10c45773d0b74b161701da398f24ba0b.ppt
- Количество слайдов: 33
 Novel Tools for (Functional) Magnetic Resonance Image Analysis Development and Implementation in the Scientific and Statistical Computing Core Robert W Cox and a cast of several
Novel Tools for (Functional) Magnetic Resonance Image Analysis Development and Implementation in the Scientific and Statistical Computing Core Robert W Cox and a cast of several
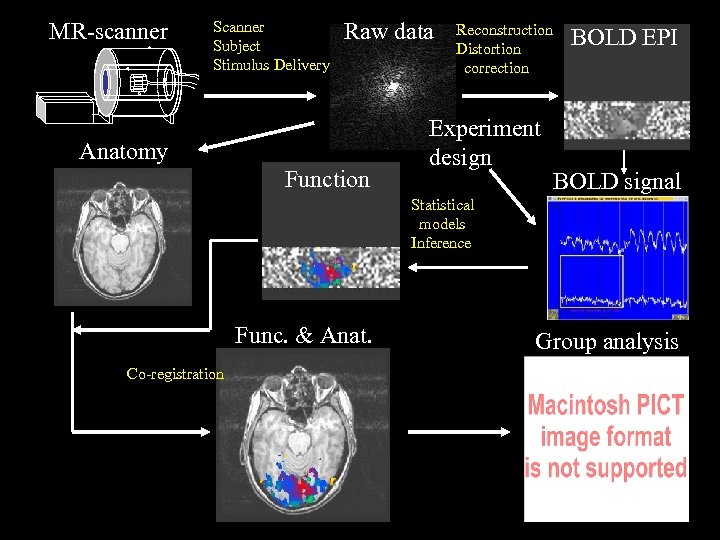 MR-scanner Subject Stimulus Delivery Raw data Anatomy Function Reconstruction Distortion correction Experiment design BOLD EPI BOLD signal Statistical models Inference Func. & Anat. Co-registration Group analysis
MR-scanner Subject Stimulus Delivery Raw data Anatomy Function Reconstruction Distortion correction Experiment design BOLD EPI BOLD signal Statistical models Inference Func. & Anat. Co-registration Group analysis
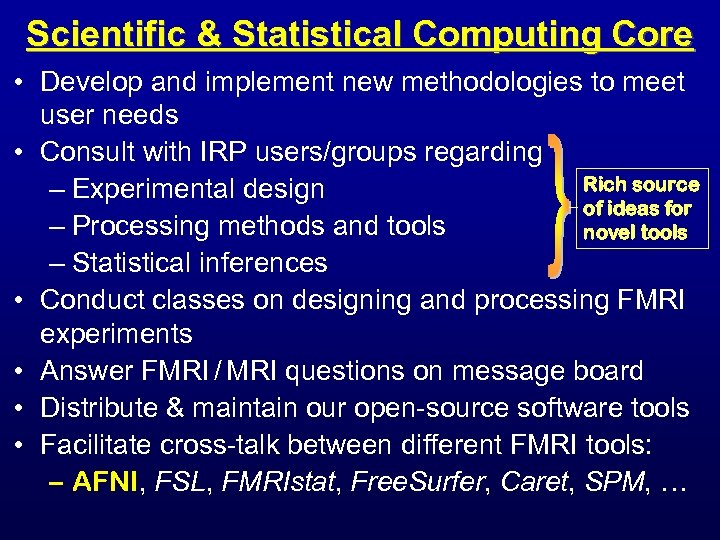 Scientific & Statistical Computing Core • Develop and implement new methodologies to meet user needs • Consult with IRP users/groups regarding Rich source – Experimental design of ideas for – Processing methods and tools novel tools – Statistical inferences • Conduct classes on designing and processing FMRI experiments • Answer FMRI / MRI questions on message board • Distribute & maintain our open-source software tools • Facilitate cross-talk between different FMRI tools: – AFNI, FSL, FMRIstat, Free. Surfer, Caret, SPM, …
Scientific & Statistical Computing Core • Develop and implement new methodologies to meet user needs • Consult with IRP users/groups regarding Rich source – Experimental design of ideas for – Processing methods and tools novel tools – Statistical inferences • Conduct classes on designing and processing FMRI experiments • Answer FMRI / MRI questions on message board • Distribute & maintain our open-source software tools • Facilitate cross-talk between different FMRI tools: – AFNI, FSL, FMRIstat, Free. Surfer, Caret, SPM, …
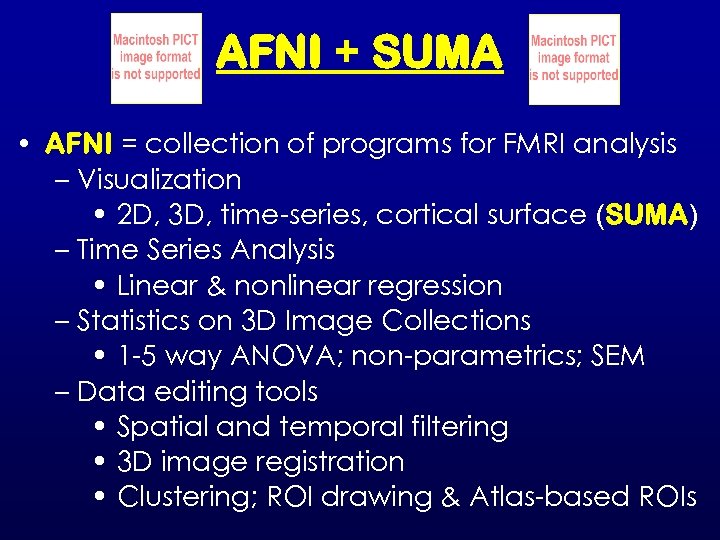 AFNI + SUMA • AFNI = collection of programs for FMRI analysis – Visualization • 2 D, 3 D, time-series, cortical surface (SUMA) – Time Series Analysis • Linear & nonlinear regression – Statistics on 3 D Image Collections • 1 -5 way ANOVA; non-parametrics; SEM – Data editing tools • Spatial and temporal filtering • 3 D image registration • Clustering; ROI drawing & Atlas-based ROIs
