 Origins of Host Specific Populations of Puccinia triticina Revealed by SNP Markers (Preliminary) M. Liu and J. A. Kolmer USDA-ARS Cereal Disease Laboratory, St. Paul, MN 55108
Origins of Host Specific Populations of Puccinia triticina Revealed by SNP Markers (Preliminary) M. Liu and J. A. Kolmer USDA-ARS Cereal Disease Laboratory, St. Paul, MN 55108
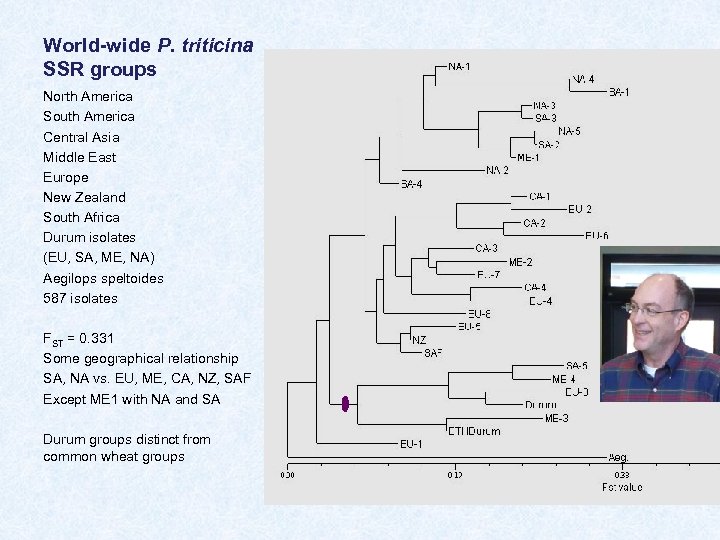 World-wide P. triticina SSR groups North America South America Central Asia Middle East Europe New Zealand South Africa Durum isolates (EU, SA, ME, NA) Aegilops speltoides 587 isolates FST = 0. 331 Some geographical relationship SA, NA vs. EU, ME, CA, NZ, SAF Except ME 1 with NA and SA Durum groups distinct from common wheat groups
World-wide P. triticina SSR groups North America South America Central Asia Middle East Europe New Zealand South Africa Durum isolates (EU, SA, ME, NA) Aegilops speltoides 587 isolates FST = 0. 331 Some geographical relationship SA, NA vs. EU, ME, CA, NZ, SAF Except ME 1 with NA and SA Durum groups distinct from common wheat groups
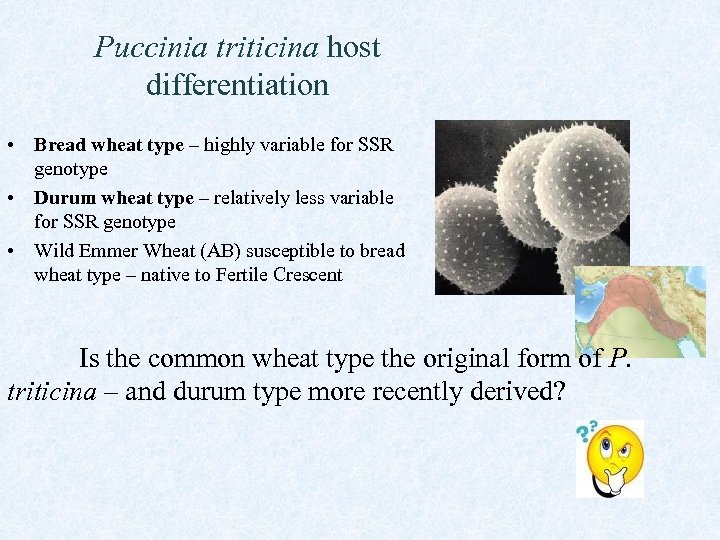 Puccinia triticina host differentiation • Bread wheat type – highly variable for SSR genotype • Durum wheat type – relatively less variable for SSR genotype • Wild Emmer Wheat (AB) susceptible to bread wheat type – native to Fertile Crescent Is the common wheat type the original form of P. triticina – and durum type more recently derived?
Puccinia triticina host differentiation • Bread wheat type – highly variable for SSR genotype • Durum wheat type – relatively less variable for SSR genotype • Wild Emmer Wheat (AB) susceptible to bread wheat type – native to Fertile Crescent Is the common wheat type the original form of P. triticina – and durum type more recently derived?
 Goal: To further infer the evolutionary relationships among populations with the aid of coalescence theory and DNA single nucleotide polymorphism (SNP) markers
Goal: To further infer the evolutionary relationships among populations with the aid of coalescence theory and DNA single nucleotide polymorphism (SNP) markers
 • Why SNP? • Development of SNP markers (poster: theme 1, #30 ) • Preliminary results
• Why SNP? • Development of SNP markers (poster: theme 1, #30 ) • Preliminary results
 • Why SNP?
