7b998c03c7dfc078afdc332a7c8411a6.ppt
- Количество слайдов: 43

Overview of Genome Databases Peter D. Karp, Ph. D. SRI International pkarp@ai. sri. com www-db. stanford. edu/dbseminar/seminar. html

Talk Overview l Definition of bioinformatics l Motivations l Issues for genome databases in building genome databases
Definition of Bioinformatics l Computational techniques for management and analysis of biological data and knowledge l Methods for disseminating, archiving, interpreting, and mining scientific information l Computational l Genome theories of biology Databases is a subfield of bioinformatics
Motivations for Bioinformatics l Growth in molecular-biology knowledge (literature) l Genomics 1. Study of genomes through DNA sequencing 2. Industrial Biology
Example Genomics Datatypes l Genome sequences l DOE Joint Genome Institute u u l Gene 511 M bases in Dec 2001 11. 97 G bases since Mar 1999 and protein expression data l Protein-protein l Protein interaction data 3 -D structures
Genome Databases l Experimental data l Archive experimental datasets l Retrieving past experimental results should be faster than repeating the experiment l Capture alternative analyses l Lots of data, simpler semantics l Computational symbolic theories l Complex theories become too large to be grasped by a single mind l The database is theory l Biology is very much concerned with qualitative relationships l Less data, more complex semantics

Bioinformatics l l Distinct intellectual field at the intersection of CS and molecular biology Distinct field because researchers in the field must know CS, biology, and bioinformatics l Spectrum from CS research to biology service l Rich source of challenging CS problems l Large, noisy, complex data-sets and knowledge-sets l Biologists and funding agencies demand working solutions
Bioinformatics Research l algorithms + data structures = programs l algorithms + databases = discoveries l Combine sophisticated algorithms with the right content: l Properly structured l Carefully curated l Relevant data fields l Proper amount of data

Reference on Major Genome Databases l Nucleic Acids Research Database Issue l http: //nar. oupjournals. org/content/vol 30/issue 1/ l 112 databases

Questions to Ask of a New Genome Database

What are Database Goals and Requirements? l What l Who problems will database be used to solve? are the users and what is their expertise?

What is its Organizing Principle? l Different DBs partition the space of genome information in different dimensions l Experimental l Organism methods (Genbank, PDB) (Eco. Cyc, Flybase)

What is its Level of Interpretation? l Laboratory l Primary l Review l Does data literature (Genbank) (Swiss. Prot, Meta. Cyc) DB model disagreement?

What are its Semantics and Content? l What l How entities and relationships does it model? does its content overlap with similar DBs? l How many entities of each type are present? l Sparseness of attributes and statistics on attribute values
What are Sources of its Data? l Potential information sources l Laboratory instruments l Scientific literature u u l Manual entry Natural-language text mining Direct submission from the scientific community u Genbank l Modification policy l DB staff only l Submission of new entries by scientific community l Update access by scientific community

What DBMS is Employed? l None l Relational l Object oriented l Frame knowledge representation system

Distribution / User Access l Multiple distribution forms enhance access l Browsing access with visualization tools l API l Portability

What Validation Approaches are Employed? l None l Declarative consistency constraints l Programmatic l Internal l What consistency checking vs external consistency checking types of systematic errors might DB contain?
Database Documentation l Schema and its semantics l Format l API l Data acquisition techniques l Validation techniques l Size of different classes l Coverage of subject matter l Sparseness of attributes l Error rates l Update frequency
Relationship of Database Field to Bioinformatics l Scientists generally unaware of basic DB principles l Complex queries vs click-at-a-time access l Data model l Defined semantics for DB fields l Controlled vocabularies l Regular syntax for flatfiles l Automated consistency checking l Most biologists take one programming class l Evolution of typical genome database l Finer points of DB research off their radar screen l Handfull of DB researchers work in bioinformatics
Database Field l For many years, the majority of bioinformatics DBs did not employ a DBMS l Flatfiles were the rule l Scientists want to see the data directly l Commercial DBMSs too expensive, too complex l DBAs too expensive l Most scientists do not understand l Differences between BA, MS, Ph. D in CS l CS research vs applications l Implications for project planning, funding, bioinformatics research
Recommendation l Teaching scientists programming is not enough l Teaching scientists how to build a DBMS is irrelevant l Teach scientists basic aspects of databases and symbolic computing l Database requirements analysis l Data models, schema design l Knowledge representation, ontologies l Formal grammars l Complex queries l Database interoperability