AFNI + SUMA • AFNI = collection of programs for FMRI analysis – Visualization • 2 D, 3 D, time-series, cortical surface (SUMA) – Time Series Analysis • Linear & nonlinear regression – Statistics on 3 D Image Collections • 1 -5 way ANOVA; non-parametrics; SEM – Data editing tools • Spatial and temporal filtering • 3 D image registration • Clustering; ROI drawing & Atlas-based ROIs
 The AFNI / SSCC Philosophy • Enable users to stay close to their data – Save intermediate results – Look at images and data in connected ways • User controls processing steps and parameters – Everyone has an opinion – Special problems need special solutions • Efficient (fast) implementations – Things that are easy and fast to do will get done more often • Help the users – Until our patience runs out
The AFNI / SSCC Philosophy • Enable users to stay close to their data – Save intermediate results – Look at images and data in connected ways • User controls processing steps and parameters – Everyone has an opinion – Special problems need special solutions • Efficient (fast) implementations – Things that are easy and fast to do will get done more often • Help the users – Until our patience runs out
 Next Set of Slides Features Added to AFNI and SUMA in Response to User Requests and / or Problems / Complaints (at least in part)
Next Set of Slides Features Added to AFNI and SUMA in Response to User Requests and / or Problems / Complaints (at least in part)
 Feature: Atlases • Problem: Navigating in a complicated folded up 3 D object (i. e. , the brain) with few easily recognized landmarks • Solution: Coordinate-based brain atlases – Accepting the 5 -10 mm uncertainty of brain coordinates • Atlas #1: Talairach-Tournoux atlas – As parsed by Peter Fox’s group at UT San Antonio • Atlas #2: Cytoarchitectonic atlases from Karl Zilles’ group at Forschungszentrum Jülich – 10 brains being sliced & diced & stained & scanned – About 40% complete at this time • Where Am I? + Jump To + Colorization + ROIs • Plans: keep up with Zilles; Animal atlases? …
Feature: Atlases • Problem: Navigating in a complicated folded up 3 D object (i. e. , the brain) with few easily recognized landmarks • Solution: Coordinate-based brain atlases – Accepting the 5 -10 mm uncertainty of brain coordinates • Atlas #1: Talairach-Tournoux atlas – As parsed by Peter Fox’s group at UT San Antonio • Atlas #2: Cytoarchitectonic atlases from Karl Zilles’ group at Forschungszentrum Jülich – 10 brains being sliced & diced & stained & scanned – About 40% complete at this time • Where Am I? + Jump To + Colorization + ROIs • Plans: keep up with Zilles; Animal atlases? …
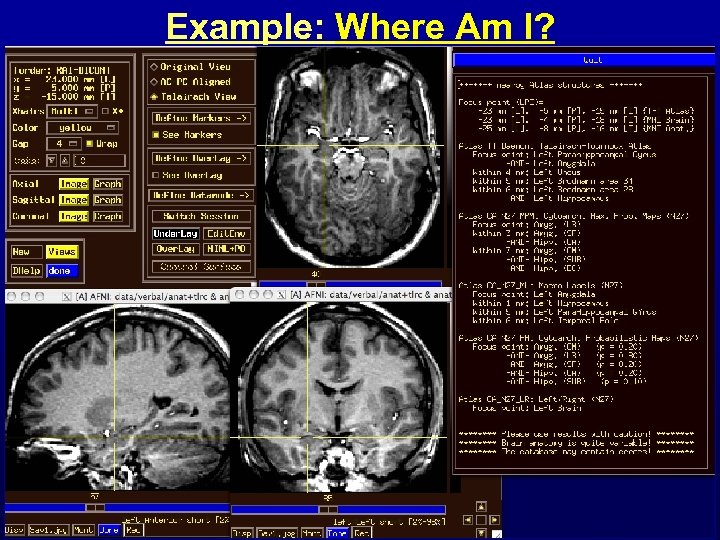 Example: Where Am I?
