76720759a142e5b225f8e5acef7ad696.ppt
- Количество слайдов: 36

OMES & OMICS in ART Moncef Benkhalifa, Ph. D. RBMG. Reproductive Biology & Genetics Technical & Development Director UNILABS. France

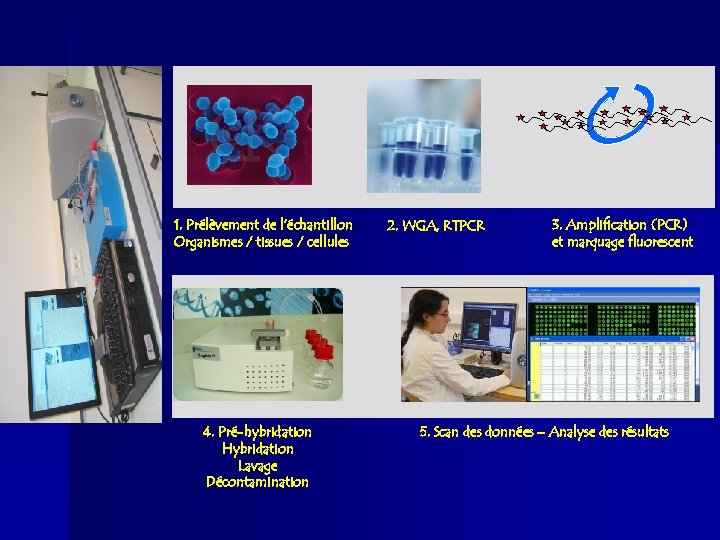
1. Prélèvement de l’échantillon Organismes / tissues / cellules 4. Pré-hybridation Hybridation Lavage Décontamination 2. WGA, RTPCR 3. Amplification (PCR) et marquage fluorescent 5. Scan des données – Analyse des résultats
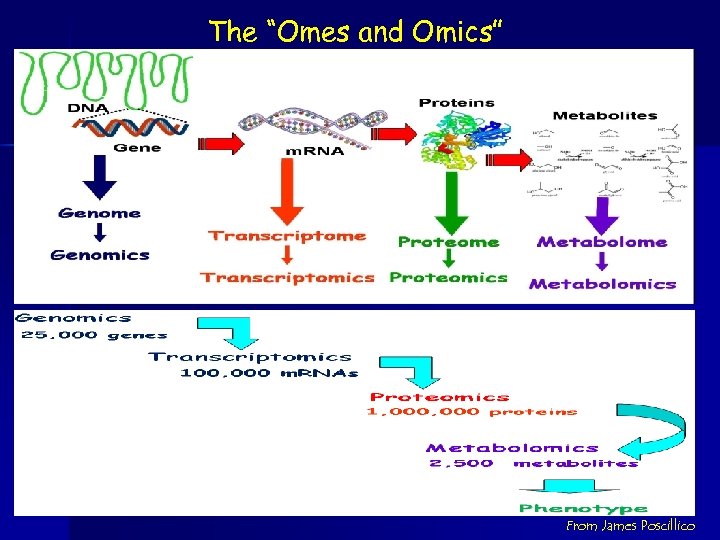
The “Omes and Omics” From James Poscillico
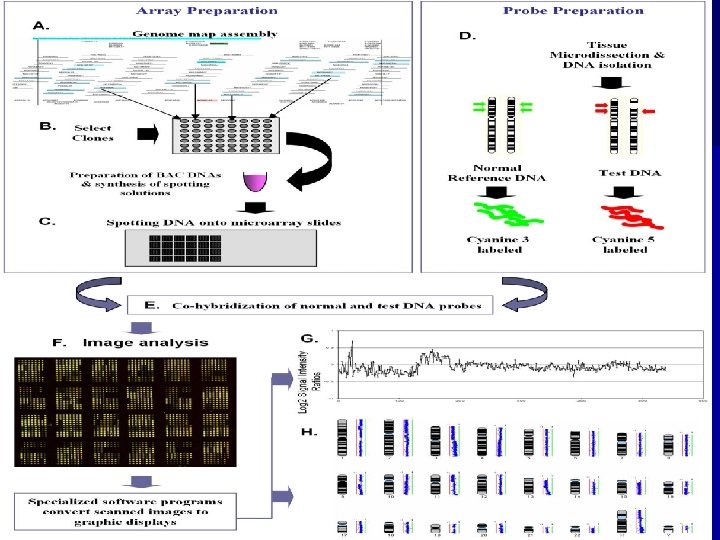
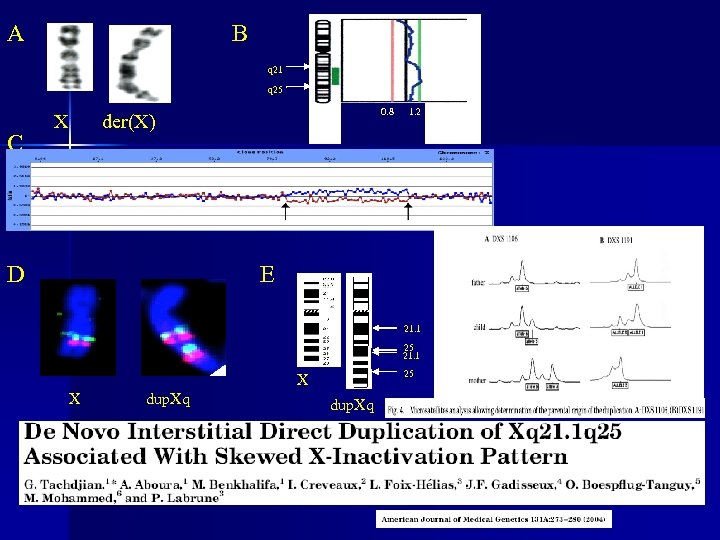
A B q 21 q 25 C 0. 8 der(X) X D 1. 2 E 21. 1 25 X X dup. Xq Tachdjian G, Aboura A, Benkhalifa M et al De novo interstitial direct duplication of Xq 21. 1 q 25 associated with skewed X-inactivation pattern.

: Prenatal Diagnosis, October 2005
