi-genome Analysis structure info SCOP, PDB Building FOLDLIB: PDB chains SCOP domains PDP domains CE matches PDB vs. SCOP 90% sequence non-identical minimum size 25 aa coverage (90%, gaps <30, ends<30) sequence info NR, PFAM 104 entries Genomes Protein sequences ~800 genomes @ 10 k-20 k per =~107 ORF’s Prediction of : signal peptides (Signal. P, PSORT) transmembrane (TMHMM, PSORT) coiled coils (COILS) low complexity regions (SEG) Create PSI-BLAST profiles for Protein sequences Structural assignment of domains by PSI-BLAST on FOLDLIB 4 CPU years 228 CPU years 3 CPU years Only sequences w/out A-prediction Structural assignment of domains by 123 D on FOLDLIB 9 CPU years Only sequences w/out A-prediction Functional assignment by PFAM, NR, PSIPred assignments FOLDLIB Domain location prediction by sequence 252 CPU years 3 CPU years Store assigned regions in the DB
 Tera. Grid application Technical aspects: • Excellent charter application for the Tera. Grid project! • Good demonstration of producing practical output from Tera. Grid computing: scientific papers and an extensive web site and services will be produced • Software pipeline now a proven technique and a sure bet • Can be implemented in the fastest possible time; project already initialized
Tera. Grid application Technical aspects: • Excellent charter application for the Tera. Grid project! • Good demonstration of producing practical output from Tera. Grid computing: scientific papers and an extensive web site and services will be produced • Software pipeline now a proven technique and a sure bet • Can be implemented in the fastest possible time; project already initialized
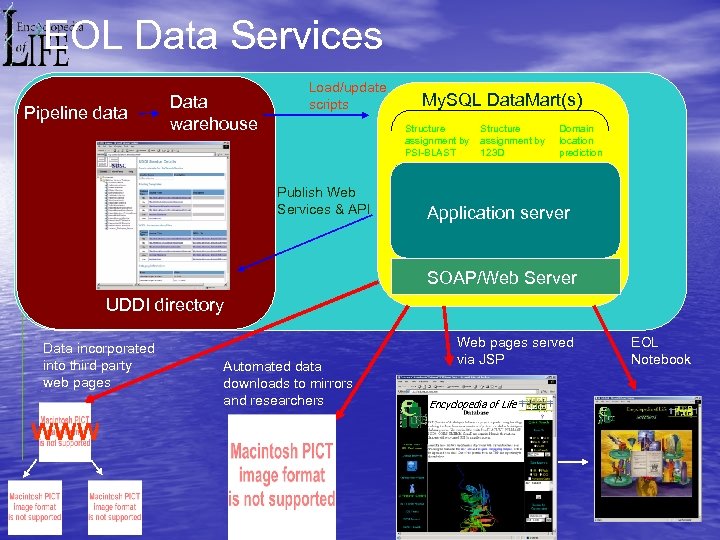 EOL Data Services Pipeline data Data warehouse Load/update scripts My. SQL Data. Mart(s) Structure assignment by PSI-BLAST Publish Web Services & API Structure assignment by 123 D Domain location prediction Application server SOAP/Web Server UDDI directory Data incorporated into third party web pages WWW Automated data downloads to mirrors and researchers Web pages served via JSP Encyclopedia of Life EOL Notebook
EOL Data Services Pipeline data Data warehouse Load/update scripts My. SQL Data. Mart(s) Structure assignment by PSI-BLAST Publish Web Services & API Structure assignment by 123 D Domain location prediction Application server SOAP/Web Server UDDI directory Data incorporated into third party web pages WWW Automated data downloads to mirrors and researchers Web pages served via JSP Encyclopedia of Life EOL Notebook
 Basic Web Interface Encyclopedia of Life MS Internet Explorer Netscape 4. 7/6. 1 Mozilla v 1. 0 Microsoft Windows Opera MS Internet Explorer Netscape 4. 7/6. 1 Mozilla v 1. 0 Opera Apple Macintosh Netscape 4. 7/6. 1 Mozilla v 1. 0 Opera Linux MS Internet Explorer Netscape 4. 7/6. 1 Mozilla v 1. 0 Opera Win-CE and pen-based devices
Basic Web Interface Encyclopedia of Life MS Internet Explorer Netscape 4. 7/6. 1 Mozilla v 1. 0 Microsoft Windows Opera MS Internet Explorer Netscape 4. 7/6. 1 Mozilla v 1. 0 Opera Apple Macintosh Netscape 4. 7/6. 1 Mozilla v 1. 0 Opera Linux MS Internet Explorer Netscape 4. 7/6. 1 Mozilla v 1. 0 Opera Win-CE and pen-based devices
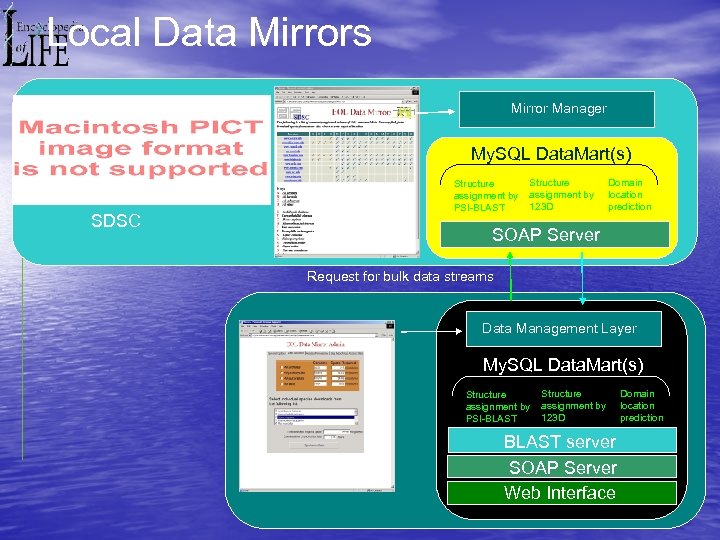 Local Data Mirrors Mirror Manager My. SQL Data. Mart(s) SDSC Structure assignment by PSI-BLAST Structure assignment by 123 D Domain location prediction SOAP Server Request for bulk data streams Data Management Layer My. SQL Data. Mart(s) Structure assignment by PSI-BLAST Structure assignment by 123 D BLAST server SOAP Server Web Interface Domain location prediction