Example: Where Am I?
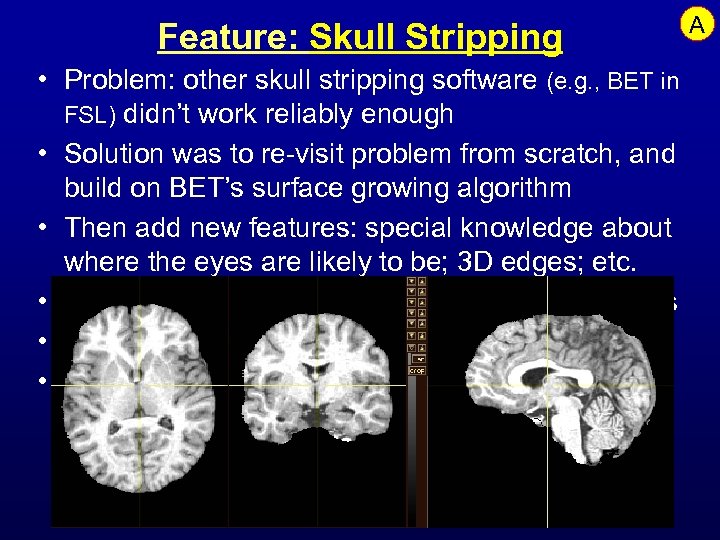 Feature: Skull Stripping • Problem: other skull stripping software (e. g. , BET in FSL) didn’t work reliably enough • Solution was to re-visit problem from scratch, and build on BET’s surface growing algorithm • Then add new features: special knowledge about where the eyes are likely to be; 3 D edges; etc. • Then test it on the hard cases from NIH (ab)users • Extra feature: extend it to monkey images • Plans: continue testing and improvements A
Feature: Skull Stripping • Problem: other skull stripping software (e. g. , BET in FSL) didn’t work reliably enough • Solution was to re-visit problem from scratch, and build on BET’s surface growing algorithm • Then add new features: special knowledge about where the eyes are likely to be; 3 D edges; etc. • Then test it on the hard cases from NIH (ab)users • Extra feature: extend it to monkey images • Plans: continue testing and improvements A
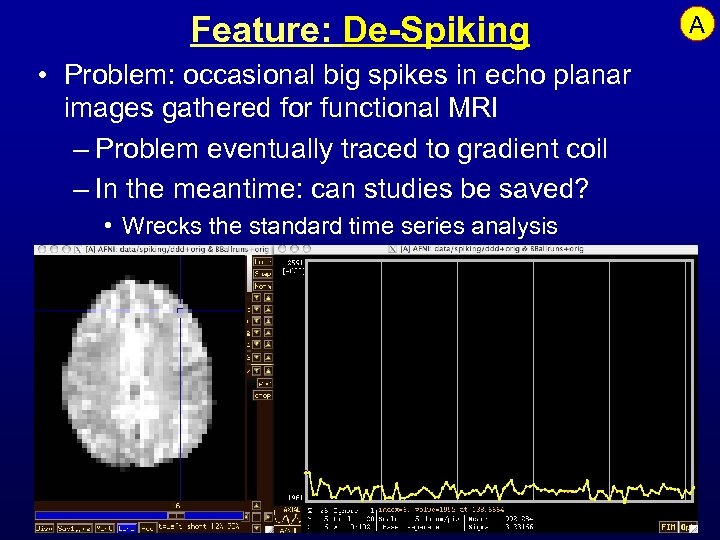 Feature: De-Spiking • Problem: occasional big spikes in echo planar images gathered for functional MRI – Problem eventually traced to gradient coil – In the meantime: can studies be saved? • Wrecks the standard time series analysis A
Feature: De-Spiking • Problem: occasional big spikes in echo planar images gathered for functional MRI – Problem eventually traced to gradient coil – In the meantime: can studies be saved? • Wrecks the standard time series analysis A
 Feature: Amplitude Modulated FMRI • Situation: Each stimulus event comes with an auxiliary parameter – May be measured (GSR, reaction time, …) or may be determined by experimenter • Want to determine if FMRI response magnitude is proportional to this auxiliary parameter • Solution was to add amplitude modulated regressors to AFNI’s 3 d. Deconvolve program – Two regressors per condition – First is: each stimulus response identical – Second is: each stimulus response proportional to auxiliary parameter for that stimulus • Plans: 2 -3 params/event; event-wise amplitudes
Feature: Amplitude Modulated FMRI • Situation: Each stimulus event comes with an auxiliary parameter – May be measured (GSR, reaction time, …) or may be determined by experimenter • Want to determine if FMRI response magnitude is proportional to this auxiliary parameter • Solution was to add amplitude modulated regressors to AFNI’s 3 d. Deconvolve program – Two regressors per condition – First is: each stimulus response identical – Second is: each stimulus response proportional to auxiliary parameter for that stimulus • Plans: 2 -3 params/event; event-wise amplitudes
