820425539d8837796bec176ca64a5183.ppt
- Количество слайдов: 17
 Non-invasive diagnosis of fetal sex using free fetal DNA: our experiences so far Rebecca Woodward, Joanne Dunlop, Stephanie Allen and Fiona Macdonald West Midlands Regional Genetics Laboratory, Birmingham
Non-invasive diagnosis of fetal sex using free fetal DNA: our experiences so far Rebecca Woodward, Joanne Dunlop, Stephanie Allen and Fiona Macdonald West Midlands Regional Genetics Laboratory, Birmingham
 Fetal sexing § Early prenatal determination of sex: fetuses at risk of X-linked disorders – Males hemizygous for X § Useful for management of CAH – Females at risk of virilisation § Invasive procedures: – CVS (10 -13 weeks) § Small but significant risk of miscarriage (~1 -3%) and limb abnormalities.
Fetal sexing § Early prenatal determination of sex: fetuses at risk of X-linked disorders – Males hemizygous for X § Useful for management of CAH – Females at risk of virilisation § Invasive procedures: – CVS (10 -13 weeks) § Small but significant risk of miscarriage (~1 -3%) and limb abnormalities.
 Free Fetal DNA (ff. DNA) § Lo et al (1997) discovered significant amounts of ff. DNA in maternal plasma – Source of ff. DNA: Placenta (majority) and Haematopoietic cells – Mechanism of release: apoptosis most likely candidate (characterised by fragmentation of genomic DNA) § Direct correlation between amount of ff. DNA in plasma and gestation – Represents 3. 4%-6. 2% of total DNA in maternal plasma § Rapid clearance from maternal circulation after delivery (half life = 4 to 30 minutes). – Unlike intact fetal cells – reported to persist for years § Fetal sexing using ff. DNA reduces need for invasive testing by 50%
Free Fetal DNA (ff. DNA) § Lo et al (1997) discovered significant amounts of ff. DNA in maternal plasma – Source of ff. DNA: Placenta (majority) and Haematopoietic cells – Mechanism of release: apoptosis most likely candidate (characterised by fragmentation of genomic DNA) § Direct correlation between amount of ff. DNA in plasma and gestation – Represents 3. 4%-6. 2% of total DNA in maternal plasma § Rapid clearance from maternal circulation after delivery (half life = 4 to 30 minutes). – Unlike intact fetal cells – reported to persist for years § Fetal sexing using ff. DNA reduces need for invasive testing by 50%
 Justification of testing § Three genetic laboratories currently offering fetal sexing using ff. DNA: – – – International Blood Reference Laboratory North East Thames Regional Genetics Laboratory Manchester Regional Genetics Service § High number of referrals for X-linked disorders and CAH within the West Midlands. § Samples currently sent to North East Thames Regional Genetics Laboratory. § Samples need to be processed quickly: sending samples away increases turn around times.
Justification of testing § Three genetic laboratories currently offering fetal sexing using ff. DNA: – – – International Blood Reference Laboratory North East Thames Regional Genetics Laboratory Manchester Regional Genetics Service § High number of referrals for X-linked disorders and CAH within the West Midlands. § Samples currently sent to North East Thames Regional Genetics Laboratory. § Samples need to be processed quickly: sending samples away increases turn around times.
