682a4e59862a165668a057921dff41cb.ppt
- Количество слайдов: 22

NSF-Relevant Challenges in Computational Intelligence Jaime Carbonell (jgc@cs. cmu. edu) & Tom Mitchell, Guy Bleloch, Randy Bryant, et al School of Computer Science Carnegie Mellon University 26 -April-2007 I) Major Computational Intelligence Research Areas II) Next-Generation Infrastructure (DISC) Carnegie Mellon School of Computer Science 1
Computational Intelligence • Machine Learning Ø Inductive learning algorithms, active leraning Ø Data mining & novel pattern detection • Language Technologies Ø Multilingual & next-veneration search engines Ø Machine translation (e. g. Arabic English) • Perception Ø Computer vision, tactile sensing (e. g. , in robotics) • Planning & optimizing Ø Reasoning & planning under uncertainty Ø Non-linear optimization (beyond O. R. ) w/uncertainty • Key scientific applications Ø Proteomics, genomics, computational biology Ø Modeling human brain functions Carnegie Mellon School of Computer Science 2
Machine Learning Speech Recognition Object recognition Data Mining • Reinforcement learning • Predictive modeling Extracting facts from text Automated Control • Pattern discovery learning • Hidden Markov models • Convex optimization • Explanation-based learning Carnegie Mellon School of Computer Science 3 • . .
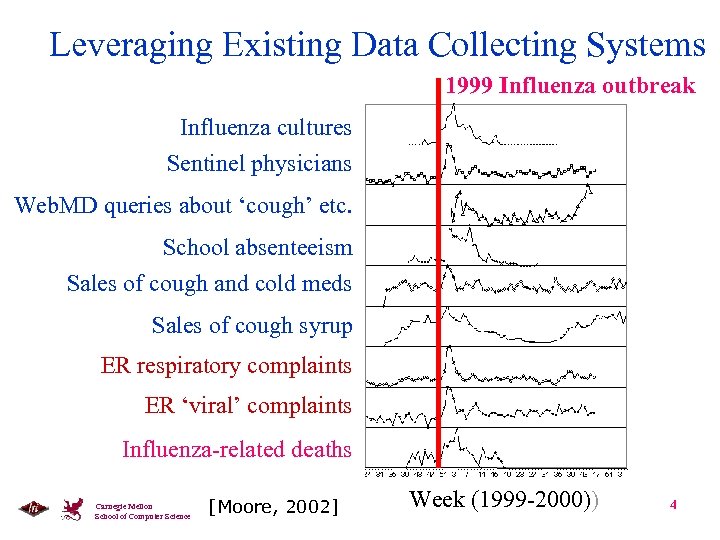
Leveraging Existing Data Collecting Systems 1999 Influenza outbreak Influenza cultures Sentinel physicians Web. MD queries about ‘cough’ etc. School absenteeism Sales of cough and cold meds Sales of cough syrup ER respiratory complaints ER ‘viral’ complaints Influenza-related deaths Carnegie Mellon School of Computer Science [Moore, 2002] Week (1999 -2000)) 4
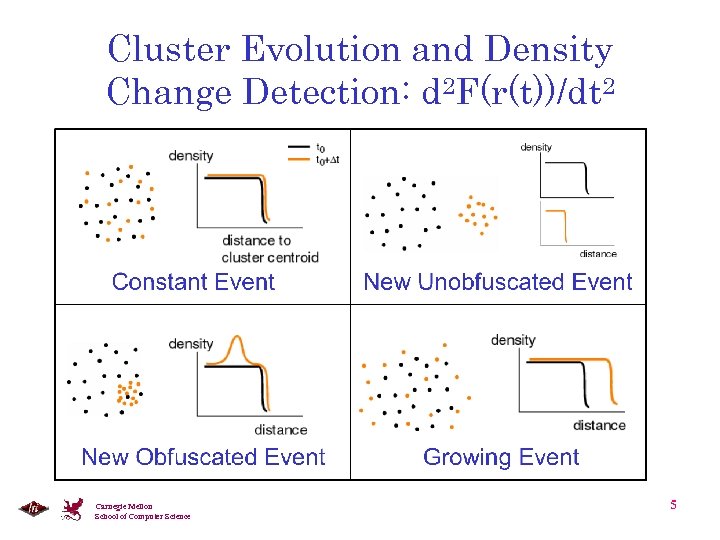
Cluster Evolution and Density Change Detection: d 2 F(r(t))/dt 2 Carnegie Mellon School of Computer Science 5
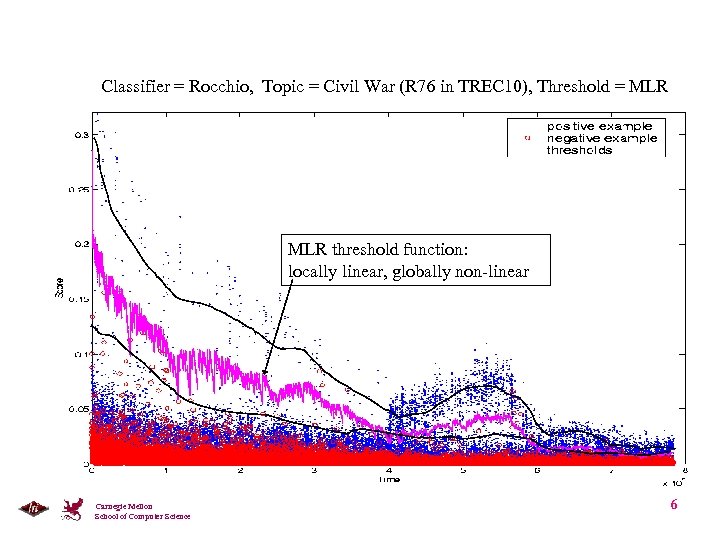
Classifier = Rocchio, Topic = Civil War (R 76 in TREC 10), Threshold = MLR threshold function: locally linear, globally non-linear Carnegie Mellon School of Computer Science 6
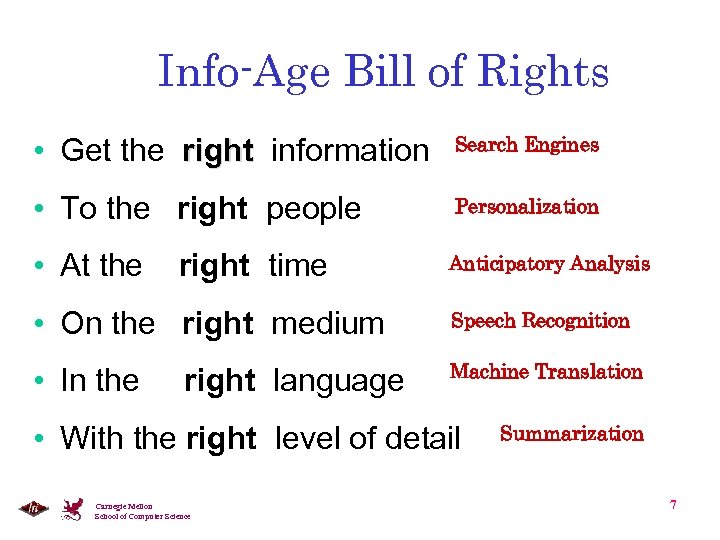
Info-Age Bill of Rights • Get the right information Search Engines • To the right people Personalization • At the Anticipatory Analysis right time • On the right medium Speech Recognition • In the Machine Translation right language • With the right level of detail Carnegie Mellon School of Computer Science Summarization 7