 Feature: Nonlinear Regression Models • Pharmacological models for time series analysis – AFNI’s nonlinear regression program 3 d. NLfim • Michaelis-Menton dynamics for BOLD FMRI with psychoactive drugs (e. g. , ethanol) • Dynamic Contrast Enhanced MRI for quantifying Gd contrast leakage through blood-brain barrier
Feature: Nonlinear Regression Models • Pharmacological models for time series analysis – AFNI’s nonlinear regression program 3 d. NLfim • Michaelis-Menton dynamics for BOLD FMRI with psychoactive drugs (e. g. , ethanol) • Dynamic Contrast Enhanced MRI for quantifying Gd contrast leakage through blood-brain barrier
 Feature: Smart Blurring • FMRI time series datasets are often smoothed (blurred) in space to – Reduce noise (by averaging) – Increase intra-subject activation “blob” overlap • Blurring brain & non-brain signals together is silly • When combining data from different scanners (i. e. , multi-center studies), image smoothness varies – Should blur images until they have the same level of smoothness so that inter-scanner combinations make statistical sense • Developed a method for blurring inside a mask that stops when image noise reaches specified level of smoothness:
Feature: Smart Blurring • FMRI time series datasets are often smoothed (blurred) in space to – Reduce noise (by averaging) – Increase intra-subject activation “blob” overlap • Blurring brain & non-brain signals together is silly • When combining data from different scanners (i. e. , multi-center studies), image smoothness varies – Should blur images until they have the same level of smoothness so that inter-scanner combinations make statistical sense • Developed a method for blurring inside a mask that stops when image noise reaches specified level of smoothness:
 Feature: Structural Equation Modeling • SEM is a form of connectivity analysis • Input: correlations between activated ROIs – Regions where the activations fluctuate in strength together will be more highly correlated • Input: connectivity diagram between ROIs • Output: strength of connections • Can also search for “better” fitting connections