Bio. SPICE Bioinformatics Database Warehouse Peter Karp, Dave Stringer-Calvert, Tom Lee, Kemal Sonmez SRI International http: //www. Bio. SPICE. org/

Project Goal l Create a toolkit for constructing bioinformatics database warehouses that collect together a set of bioinformatics databases into one physical DBMS
Motivations l l l Important bioinformatics problems require access to multiple bioinformatics databases Hundreds of bioinformatics databases exist l Nucleic Acids Research 30(1) 2002 – DB issue l Nucleic Acids Research DB list: 350 DBs at http: //www 3. oup. co. uk/nar/database/a/ Different problems require different sets of databases

Motivations l Combining multiple databases allows for data verification and complementation l Simulation problems require access to data on pathways, enzymes, reactions, genetic regulation

Why is the Multidatabase Approach Not Sufficient? l l l Multidatabase query approaches assume databases are in a DBMS Internet bandwidth limits query throughput Most sites that do operate DBMSs do not allow remote SQL access because of security and loading concerns Control data stability Need to capture, integrate and publish locally produced data of different types Multidatabase and Warehouse approaches complementary
Scenario 1 l Bio. SPICE scientist wants to model multiple metabolic pathways in a given organism l Enumerate pathways and reactions l What enzymes catalyze each reaction? l What genes code for each enzyme? l What control regions regulate each gene?

Approach l l Oracle and My. SQL implementations Warehouse schema defines many bioinformatics datatypes Create loaders for public bioinformatics DBs l Parse file format for the DB l Semantic transformations l Insert database into warehouse tables Warehouse query access mechanisms l SQL queries via Perl, ODBC, OAA

Example: Swiss-Prot DB l l Version 40. 0 describes 101 K proteins in a 320 MB file Each protein described as one block of records (an entry) in a large text file Loader tool parses file one entry at a time Creates new entries in a set of warehouse tables

Warehouse Schema l l Manages many bioinformatics datatypes simultaneously l Pathways, Reactions, Chemicals l Proteins, Genes, Replicons l Citations, Organisms l Links to external databases Each type of warehouse object implemented through one or more relational tables (currently 43)

Warehouse Schema l Databases on our wish list: l Genbank (nucleotide sequences) l Protein expression database l Protein-protein interactions database l Gene expression database l NCBI Taxonomy database l Gene Ontology l CMR

Warehouse Schema l l l Manages multiple datasets simultaneously l Dataset = Single version of a database Support alternative measurements and viewpoints Version comparison Multiple software tools or experiments that require access to different versions Each dataset is a warehouse entity Every warehouse object is registered in a dataset

Warehouse Schema l l l Different databases storing the same biological types are coerced into same warehouse tables Design of most datatypes inspired by multiple databases Representational tricks to decrease schema bloat l Single space of primary keys l Single set of satellite tables such as for synonyms, citations, comments, etc.