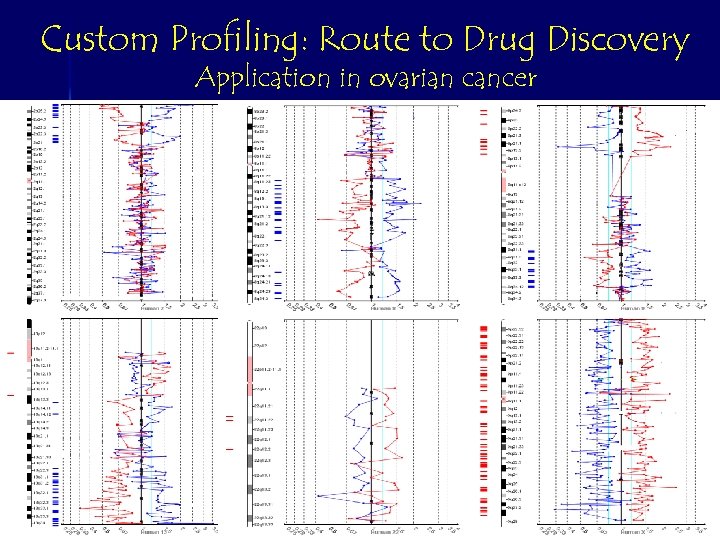
Custom Profiling: Route to Drug Discovery Application in ovarian cancer Chromosome segment 8 q Sample 2 Chromosome 13
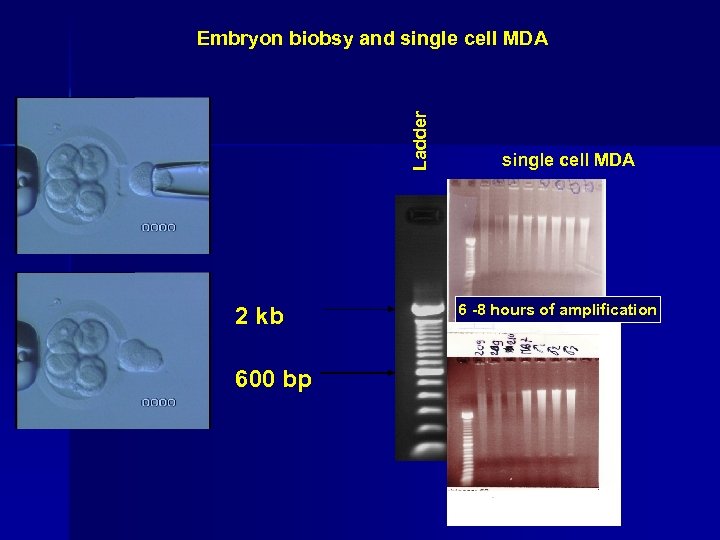
Ladder Embryon biobsy and single cell MDA 2 kb 600 bp single cell MDA 6 -8 hours of amplification
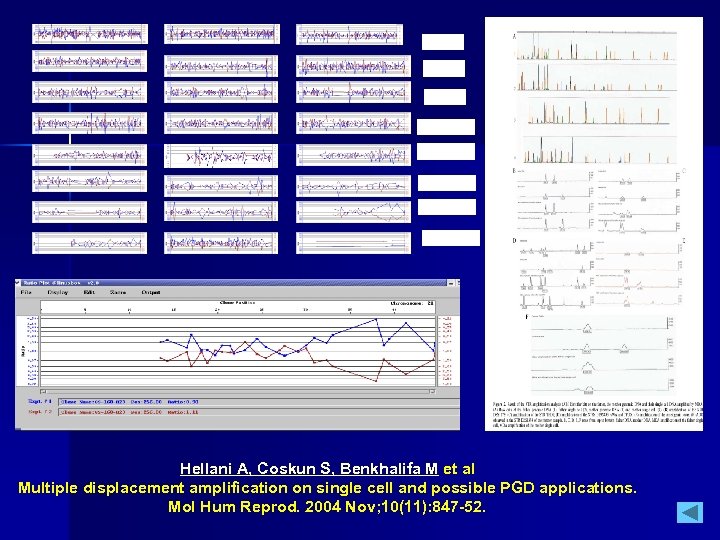
1; 2; 3 4; 5; 6 7; 8; 9 10; 11; 12 13; 14; 15 16; 17; 18 19; 20; 21 22; X; Y Hellani A, Coskun S, Benkhalifa M et al Multiple displacement amplification on single cell and possible PGD applications. Mol Hum Reprod. 2004 Nov; 10(11): 847 -52.
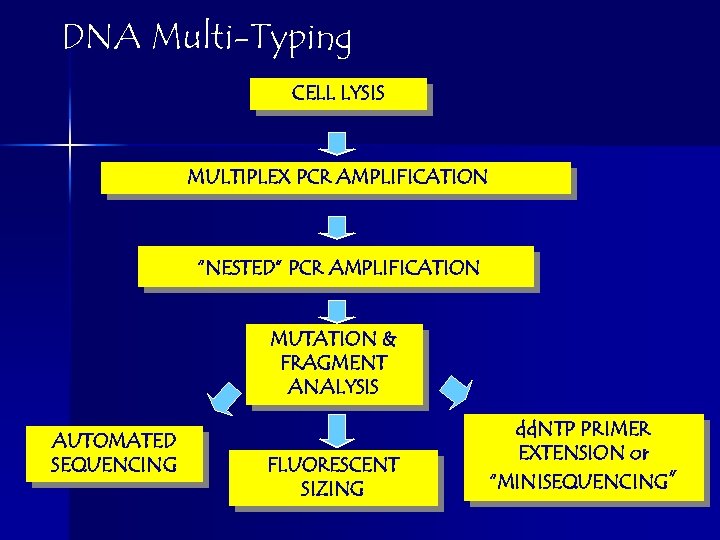
DNA Multi-Typing CELL LYSIS MULTIPLEX PCR AMPLIFICATION “NESTED” PCR AMPLIFICATION MUTATION & FRAGMENT ANALYSIS AUTOMATED SEQUENCING FLUORESCENT SIZING dd. NTP PRIMER EXTENSION or “MINISEQUENCING”
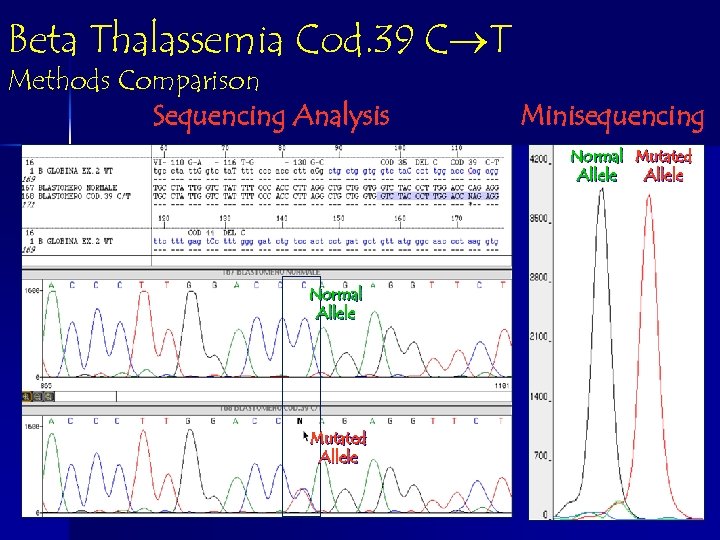