Feature: Structural Equation Modeling • SEM is a form of connectivity analysis • Input: correlations between activated ROIs – Regions where the activations fluctuate in strength together will be more highly correlated • Input: connectivity diagram between ROIs • Output: strength of connections • Can also search for “better” fitting connections
 Feature: All-in-One Analysis Program • Common complaint: “AFNI is tooooooo hard to use” • Analysis of single subject data involves several steps, each instantiated in separate programs – Registration, smoothing, normalizing, model analysis • Solution is a program afni_proc. py that will run all these programs in a coherent sequence – Intermediate results are saved to make it possible to track backwards when results are confusing • This script is not intended to let the user avoid understanding the data analysis process!
Feature: All-in-One Analysis Program • Common complaint: “AFNI is tooooooo hard to use” • Analysis of single subject data involves several steps, each instantiated in separate programs – Registration, smoothing, normalizing, model analysis • Solution is a program afni_proc. py that will run all these programs in a coherent sequence – Intermediate results are saved to make it possible to track backwards when results are confusing • This script is not intended to let the user avoid understanding the data analysis process!
 Feature: Diffusion Tensor Analysis • Goal: Compute the Diffusion Tensor (etc. ) from Diffusion Weighted image collections – Problem #1: log+linear method is inaccurate in highly anisotropic locations (the cool places to be) – Problem #2: published nonlinear solution methods not available in open-source software • Solution was to create and implement an efficient robust nonlinear method for finding the diffusion tensor D in each voxel – Also, a optional nonlinear image smoother (2 D and 3 D) to reduce noise in homogenous areas • Our code now incorporated into DTI Query, an open-source tractography program from Stanford
Feature: Diffusion Tensor Analysis • Goal: Compute the Diffusion Tensor (etc. ) from Diffusion Weighted image collections – Problem #1: log+linear method is inaccurate in highly anisotropic locations (the cool places to be) – Problem #2: published nonlinear solution methods not available in open-source software • Solution was to create and implement an efficient robust nonlinear method for finding the diffusion tensor D in each voxel – Also, a optional nonlinear image smoother (2 D and 3 D) to reduce noise in homogenous areas • Our code now incorporated into DTI Query, an open-source tractography program from Stanford