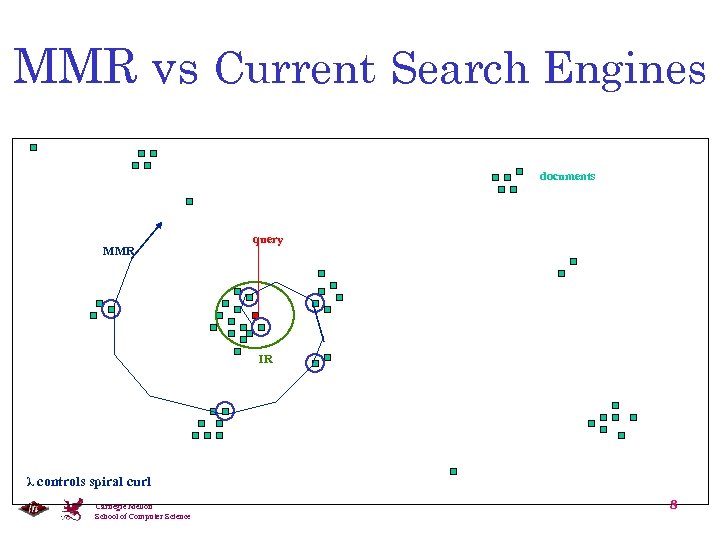
MMR vs Current Search Engines documents MMR query IR λ controls spiral curl Carnegie Mellon School of Computer Science 8
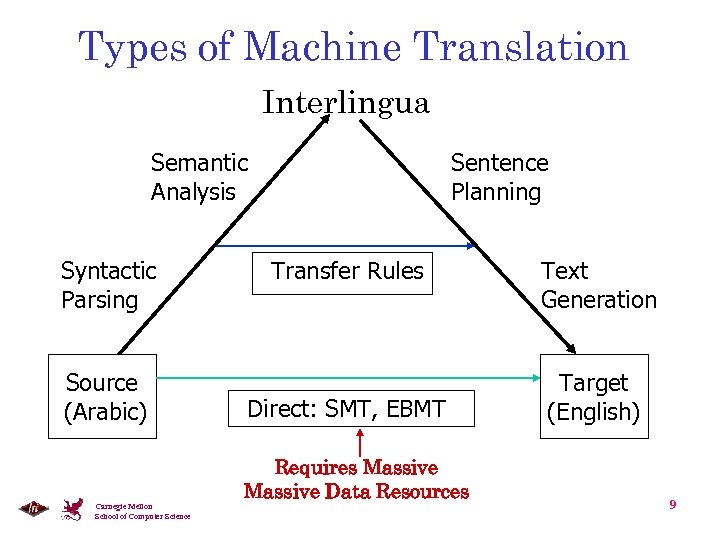
Types of Machine Translation Interlingua Semantic Analysis Syntactic Parsing Source (Arabic) Carnegie Mellon School of Computer Science Sentence Planning Transfer Rules Direct: SMT, EBMT Requires Massive Data Resources Text Generation Target (English) 9
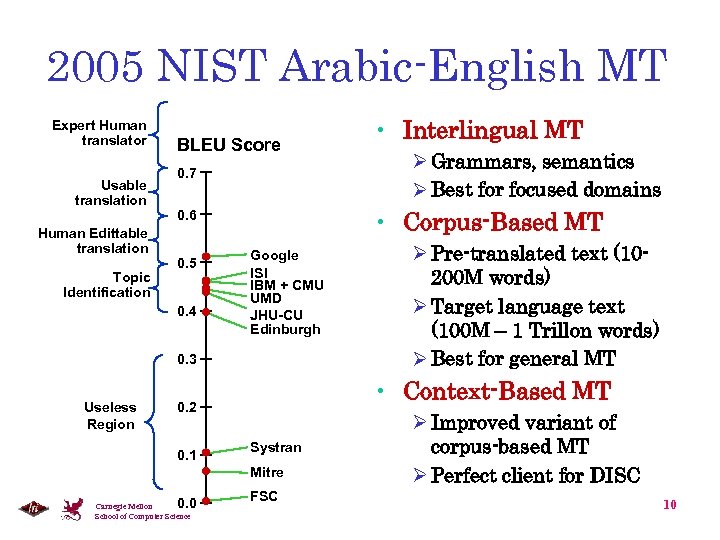
2005 NIST Arabic-English MT Expert Human translator Usable translation Human Edittable translation Topic Identification BLEU Score 0. 7 0. 4 Google ISI IBM + CMU UMD JHU-CU Edinburgh 0. 3 Useless Region Systran Mitre 0. 0 Carnegie Mellon School of Computer Science Ø Pre-translated text (10200 M words) Ø Target language text (100 M – 1 Trillon words) Ø Best for general MT • Context-Based MT 0. 2 0. 1 Ø Grammars, semantics Ø Best for focused domains • Corpus-Based MT 0. 6 0. 5 • Interlingual MT FSC Ø Improved variant of corpus-based MT Ø Perfect client for DISC 10