Warehouse Schema l Examples Protein data from Swiss-Prot, Tr. EMBL, KEGG, and Eco. Cyc all loaded into same relational tables l Pathway data from Meta. Cyc and KEGG are loaded into the same relational tables l
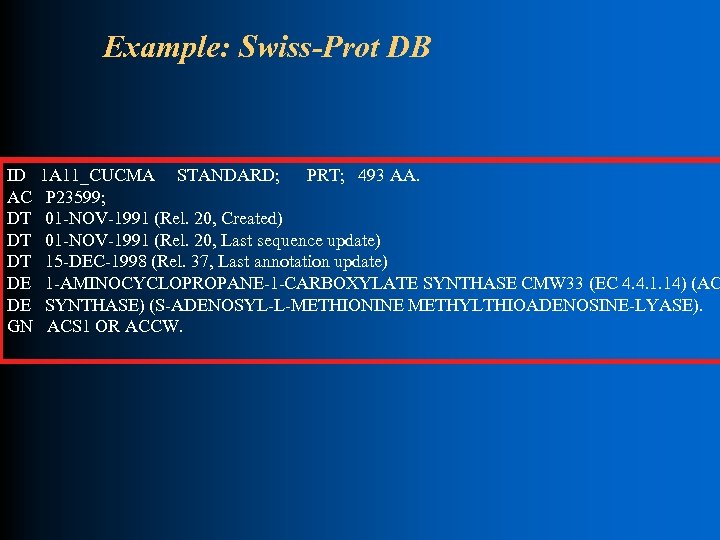
Example: Swiss-Prot DB ID AC DT DT DT DE DE GN 1 A 11_CUCMA STANDARD; PRT; 493 AA. P 23599; 01 -NOV-1991 (Rel. 20, Created) 01 -NOV-1991 (Rel. 20, Last sequence update) 15 -DEC-1998 (Rel. 37, Last annotation update) 1 -AMINOCYCLOPROPANE-1 -CARBOXYLATE SYNTHASE CMW 33 (EC 4. 4. 1. 14) (AC SYNTHASE) (S-ADENOSYL-L-METHIONINE METHYLTHIOADENOSINE-LYASE). ACS 1 OR ACCW.

How Swiss-Prot is Loaded into The Warehouse l Register Swiss-Prot in Datasets table l Create entry in Entry and Protein tables for each Swiss-Prot protein l Satellite tables store l Protein synonyms, citations, comments, accession numbers, organism, sequence features, subunits/complexes, DB links

Protein Table CREATE TABLE Protein ( WID Name AASequence Charge Fragment Molecular. Weight. Calc Molecular. Weight. Exp PICalc PIExp Data. Set. WID ); NUMBER --The warehouse ID of this protein VARCHAR 2(500) --Common name of the protein VARCHAR 2(4000), --Amino-acid sequence for this protei NUMBER, --Charge of the chemical CHAR(1), --Is this protein a fragment or not, NUMBER, --Molecular weight calculated from se NUMBER, --Molecular Weight determined through VARCHAR 2(50), --p. I calculated from its sqeuence. VARCHAR 2(50), --p. I value determined through experim NUMBER --Reference to the data set from whic