 Feature: Inter-Modality Registration • Goal: Efficiently align 3 D volumes acquired with different imaging contrasts • Solution is a general program using histogrambased measurements of image matching (e. g. , mutual information) • This one is still very much a work-in-progress – Works pretty well on “simple” cases (e. g. , wholebrain to whole-brain) – Dealing with partial-brain to whole-brain and with brain images that have holes in them is less reliable right now – Also want to add non-affine warping capabilities
Feature: Inter-Modality Registration • Goal: Efficiently align 3 D volumes acquired with different imaging contrasts • Solution is a general program using histogrambased measurements of image matching (e. g. , mutual information) • This one is still very much a work-in-progress – Works pretty well on “simple” cases (e. g. , wholebrain to whole-brain) – Dealing with partial-brain to whole-brain and with brain images that have holes in them is less reliable right now – Also want to add non-affine warping capabilities
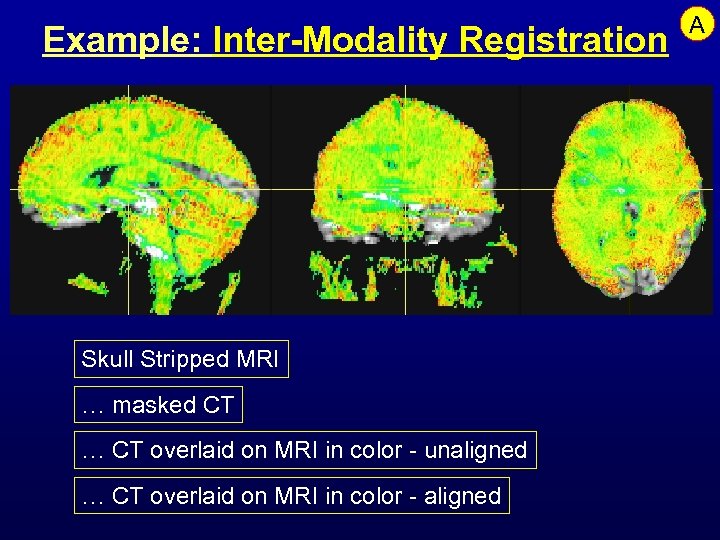 Example: Inter-Modality Registration Skull Stripped MRI … masked CT … CT overlaid on MRI in color - unaligned … CT overlaid on MRI in color - aligned A
Example: Inter-Modality Registration Skull Stripped MRI … masked CT … CT overlaid on MRI in color - unaligned … CT overlaid on MRI in color - aligned A
 Feature: Analysis of Mn Contrast MRI • Mn is an MRI contrast agent and a calcium analog • Goal: time-dependent in vivo tract tracing in monkeys • Problems abound: – Like FMRI, signal changes are small – Other artifacts from day-to-day scanning are larger – Simple image subtraction isn’t reliable • Next 3 slides: some data and results …
Feature: Analysis of Mn Contrast MRI • Mn is an MRI contrast agent and a calcium analog • Goal: time-dependent in vivo tract tracing in monkeys • Problems abound: – Like FMRI, signal changes are small – Other artifacts from day-to-day scanning are larger – Simple image subtraction isn’t reliable • Next 3 slides: some data and results …
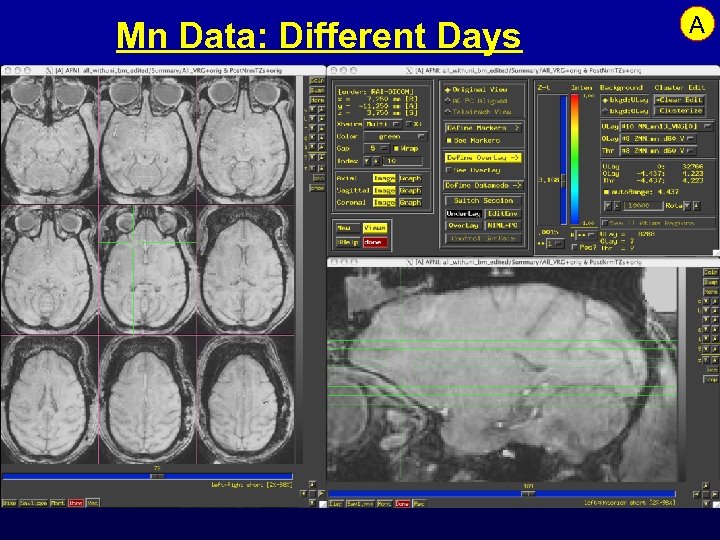 Mn Data: Different Days A
Mn Data: Different Days A
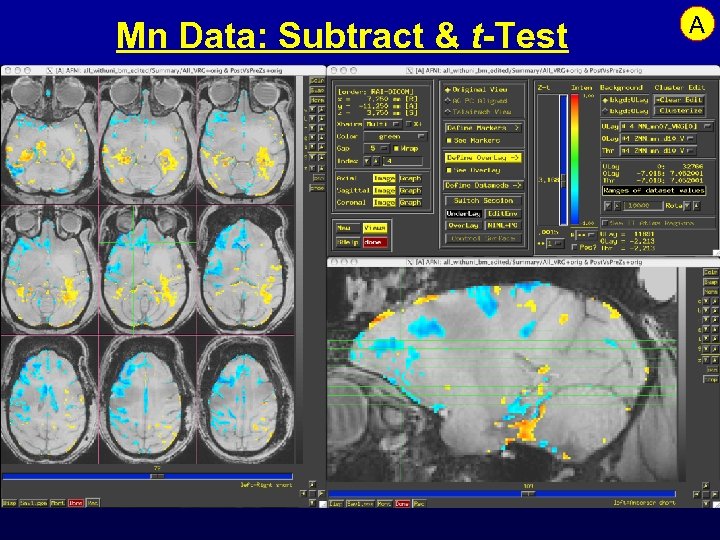 Mn Data: Subtract & t-Test A