 Testing strategy § Testing strategy involves: 1. Separation of plasma from cellular components 1. Extraction of ff. DNA from maternal plasma 1. Detection of Y specific sequences from male fetuses § Pyrophosphorolysis-activated polymerisation (PAP) § Real-Time PCR
Testing strategy § Testing strategy involves: 1. Separation of plasma from cellular components 1. Extraction of ff. DNA from maternal plasma 1. Detection of Y specific sequences from male fetuses § Pyrophosphorolysis-activated polymerisation (PAP) § Real-Time PCR
 Isolation of ff. DNA from maternal plasma § Plasma separated by centrifugation within 48 hrs (3000 rpm; 10 mins) § Further micro-centrifugation prior to extraction to remove any remaining intact cells (persist from previous pregnancies) § ff. DNA extracted using EZ 1 Virus minikit v 2 (QIAGEN) and the EZ 1 Bio. Robot workstation: – Majority ff. DNA fragments <300 bp: method optimised for viral DNA is ideal § Once DNA extracted – used within half a day
Isolation of ff. DNA from maternal plasma § Plasma separated by centrifugation within 48 hrs (3000 rpm; 10 mins) § Further micro-centrifugation prior to extraction to remove any remaining intact cells (persist from previous pregnancies) § ff. DNA extracted using EZ 1 Virus minikit v 2 (QIAGEN) and the EZ 1 Bio. Robot workstation: – Majority ff. DNA fragments <300 bp: method optimised for viral DNA is ideal § Once DNA extracted – used within half a day
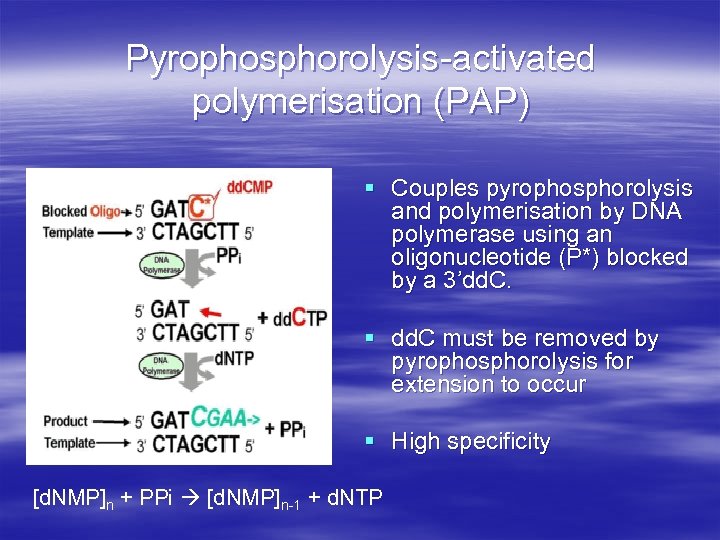 Pyrophosphorolysis-activated polymerisation (PAP) § Couples pyrophosphorolysis and polymerisation by DNA polymerase using an oligonucleotide (P*) blocked by a 3’dd. C. § dd. C must be removed by pyrophosphorolysis for extension to occur § High specificity [d. NMP]n + PPi [d. NMP]n-1 + d. NTP
Pyrophosphorolysis-activated polymerisation (PAP) § Couples pyrophosphorolysis and polymerisation by DNA polymerase using an oligonucleotide (P*) blocked by a 3’dd. C. § dd. C must be removed by pyrophosphorolysis for extension to occur § High specificity [d. NMP]n + PPi [d. NMP]n-1 + d. NTP
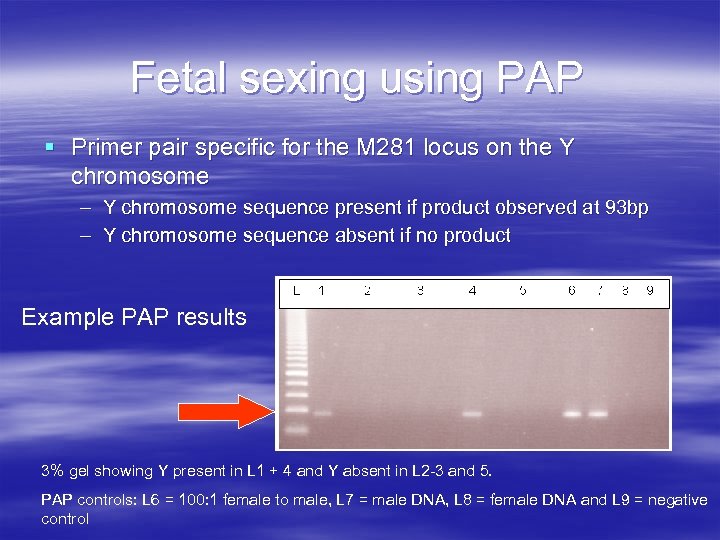 Fetal sexing using PAP § Primer pair specific for the M 281 locus on the Y chromosome – Y chromosome sequence present if product observed at 93 bp – Y chromosome sequence absent if no product Example PAP results 3% gel showing Y present in L 1 + 4 and Y absent in L 2 -3 and 5. PAP controls: L 6 = 100: 1 female to male, L 7 = male DNA, L 8 = female DNA and L 9 = negative control
Fetal sexing using PAP § Primer pair specific for the M 281 locus on the Y chromosome – Y chromosome sequence present if product observed at 93 bp – Y chromosome sequence absent if no product Example PAP results 3% gel showing Y present in L 1 + 4 and Y absent in L 2 -3 and 5. PAP controls: L 6 = 100: 1 female to male, L 7 = male DNA, L 8 = female DNA and L 9 = negative control