Beta Thalassemia Cod. 39 C T Methods Comparison Sequencing Analysis Minisequencing Normal Mutated Allele Normal Allele Mutated Allele
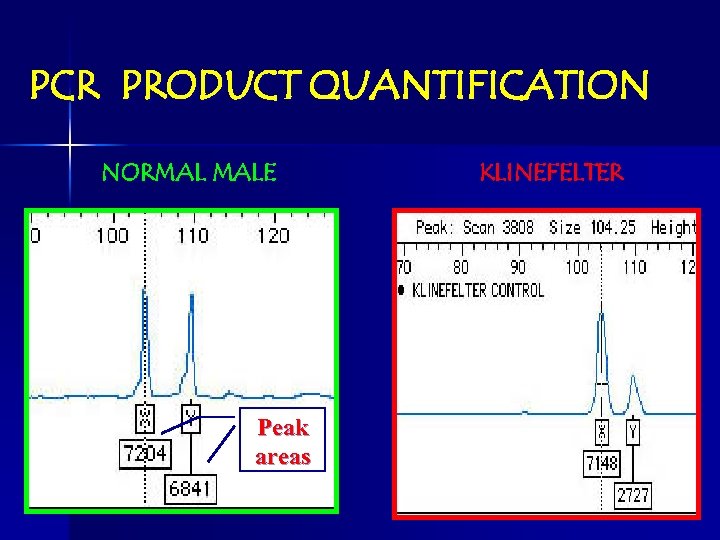
PCR PRODUCT QUANTIFICATION NORMAL MALE Peak areas KLINEFELTER
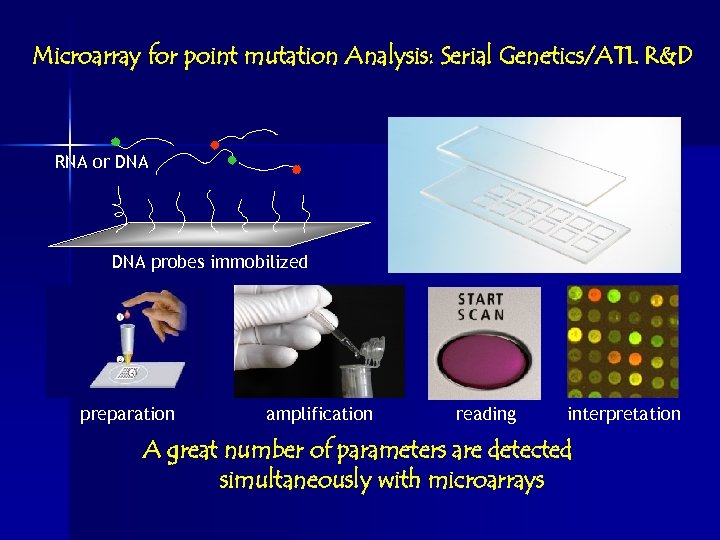
Microarray for point mutation Analysis: Serial Genetics/ATL R&D RNA or DNA probes immobilized preparation amplification reading interpretation A great number of parameters are detected simultaneously with microarrays
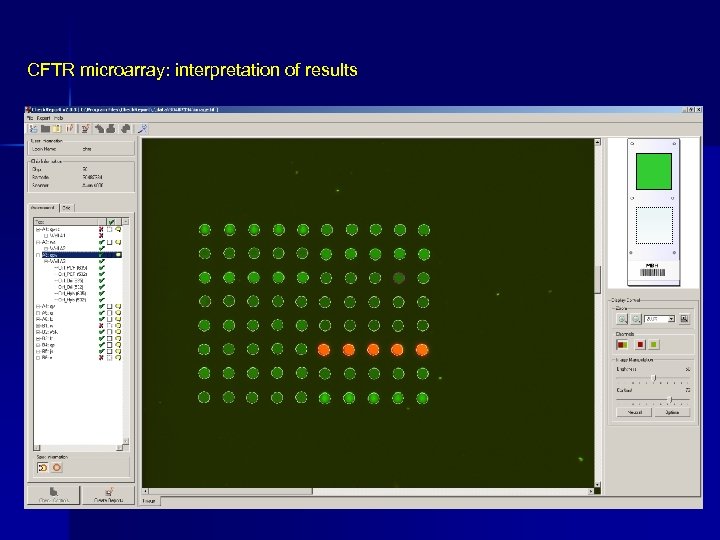
CFTR microarray: interpretation of results
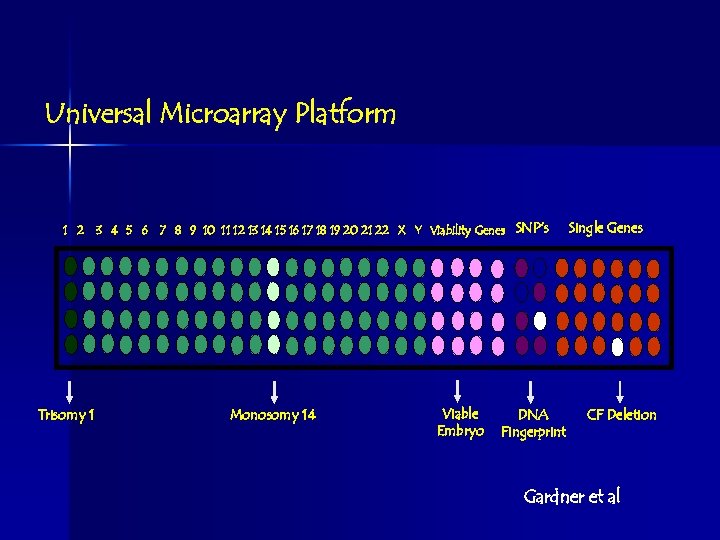
Universal Microarray Platform 1 2 Trisomy 1 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 X Y Viability Genes SNP’s Monosomy 14 Viable Embryo DNA Fingerprint Single Genes CF Deletion Gardner et al
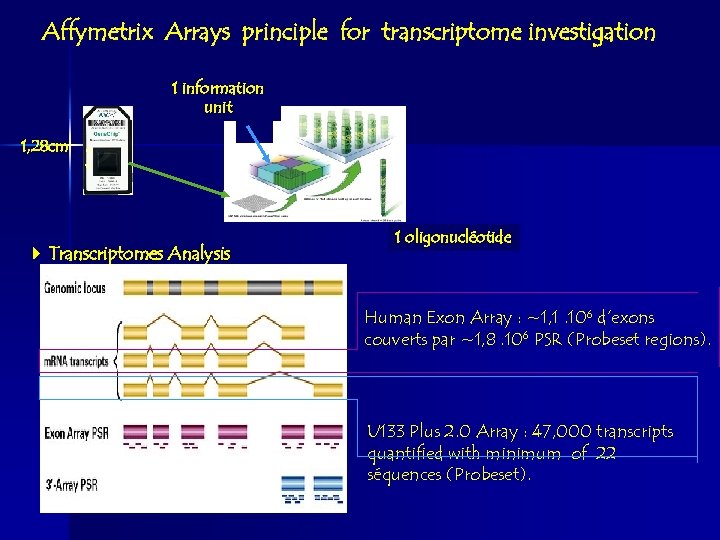