Mn Data: Subtract & t-Test A
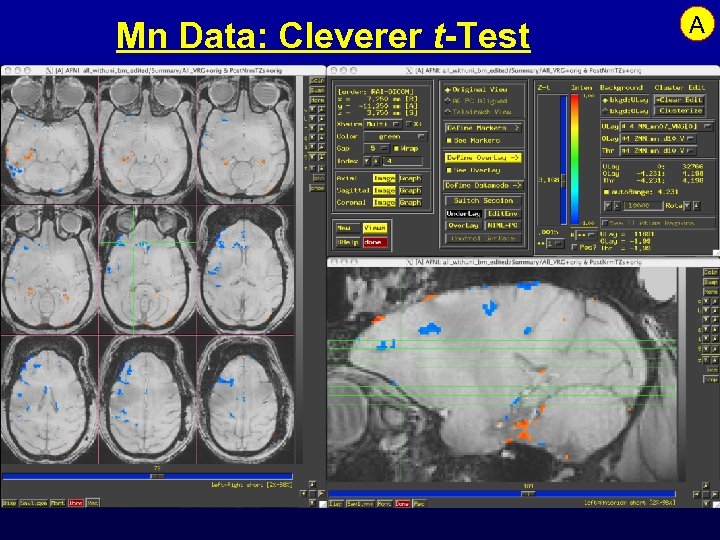 Mn Data: Cleverer t-Test A
Mn Data: Cleverer t-Test A
 Next Set of Slides Features Added to AFNI and SUMA in Response to Our Own Crazy Thoughts (mostly)
Next Set of Slides Features Added to AFNI and SUMA in Response to Our Own Crazy Thoughts (mostly)
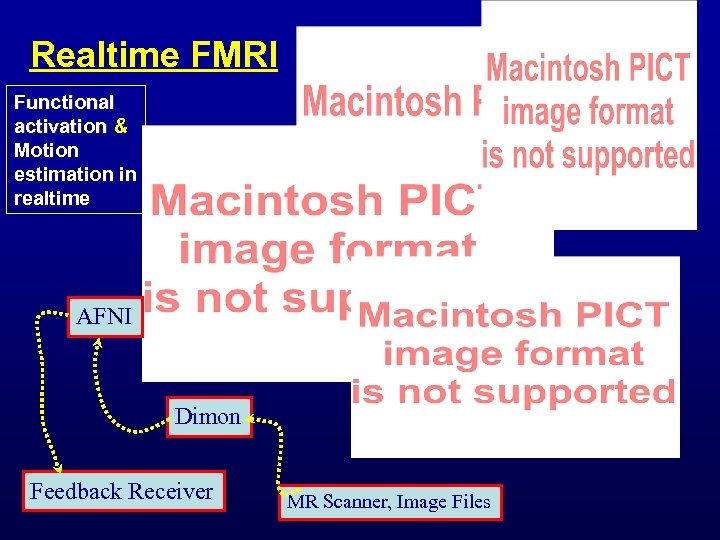 Realtime FMRI Functional activation & Motion estimation in realtime AFNI Dimon Feedback Receiver MR Scanner, Image Files
Realtime FMRI Functional activation & Motion estimation in realtime AFNI Dimon Feedback Receiver MR Scanner, Image Files
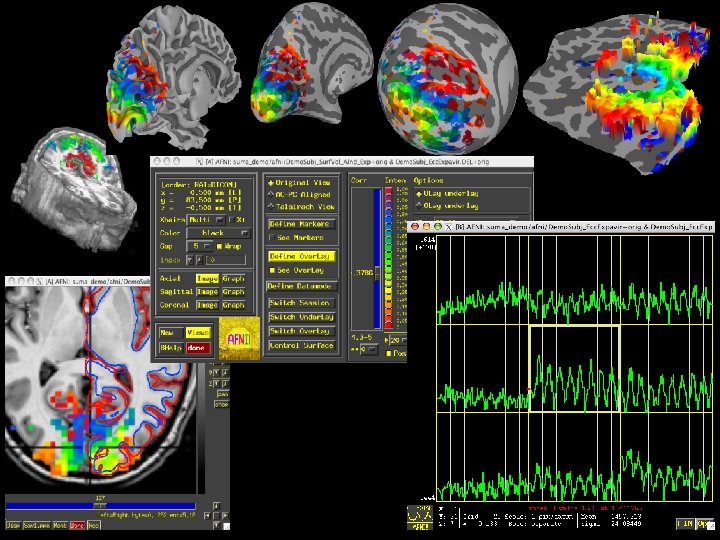
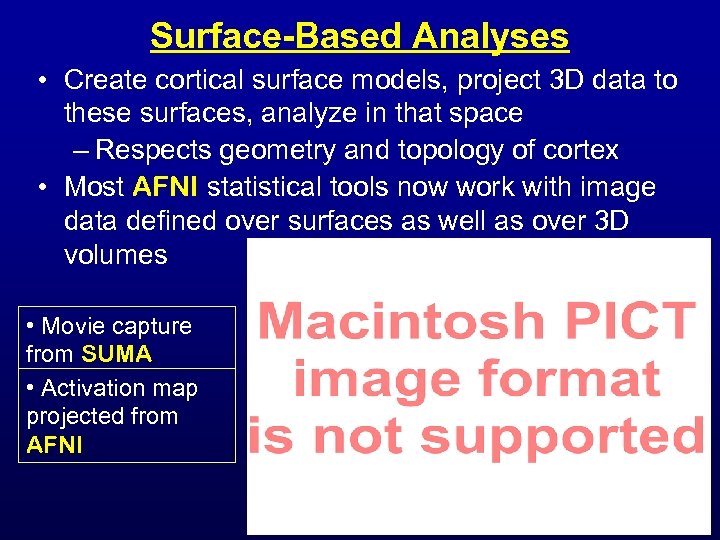 Surface-Based Analyses • Create cortical surface models, project 3 D data to these surfaces, analyze in that space – Respects geometry and topology of cortex • Most AFNI statistical tools now work with image data defined over surfaces as well as over 3 D volumes • Movie capture from SUMA • Activation map projected from AFNI
Surface-Based Analyses • Create cortical surface models, project 3 D data to these surfaces, analyze in that space – Respects geometry and topology of cortex • Most AFNI statistical tools now work with image data defined over surfaces as well as over 3 D volumes • Movie capture from SUMA • Activation map projected from AFNI
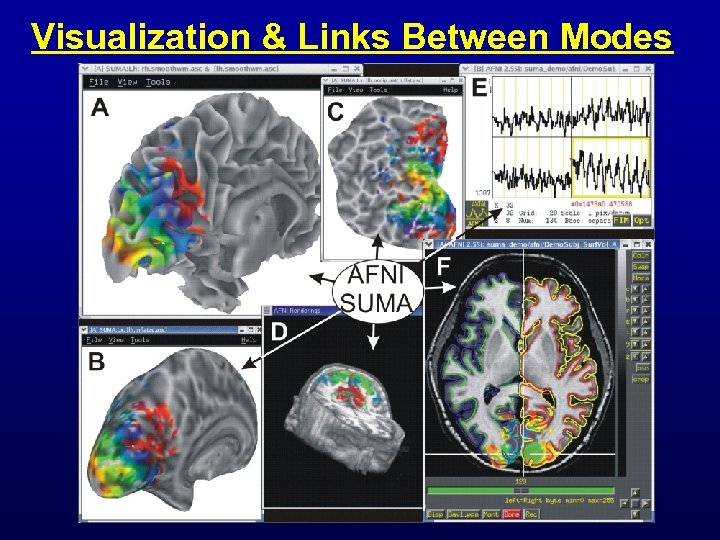 Visualization & Links Between Modes
Visualization & Links Between Modes
 NIf. TI Neuroimaging Informatics Technology Initiative • Goal: facilitate inter-operability of FMRI data analysis software • First fruit: NIf. TI-1. 1 standard for storing datasets defined over 3 D volumes (plus time axis) – Works with AFNI, FSL, SPM, Brain. Voyager, … • Agreement is not a one-time thing – Ongoing process is needed to deal with compatibility, extensions, new ideas along the same line, … • Efforts underway: – NIf. TI-G: standard for storing cortical surface models (and associated data) – NIf. TI-W: standard for storing non-affine spatial warps
NIf. TI Neuroimaging Informatics Technology Initiative • Goal: facilitate inter-operability of FMRI data analysis software • First fruit: NIf. TI-1. 1 standard for storing datasets defined over 3 D volumes (plus time axis) – Works with AFNI, FSL, SPM, Brain. Voyager, … • Agreement is not a one-time thing – Ongoing process is needed to deal with compatibility, extensions, new ideas along the same line, … • Efforts underway: – NIf. TI-G: standard for storing cortical surface models (and associated data) – NIf. TI-W: standard for storing non-affine spatial warps