 Real-time PCR § Primers and probes specific to: – SRY: Y chromosome specific probe (8 replicates) – CCR 5: ‘Housekeeping gene’ located on chromosome 3 (2 replicates) § Confirms success of extraction (maternal and fetal DNA) § Terminology – CT value: The cycle at which the fluorescence passes the threshold § Higher the CT, the lower the amount of PCR product produced – Threshold: the line whose intersection with the amplification plot defines the CT value § Analysis parameters: – SRY present: CT<40 in ≥ 5/8 or 6/8 replicates § 47 samples audited: no result rate decreased from 29. 8% to 23. 4% using ≥ 5/8 replicates – SRY absent: CT=45 (no amplification) in 8/8 replicates
Real-time PCR § Primers and probes specific to: – SRY: Y chromosome specific probe (8 replicates) – CCR 5: ‘Housekeeping gene’ located on chromosome 3 (2 replicates) § Confirms success of extraction (maternal and fetal DNA) § Terminology – CT value: The cycle at which the fluorescence passes the threshold § Higher the CT, the lower the amount of PCR product produced – Threshold: the line whose intersection with the amplification plot defines the CT value § Analysis parameters: – SRY present: CT<40 in ≥ 5/8 or 6/8 replicates § 47 samples audited: no result rate decreased from 29. 8% to 23. 4% using ≥ 5/8 replicates – SRY absent: CT=45 (no amplification) in 8/8 replicates
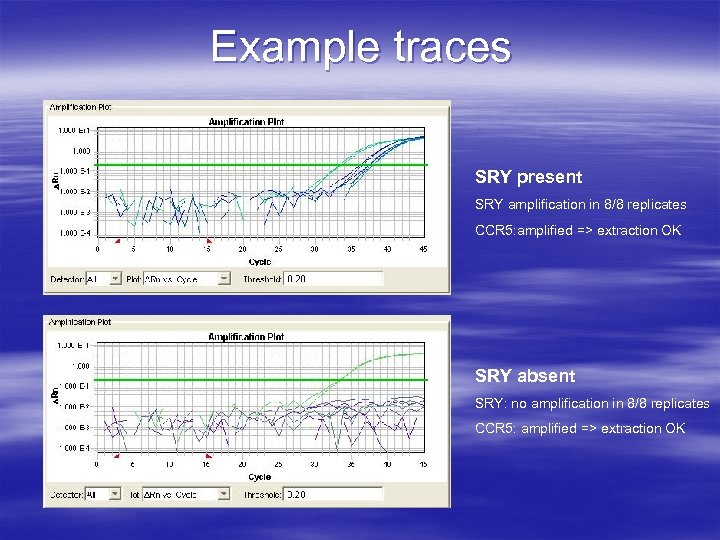 Example traces SRY present SRY amplification in 8/8 replicates CCR 5: amplified => extraction OK SRY absent SRY: no amplification in 8/8 replicates CCR 5: amplified => extraction OK
Example traces SRY present SRY amplification in 8/8 replicates CCR 5: amplified => extraction OK SRY absent SRY: no amplification in 8/8 replicates CCR 5: amplified => extraction OK
 Validation § Testing strategy validated using 78 samples: – Single frozen plasma aliquots (47) § Manchester Regional Genetics Service § International Blood Reference Laboratory (Bristol) § University College London – Maternal blood samples (31) collected in house – Mean gestation of samples = 11+6 weeks. § PAP and Real-time PCR performed in parallel using the same plasma sample – Samples scored using each method separately and in combination to access the reliability and robustness of each method § Where multiple aliquots of plasma were available, test repeated up to 3 x if calling criteria was not met – No result after 3 attempts
Validation § Testing strategy validated using 78 samples: – Single frozen plasma aliquots (47) § Manchester Regional Genetics Service § International Blood Reference Laboratory (Bristol) § University College London – Maternal blood samples (31) collected in house – Mean gestation of samples = 11+6 weeks. § PAP and Real-time PCR performed in parallel using the same plasma sample – Samples scored using each method separately and in combination to access the reliability and robustness of each method § Where multiple aliquots of plasma were available, test repeated up to 3 x if calling criteria was not met – No result after 3 attempts