Arabic Statistical-MT Output ﺑﻜﻴﻦ 71 ﻳﻨﺎﻳﺮ / ﺷﻴﻨﺨﻮﺍ / ﺣﺚ ﻣﺴﺌﻮﻟﻮﻥ ﺻﻴﻨﻴﻮﻥ ﻭﺭﻭﺱ ﺟﻤﻴﻊ ﺍﻻﻃﺮﺍﻑ ﺍﻟﻤﻌﻨﻴﺔ ﻋﻠﻲ " ﺍﻟﺘﺰﺍﻡ ﺍﻟﻬﺪﻭﺀ ﻭﻣﻤﺎﺭﺳﺔ ﺿﺒﻂ ﺍﻟﻨﻔﺲ " ﺑﺸﺎﻥ ﺍﻟﻘﻀﻴﺔ ﺍﻟﻨﻮﻭﻳﺔ ﺍﻟﺨﺎﺻﺔ ﺑﺠﻤﻬﻮﺭﻳﺔ ﻛﻮﺭﻳﺎ . ﺍﻟﺪﻳﻤﻘﺮﺍﻃﻴﺔ ﺍﻟﺸﻌﺒﻴﺔ ﻭﻗﺪ ﺍﻟﺘﻘﻲ ﻧﺎﺋﺐ ﻭﺯﻳﺮ ﺍﻟﺨﺎﺭﺟﻴﺔ ﺍﻟﺼﻴﻨﻲ ﻳﺎﻧﻎ ﻭﻥ ﺗﺸﺎﻧﻎ ﻭﻧﺎﺋﺐ ﻭﺯﻳﺮ ﺍﻟﺨﺎﺭﺟﻴﺔ ﺍﻟﺮﻭﺳﻲ ﺍﻟﻜﺴﻨﺪﺭ ﻟﻮﺳﻴﻮﻛﻮﻑ ﻋﻠﻲ ﻣﺎﺩﺑﺔ ﻏﺪﺍﺀ ﺣﻴﺚ ﺩﻋﻴﺎ ﺍﻻﻃﺮﺍﻑ ﺍﻟﻤﻌﻨﻴﺔ ﺍﻟﻲ ﻣﻮﺍﺻﻠﺔ ﺍﻟﺴﻌﻲ ﻣﻦ ﺍﺟﻞ ﺍﻟﺤﻞ ﺍﻟﺴﻠﻤﻲ . ﻣﻦ ﺧﻼﻝ ﺍﻟﺤﻮﺍﺭ ﻓﻲ ﻇﻞ ﺍﻟﻮﺿﻊ ﺍﻟﻤﻌﻘﺪ ﺍﻟﺤﺎﻟﻲ Beijing January 17 / Shinhua / the Chinese and Russian officials urged all parties concerned to " remain calm and exercise restraint " over the nuclear issue of the Democratic People's Republic of Korea. He met with vice Chinese foreign minister Yang Chang won the deputy of the Russian foreign minister Alexander Losyukov at a lunch with invited interested parties to continue the search for a peaceful solution through dialogue under the current complicated situation. Carnegie Mellon School of Computer Science BLEU =. 64 11
What About Minor Languages or Dialects without Massive Data? Carnegie Mellon School of Computer Science 12
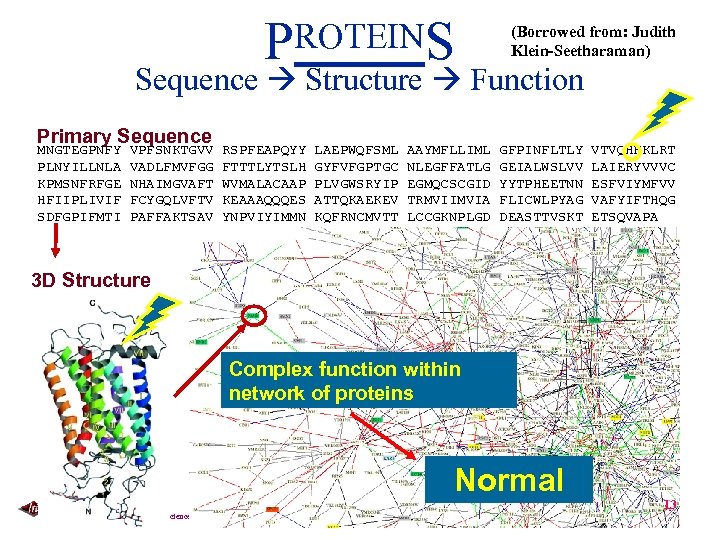