Affymetrix Arrays principle for transcriptome investigation 1 information unit 1, 28 cm Transcriptomes Analysis 1 oligonucléotide Human Exon Array : ~1, 1. 106 d’exons couverts par ~1, 8. 106 PSR (Probeset regions). U 133 Plus 2. 0 Array : 47, 000 transcripts quantified with minimum of 22 séquences (Probeset).

Experiment Total RNAs extraction quality control c. DNA quality control Données contrôlées Data Control
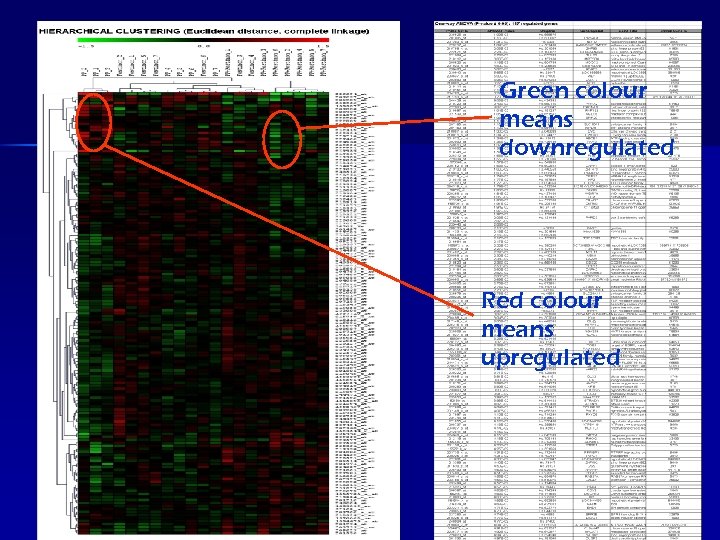
Green colour means downregulated Red colour means upregulated
What is Proteomics? n n n Proteomics is the large-scale study of proteins, particularly their structures and functions. Relatively little is known regarding the proteome of the human biological cell system Unlike the genome, the proteome is dynamic constantly changing through its biochemical interactions with the genome and its response to internal and external stimuli