• Why SNP?
 • Why SNP? ü Ubiquitous — accessible, representative
• Why SNP? ü Ubiquitous — accessible, representative
 • Why SNP? ü Ubiquitous — accessible, representative ü Variable mutation rates ü Suitable to automatic genotyping ü Amiable to sequence-based analytical tools ü Alternative approach
• Why SNP? ü Ubiquitous — accessible, representative ü Variable mutation rates ü Suitable to automatic genotyping ü Amiable to sequence-based analytical tools ü Alternative approach
 • Why SNP? • Development of SNP markers (poster: theme 1, #30 ) • Preliminary results
• Why SNP? • Development of SNP markers (poster: theme 1, #30 ) • Preliminary results
 • Sampling 7
• Sampling 7
 • Preliminary results 100 79 100 7 <50 Clusters of P. triticina populations based on 94 SNPs from three house keeping genes, seven SSR flanking regions and six IGV selected anonymous loci. L=134 CI=0. 709 RI=0. 884
• Preliminary results 100 79 100 7 <50 Clusters of P. triticina populations based on 94 SNPs from three house keeping genes, seven SSR flanking regions and six IGV selected anonymous loci. L=134 CI=0. 709 RI=0. 884
 • Preliminary results Aeg ETH durum 100 78 Durum 99 SA-5 NZ 100 SAF EU-7 CA-3 NA-4 ME-1 NA-1 One of 258367 most parsimonious phylograms based on SNPs from three house keeping genes and six IGV selected anonymous loci. Isolates on durum wheat formed clade. L=271 CI=0. 915 RI=0. 955
• Preliminary results Aeg ETH durum 100 78 Durum 99 SA-5 NZ 100 SAF EU-7 CA-3 NA-4 ME-1 NA-1 One of 258367 most parsimonious phylograms based on SNPs from three house keeping genes and six IGV selected anonymous loci. Isolates on durum wheat formed clade. L=271 CI=0. 915 RI=0. 955
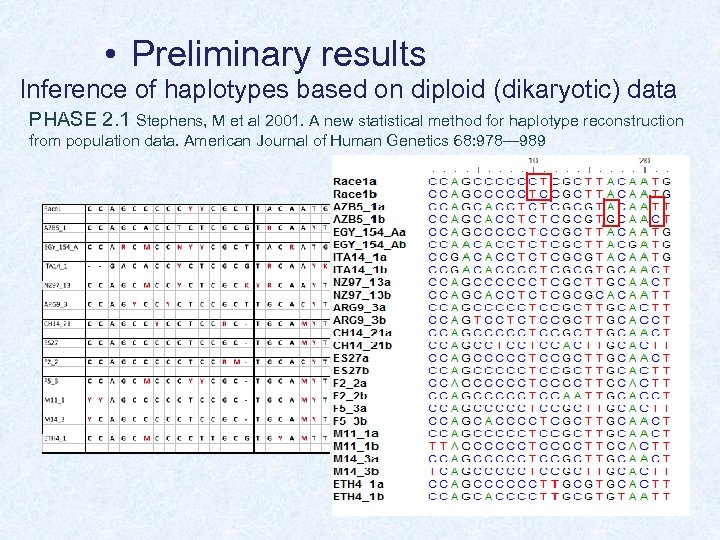 • Preliminary results Inference of haplotypes based on diploid (dikaryotic) data PHASE 2. 1 Stephens, M et al 2001. A new statistical method for haplotype reconstruction from population data. American Journal of Human Genetics 68: 978— 989
• Preliminary results Inference of haplotypes based on diploid (dikaryotic) data PHASE 2. 1 Stephens, M et al 2001. A new statistical method for haplotype reconstruction from population data. American Journal of Human Genetics 68: 978— 989
 Coalescence analysis Carbone Lab IM, IMa 2 Department of Genome Science, U of Washington
Coalescence analysis Carbone Lab IM, IMa 2 Department of Genome Science, U of Washington
 ACKNOWLEDGEMENTS We thank Drs. Les Szabo, John Fellers and Christina Cuomo for facilitating ML to access IGV and Pt whole genome database; Kun Xiao, Jerry Johnson and Kim Phuong Nguyen for technical help.
ACKNOWLEDGEMENTS We thank Drs. Les Szabo, John Fellers and Christina Cuomo for facilitating ML to access IGV and Pt whole genome database; Kun Xiao, Jerry Johnson and Kim Phuong Nguyen for technical help.