PROTEINS (Borrowed from: Judith Klein-Seetharaman) Sequence Structure Function Primary Sequence MNGTEGPNFY PLNYILLNLA KPMSNFRFGE HFIIPLIVIF SDFGPIFMTI VPFSNKTGVV VADLFMVFGG NHAIMGVAFT FCYGQLVFTV PAFFAKTSAV RSPFEAPQYY FTTTLYTSLH WVMALACAAP KEAAAQQQES YNPVIYIMMN LAEPWQFSML GYFVFGPTGC PLVGWSRYIP ATTQKAEKEV KQFRNCMVTT AAYMFLLIML NLEGFFATLG EGMQCSCGID TRMVIIMVIA LCCGKNPLGD GFPINFLTLY GEIALWSLVV YYTPHEETNN FLICWLPYAG DEASTTVSKT VTVQHKKLRT LAIERYVVVC ESFVIYMFVV VAFYIFTHQG ETSQVAPA Folding 3 D Structure Complex function within network of proteins Normal Carnegie Mellon School of Computer Science 13
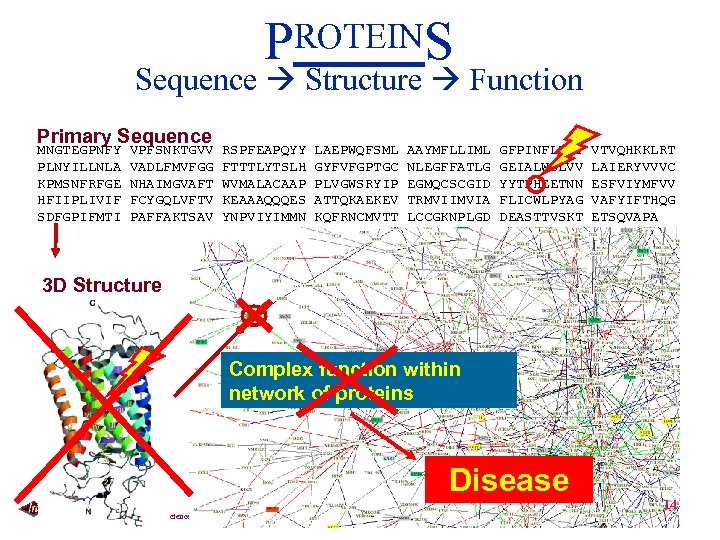
PROTEINS Sequence Structure Function Primary Sequence MNGTEGPNFY PLNYILLNLA KPMSNFRFGE HFIIPLIVIF SDFGPIFMTI VPFSNKTGVV VADLFMVFGG NHAIMGVAFT FCYGQLVFTV PAFFAKTSAV RSPFEAPQYY FTTTLYTSLH WVMALACAAP KEAAAQQQES YNPVIYIMMN LAEPWQFSML GYFVFGPTGC PLVGWSRYIP ATTQKAEKEV KQFRNCMVTT AAYMFLLIML NLEGFFATLG EGMQCSCGID TRMVIIMVIA LCCGKNPLGD GFPINFLTLY GEIALWSLVV YYTPHEETNN FLICWLPYAG DEASTTVSKT VTVQHKKLRT LAIERYVVVC ESFVIYMFVV VAFYIFTHQG ETSQVAPA Folding 3 D Structure Complex function within network of proteins Disease Carnegie Mellon School of Computer Science 14