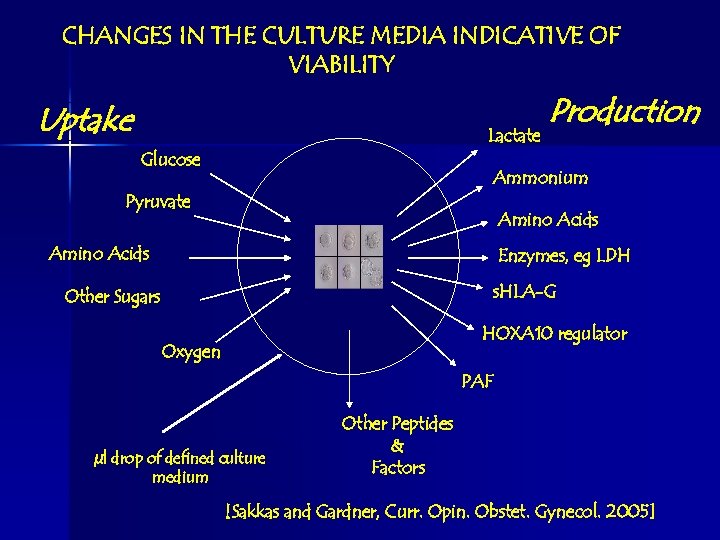
CHANGES IN THE CULTURE MEDIA INDICATIVE OF VIABILITY Uptake Lactate Glucose Production Ammonium Pyruvate Amino Acids Enzymes, eg LDH s. HLA-G Other Sugars HOXA 10 regulator Oxygen PAF µl drop of defined culture medium Other Peptides & Factors [Sakkas and Gardner, Curr. Opin. Obstet. Gynecol. 2005]
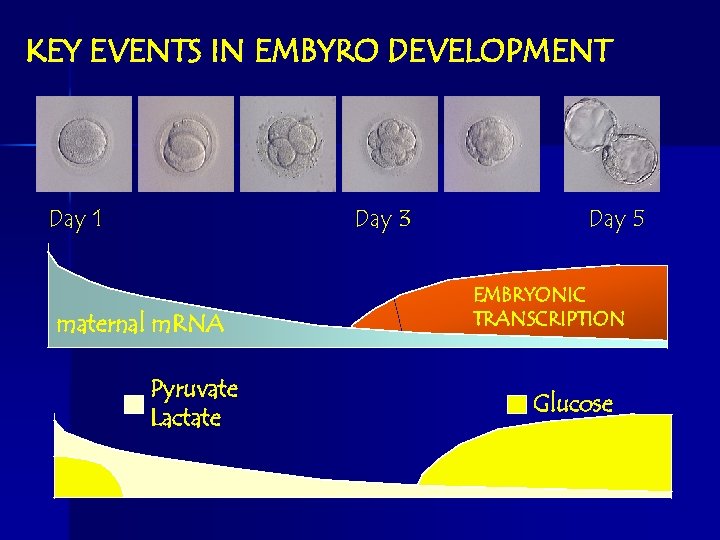
KEY EVENTS IN EMBYRO DEVELOPMENT Day 1 Day 3 maternal m. RNA Pyruvate Lactate Day 5 EMBRYONIC TRANSCRIPTION Glucose
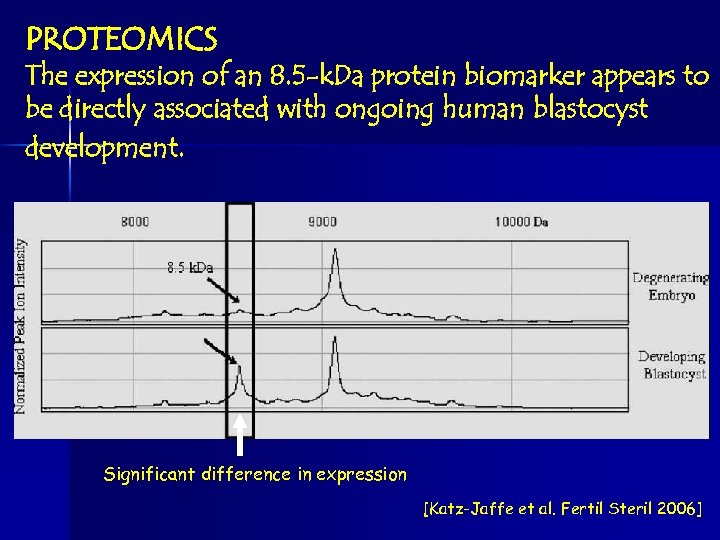
PROTEOMICS The expression of an 8. 5 -k. Da protein biomarker appears to be directly associated with ongoing human blastocyst development. Significant difference in expression [Katz-Jaffe et al. Fertil Steril 2006]