Database Loaders l l l Loader tool defined for each DB to be loaded into Warehouse Example loaders available in several languages Loaders l KEGG (C) l Bio. Cyc collection of 15 pathway DBs (C) l Swiss-Prot (Java) l ENZYME (Java)
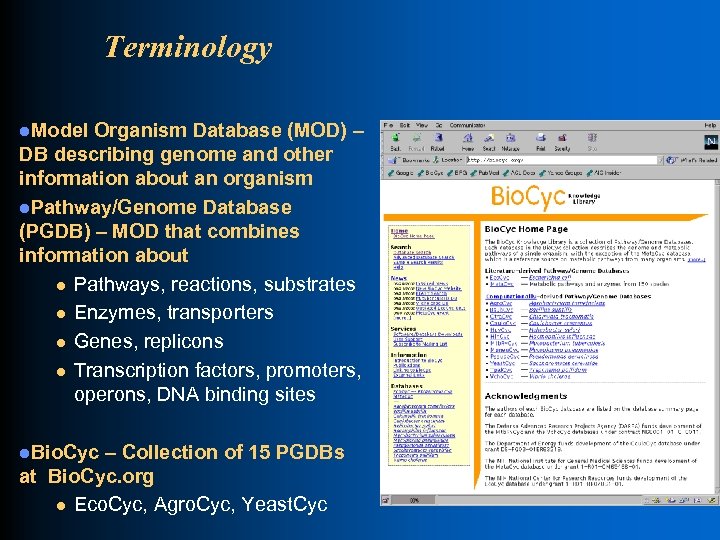
Terminology l. Model Organism Database (MOD) – DB describing genome and other information about an organism l. Pathway/Genome Database (PGDB) – MOD that combines information about l Pathways, reactions, substrates l Enzymes, transporters l Genes, replicons l Transcription factors, promoters, operons, DNA binding sites l. Bio. Cyc – Collection of 15 PGDBs at Bio. Cyc. org l Eco. Cyc, Agro. Cyc, Yeast. Cyc
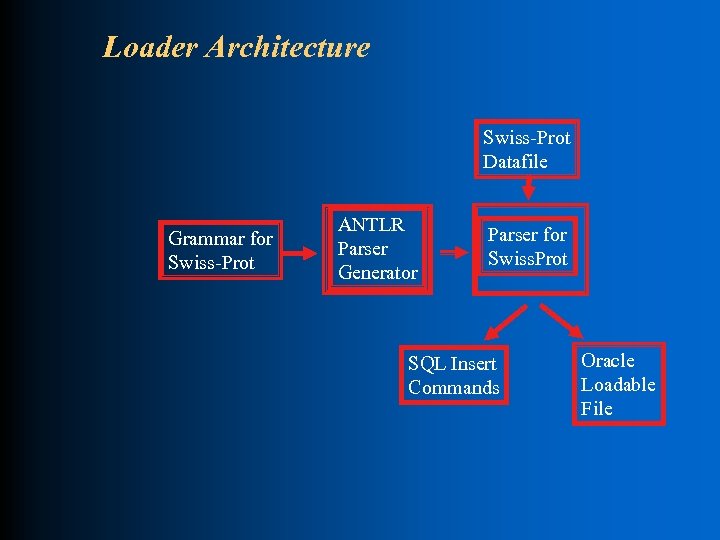
Loader Architecture Swiss-Prot Datafile Grammar for Swiss-Prot ANTLR Parser Generator Parser for Swiss. Prot SQL Insert Commands Oracle Loadable File
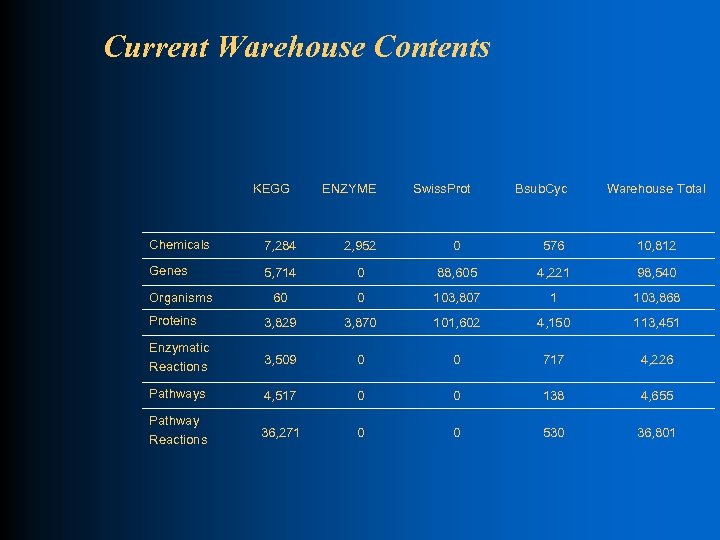
Current Warehouse Contents KEGG ENZYME Swiss. Prot Bsub. Cyc Warehouse Total Chemicals 7, 284 2, 952 0 576 10, 812 Genes 5, 714 0 88, 605 4, 221 98, 540 60 0 103, 807 1 103, 868 Proteins 3, 829 3, 870 101, 602 4, 150 113, 451 Enzymatic Reactions 3, 509 0 0 717 4, 226 Pathways 4, 517 0 0 138 4, 655 Pathway Reactions 36, 271 0 0 530 36, 801 Organisms

Example Warehouse Uses l Check completeness of data sources Count reactions in ENZYME database with (and without) associated protein sequences in SWISS-PROT database: 3870 reactions in ENZYME 1662 reactions (43%) with a sequence in SWISS-PROT 2208 reactions (57%) without a sequence in SWISS-PROT Count #of distinct non-partial EC numbers in SWISS-PROT: 1554 distinct EC numbers in SWISS-PROT (non-partial)
7b998c03c7dfc078afdc332a7c8411a6.ppt