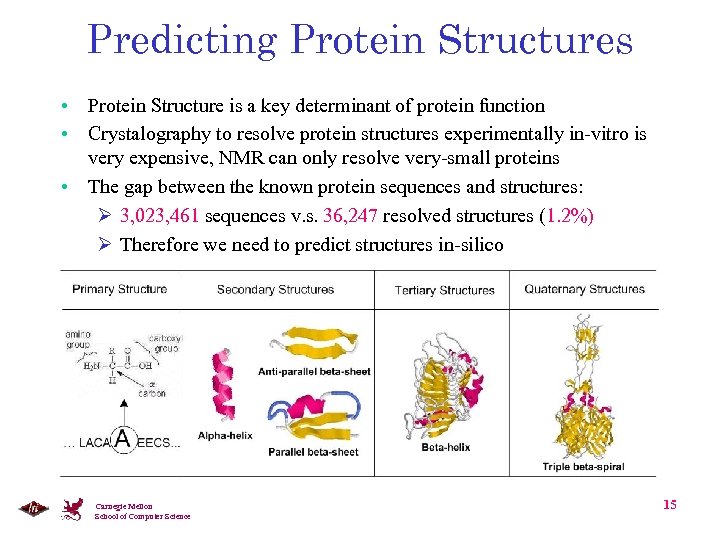
Predicting Protein Structures • Protein Structure is a key determinant of protein function • Crystalography to resolve protein structures experimentally in-vitro is very expensive, NMR can only resolve very-small proteins • The gap between the known protein sequences and structures: Ø 3, 023, 461 sequences v. s. 36, 247 resolved structures (1. 2%) Ø Therefore we need to predict structures in-silico Carnegie Mellon School of Computer Science 15
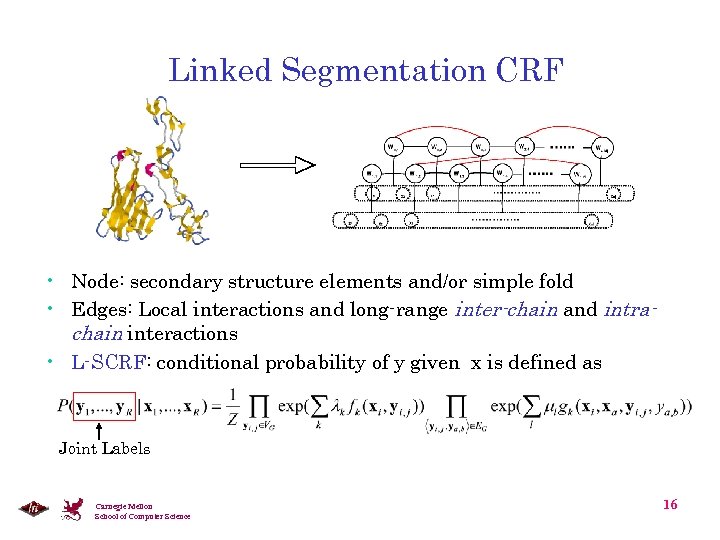
Linked Segmentation CRF • Node: secondary structure elements and/or simple fold • Edges: Local interactions and long-range inter-chain and intrachain interactions • L-SCRF: conditional probability of y given x is defined as Joint Labels Carnegie Mellon School of Computer Science 16
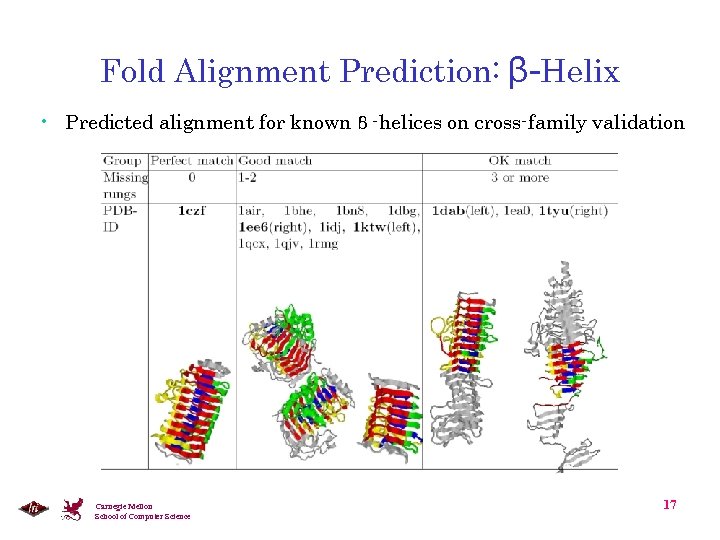
Fold Alignment Prediction: β-Helix • Predicted alignment for known β -helices on cross-family validation Carnegie Mellon School of Computer Science 17
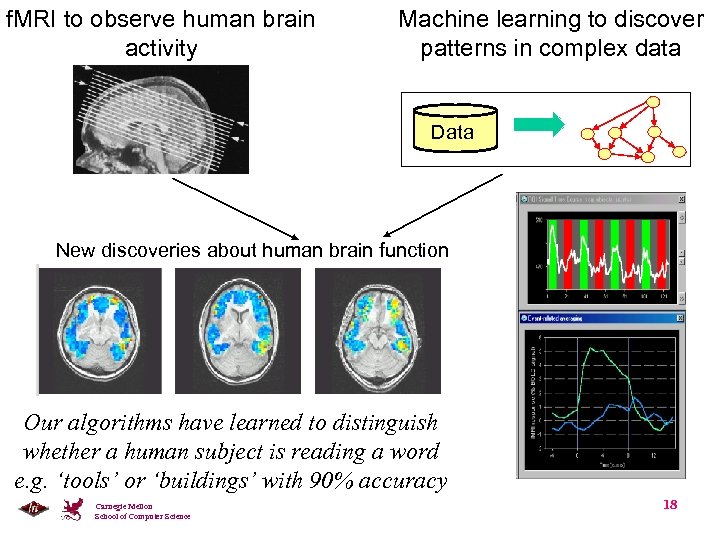