Metabolomics n Metabolomics can be thought of as the interface between genomics and a functional phenotype n The metabolome represents the final products of gene expression n Assessed by investigating inventory of small molecule biomarkers (LMW)

What is Measured? n Clinically – How the embryo modifies its environment n Biologically – Changes in concentrations of: Functional Groups • CH • NH • OH • SH • C=C Constituents • Albumin • Lactate • Pyruvate • Glutamate • Glucose

Spent Culture Media Analyses Specific Targets (hypotheses-based) Utilization of media component Targeted secretion products Protein profiling Metabolite profiling Profile Analyses (systems-based)
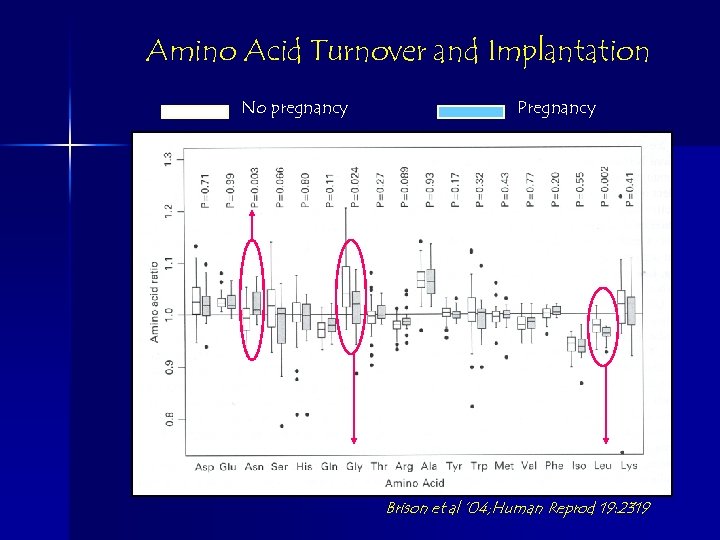
Amino Acid Turnover and Implantation No pregnancy Pregnancy Brison et al ’ 04; Human Reprod 19: 2319
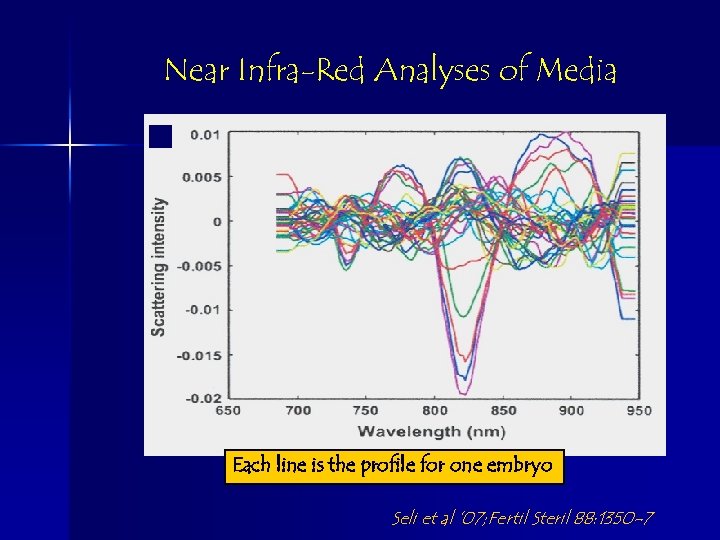
Near Infra-Red Analyses of Media Each line is the profile for one embryo Seli et al ’ 07; Fertil Steril 88: 1350 -7