Local Data Mirrors Mirror Manager My. SQL Data. Mart(s) SDSC Structure assignment by PSI-BLAST Structure assignment by 123 D Domain location prediction SOAP Server Request for bulk data streams Data Management Layer My. SQL Data. Mart(s) Structure assignment by PSI-BLAST Structure assignment by 123 D BLAST server SOAP Server Web Interface Domain location prediction
 Local Data Mirrors • Support for server platforms, i. e. • • – Sparc Solaris – IRIX – Linux Based on My. SQL + Apache because of availability Automated mirror registration and listing User-friendly admin for mirror maintenance Means of metering of data usage per species data stream to generate revenue from industry
Local Data Mirrors • Support for server platforms, i. e. • • – Sparc Solaris – IRIX – Linux Based on My. SQL + Apache because of availability Automated mirror registration and listing User-friendly admin for mirror maintenance Means of metering of data usage per species data stream to generate revenue from industry
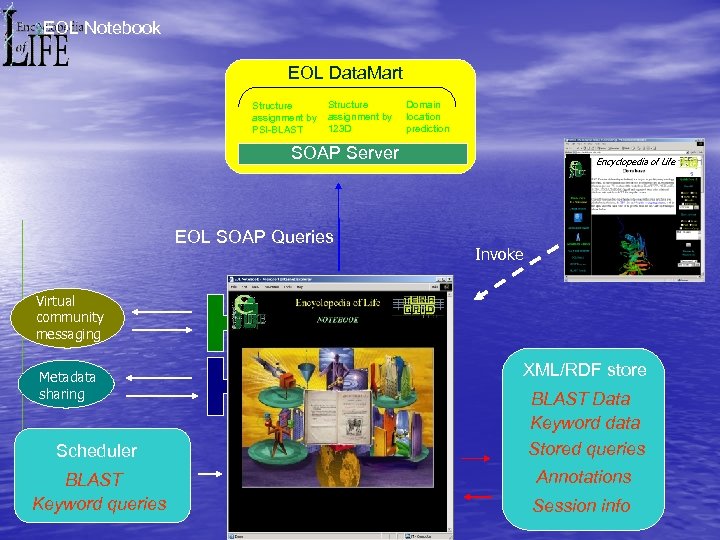 EOL Notebook EOL Data. Mart Structure assignment by PSI-BLAST Structure assignment by 123 D Domain location prediction SOAP Server EOL SOAP Queries Encyclopedia of Life Invoke Virtual community messaging Metadata sharing Scheduler BLAST Keyword queries XML/RDF store BLAST Data Keyword data Stored queries Annotations Session info
EOL Notebook EOL Data. Mart Structure assignment by PSI-BLAST Structure assignment by 123 D Domain location prediction SOAP Server EOL SOAP Queries Encyclopedia of Life Invoke Virtual community messaging Metadata sharing Scheduler BLAST Keyword queries XML/RDF store BLAST Data Keyword data Stored queries Annotations Session info
 EOL Notebook • Provides a consistent, advanced, cross-platform GUI to view returned data from queries to the EOL database via Web Services. • Provide persistence of both queries and returned data via local XML database • Provide mechanism to enable unattended, scheduled, periodic queries • Provides means to annotate data and results and share those with others, in effect a scientific Napster • Provide means to create virtual community(s)
EOL Notebook • Provides a consistent, advanced, cross-platform GUI to view returned data from queries to the EOL database via Web Services. • Provide persistence of both queries and returned data via local XML database • Provide mechanism to enable unattended, scheduled, periodic queries • Provides means to annotate data and results and share those with others, in effect a scientific Napster • Provide means to create virtual community(s)
 Summary 1. EOL is a large-scale data analysis project, one of the largest biological computations attempted, whose results will be eagerly awaited by an enormous number of biologists 2. Core scientific analysis techniques well-proven in existing arabidopsis project 3. It’s a perfect choice as a charter application for the Tera. Grid • Very large scale computation • Pipeline-type computations well suited to the Grid platform • High visibility and very practical use of Tera. Grid results • Tera. Grid name will become associated with high quality data analysis
Summary 1. EOL is a large-scale data analysis project, one of the largest biological computations attempted, whose results will be eagerly awaited by an enormous number of biologists 2. Core scientific analysis techniques well-proven in existing arabidopsis project 3. It’s a perfect choice as a charter application for the Tera. Grid • Very large scale computation • Pipeline-type computations well suited to the Grid platform • High visibility and very practical use of Tera. Grid results • Tera. Grid name will become associated with high quality data analysis