f. MRI to observe human brain activity Machine learning to discover patterns in complex data Data New discoveries about human brain function Our algorithms have learned to distinguish whether a human subject is reading a word e. g. ‘tools’ or ‘buildings’ with 90% accuracy Carnegie Mellon School of Computer Science 18
Requisite Infrastructure • Data Intensive Super. Computing (DISC) for tera-scale and peta-scale data repositories • Advanced algorithms research Ø Massively-parallel decomposition Ø Scalability in analytics & learning Ø Extracting compact models for run-time Ø Planning, reasoning, learning w/uncertainty) Ø Active Learning (maximally reducing uncertainty) • Domain expertise (e. g. proteomics, neural sciences, astronomy, network security, …) Carnegie Mellon School of Computer Science 19
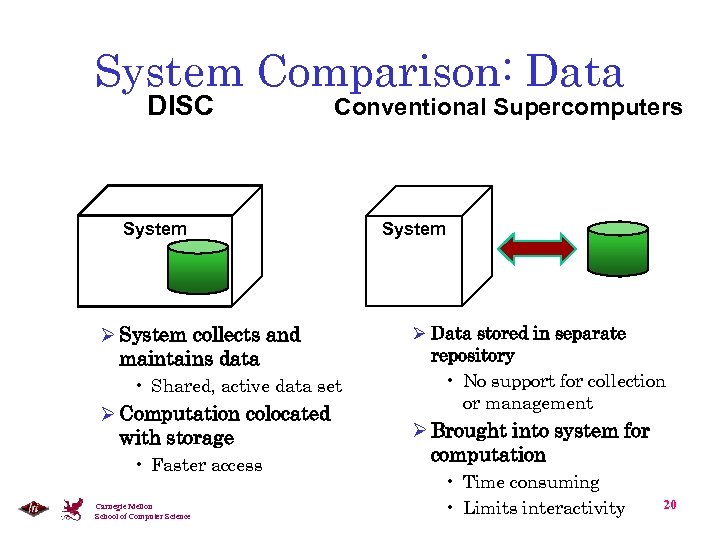
System Comparison: Data DISC Conventional Supercomputers System Ø System collects and maintains data • Shared, active data set Ø Computation colocated with storage • Faster access Carnegie Mellon School of Computer Science System Ø Data stored in separate repository • No support for collection or management Ø Brought into system for computation • Time consuming • Limits interactivity 20