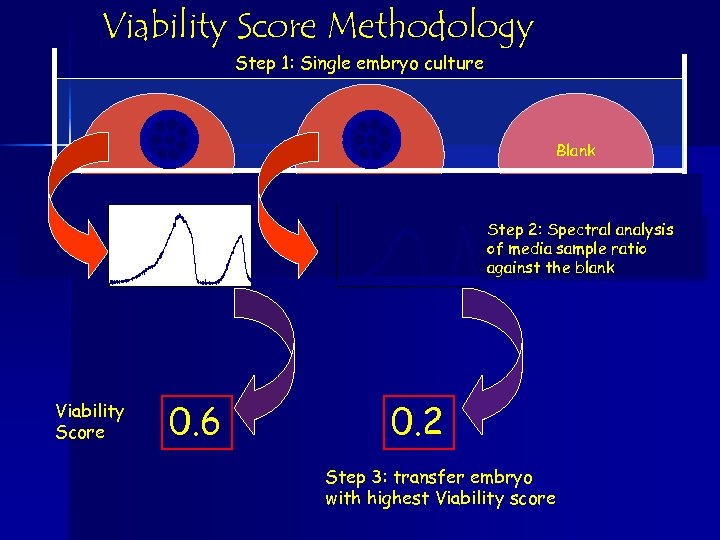
Viability Score Methodology Step 1: Single embryo culture Blank Step 2: Spectral analysis of media sample ratio against the blank Viability Score 0. 6 0. 2 Step 3: transfer embryo with highest Viability score
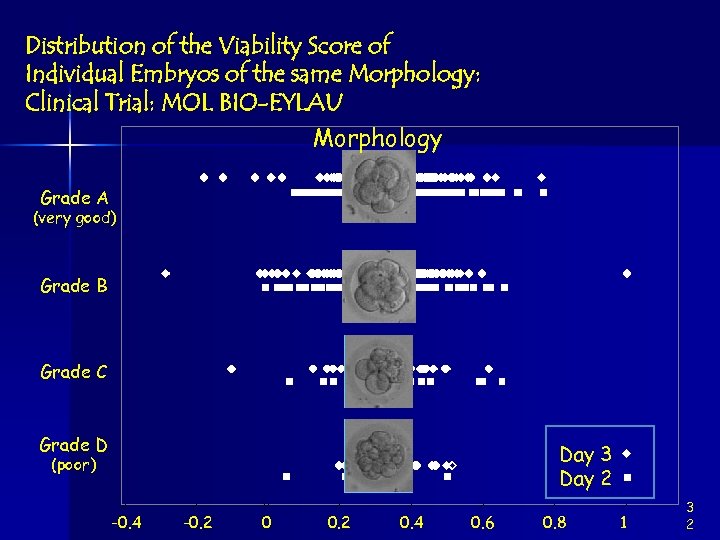
Distribution of the Viability Score of Individual Embryos of the same Morphology: Clinical Trial: MOL BIO-EYLAU Morphology Grade A (very good) Grade B Grade C Grade D Day 3 Day 2 (poor) -0. 4 -0. 2 0. 4 0. 6 0. 8 1 3 2
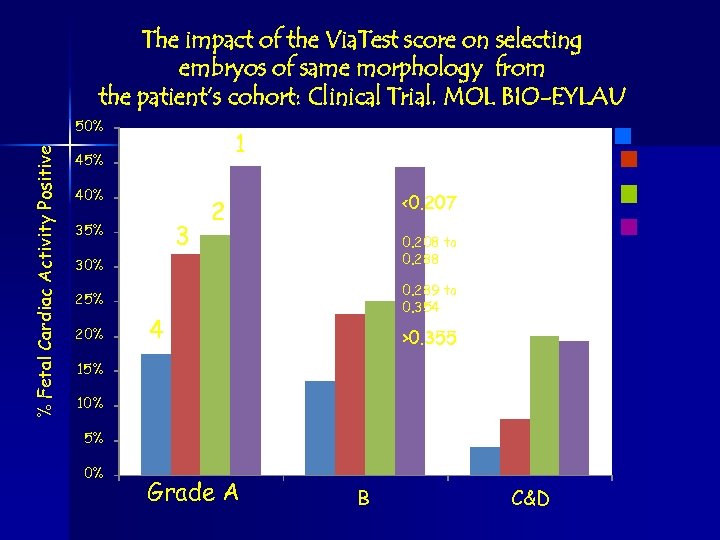
The impact of the Via. Test score on selecting embryos of same morphology from the patient’s cohort: Clinical Trial. MOL BIO-EYLAU % Fetal Cardiac Activity Positive 50% 1 45% 40% 3 35% <0. 207 2 0. 208 to 0. 288 30% 0. 289 to 0. 354 25% 20% 4 >0. 355 15% 10% 5% 0% Grade A B C&D
n. Genomics n. Proteomics n. Metabolomics n. Economics

Conclusions n n n Developments in genomics will serve to improve analysis of entire genome and facilitate genetic fingerprinting Analysis of the secretome, plausibly combined with carbohydrate utilization and amino acid turnover, and a general analysis of metabolic footprint, will serve to form the base of new assays of human viability of any biological process. Simplification of proteomics and metabolic analysis through microfluidic devices may serve as an economic methodology in clinical practice in the future.

Eylau. Laboratory/ Unilabs. Paris-Geneva Paul Cohen Bacrie Stéphanie Belloc Martine Dumont Anne Marie Junca Philippe Renard Pr Yyves Menezo Pr Alain Dalleac Gurgan CLINIC. IVF & Genetic Dept. Ankara. Turkey Pr T Gurgan Dr A Demirol T Sari
76720759a142e5b225f8e5acef7ad696.ppt