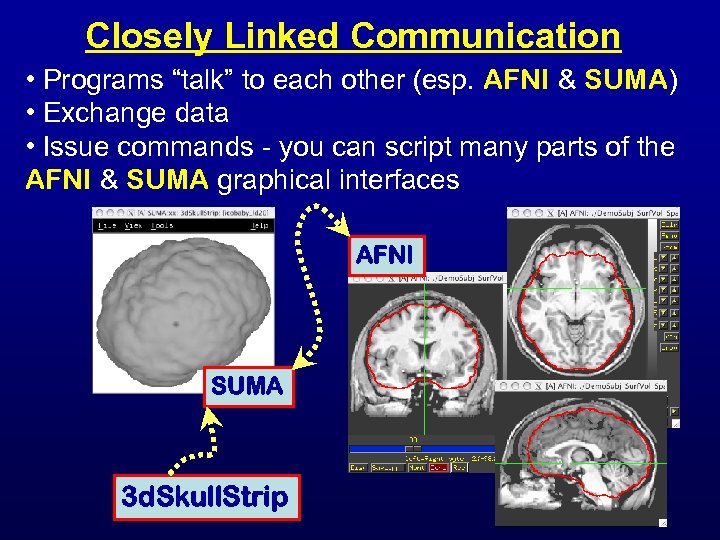 Closely Linked Communication • Programs “talk” to each other (esp. AFNI & SUMA) • Exchange data • Issue commands - you can script many parts of the AFNI & SUMA graphical interfaces AFNI SUMA 3 d. Skull. Strip
Closely Linked Communication • Programs “talk” to each other (esp. AFNI & SUMA) • Exchange data • Issue commands - you can script many parts of the AFNI & SUMA graphical interfaces AFNI SUMA 3 d. Skull. Strip
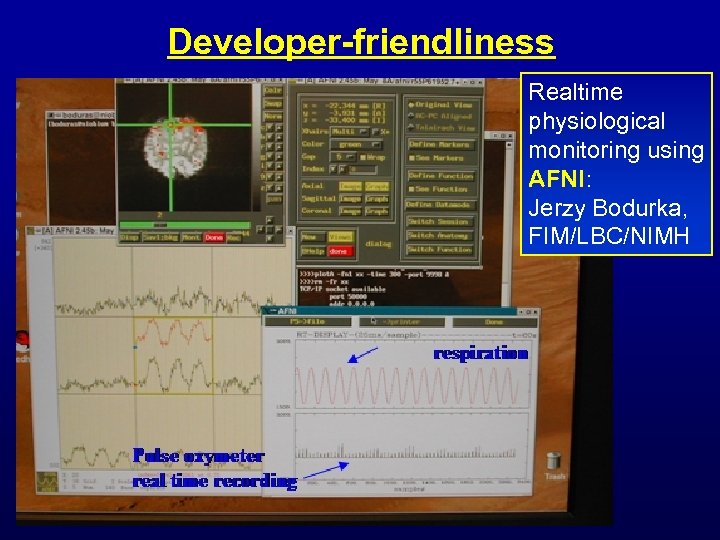 Developer-friendliness Realtime physiological monitoring using AFNI: Jerzy Bodurka, FIM/LBC/NIMH
Developer-friendliness Realtime physiological monitoring using AFNI: Jerzy Bodurka, FIM/LBC/NIMH
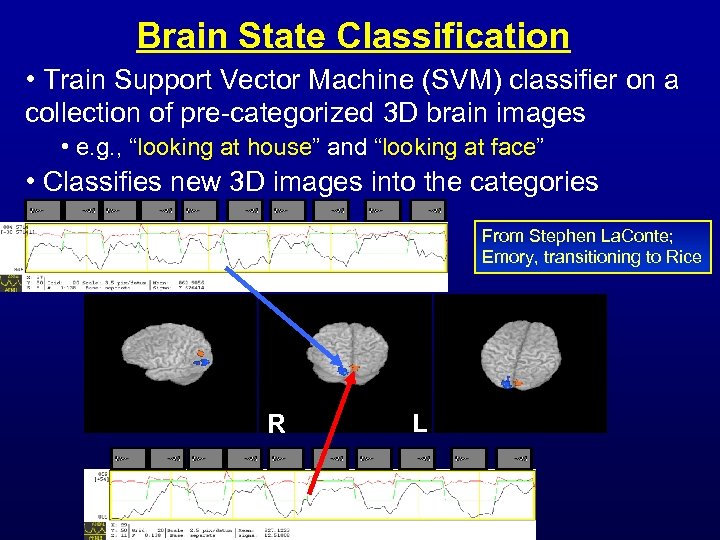 Brain State Classification • Train Support Vector Machine (SVM) classifier on a collection of pre-categorized 3 D brain images • e. g. , “looking at house” and “looking at face” • Classifies new 3 D images into the categories From Stephen La. Conte; Emory, transitioning to Rice R L
Brain State Classification • Train Support Vector Machine (SVM) classifier on a collection of pre-categorized 3 D brain images • e. g. , “looking at house” and “looking at face” • Classifies new 3 D images into the categories From Stephen La. Conte; Emory, transitioning to Rice R L
 Penultimate Slide • Much of our most fruitful and satisfying work comes from close and ongoing interactions with investigators that have interesting problems – Derived from studies that are pushing the envelope of deriving information from MRI • We are here to provide solutions to problems (of image analysis) – Your current short-term problems (lots of these!) – Your actual longer-term problems – What we think your future needs will be
Penultimate Slide • Much of our most fruitful and satisfying work comes from close and ongoing interactions with investigators that have interesting problems – Derived from studies that are pushing the envelope of deriving information from MRI • We are here to provide solutions to problems (of image analysis) – Your current short-term problems (lots of these!) – Your actual longer-term problems – What we think your future needs will be
 Ultimate Slide
Ultimate Slide