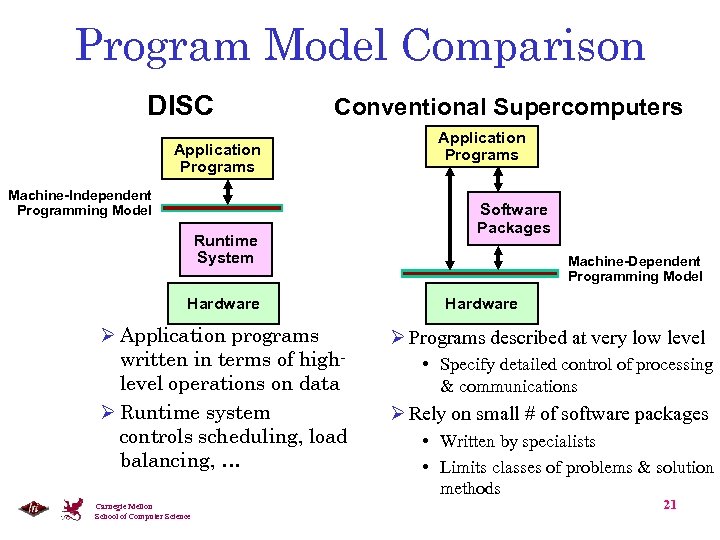
Program Model Comparison DISC Conventional Supercomputers Application Programs Machine-Independent Programming Model Runtime System Hardware Ø Application programs written in terms of highlevel operations on data Ø Runtime system controls scheduling, load balancing, … Carnegie Mellon School of Computer Science Application Programs Software Packages Machine-Dependent Programming Model Hardware Ø Programs described at very low level • Specify detailed control of processing & communications Ø Rely on small # of software packages • Written by specialists • Limits classes of problems & solution methods 21

Final Thoughts • Opportunities in Computational Intelligence Ø Machine learning for tough problems: relevant novelty detection, structural learning, active learning Ø Scientific applications: Computational X (X=biology, linguistics, astrophysics, chemistry, …) • Next generation computational infrastructure Ø DISC principle (beyond HPC, beyond grid, …) Ø Algorithmic fundamentals • International programs (on common problems) Carnegie Mellon School of Computer Science 22
682a4e59862a165668a057921dff41cb.ppt