 Results: PAP § § § Y present: 93 bp PCR product Y absent: No PCR product Faint bands were scored as a no result § Sensitivity (false –ve) = 97. 4% § Specificity (false +ve) = 96. 2% § Failure rate = 0%
Results: PAP § § § Y present: 93 bp PCR product Y absent: No PCR product Faint bands were scored as a no result § Sensitivity (false –ve) = 97. 4% § Specificity (false +ve) = 96. 2% § Failure rate = 0%
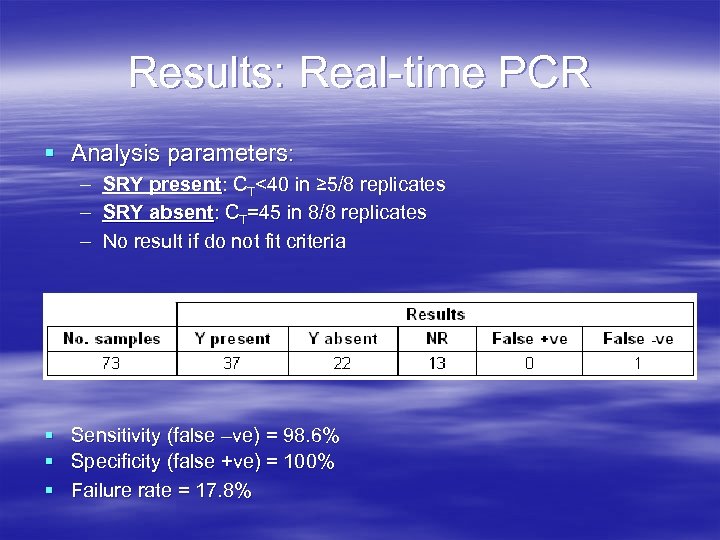 Results: Real-time PCR § Analysis parameters: – – – SRY present: CT<40 in ≥ 5/8 replicates SRY absent: CT=45 in 8/8 replicates No result if do not fit criteria § Sensitivity (false –ve) = 98. 6% § Specificity (false +ve) = 100% § Failure rate = 17. 8%
Results: Real-time PCR § Analysis parameters: – – – SRY present: CT<40 in ≥ 5/8 replicates SRY absent: CT=45 in 8/8 replicates No result if do not fit criteria § Sensitivity (false –ve) = 98. 6% § Specificity (false +ve) = 100% § Failure rate = 17. 8%
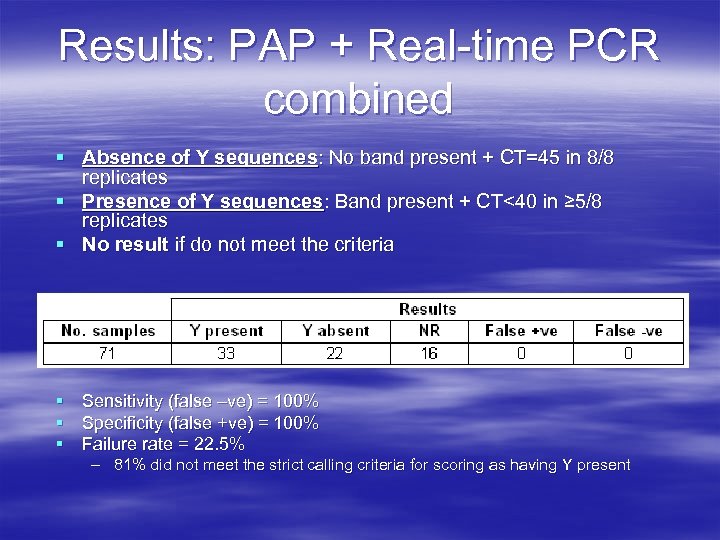 Results: PAP + Real-time PCR combined § Absence of Y sequences: No band present + CT=45 in 8/8 replicates § Presence of Y sequences: Band present + CT<40 in ≥ 5/8 replicates § No result if do not meet the criteria § Sensitivity (false –ve) = 100% § Specificity (false +ve) = 100% § Failure rate = 22. 5% – 81% did not meet the strict calling criteria for scoring as having Y present
Results: PAP + Real-time PCR combined § Absence of Y sequences: No band present + CT=45 in 8/8 replicates § Presence of Y sequences: Band present + CT<40 in ≥ 5/8 replicates § No result if do not meet the criteria § Sensitivity (false –ve) = 100% § Specificity (false +ve) = 100% § Failure rate = 22. 5% – 81% did not meet the strict calling criteria for scoring as having Y present
 Confirming the presence of ff. DNA § If SRY is absent in a sample: – ? Fetal sex is female – ? Absence of ff. DNA § Need a method to confirm the presence of ff. DNA – Non-Y-associated gene inherited from the father, not present in the maternal genome § 8 -10 polymorphic biallelic markers – Other methods being developed - methylation based § Biallelic markers NOT validated: reported to be informative in only ~40% of patients
Confirming the presence of ff. DNA § If SRY is absent in a sample: – ? Fetal sex is female – ? Absence of ff. DNA § Need a method to confirm the presence of ff. DNA – Non-Y-associated gene inherited from the father, not present in the maternal genome § 8 -10 polymorphic biallelic markers – Other methods being developed - methylation based § Biallelic markers NOT validated: reported to be informative in only ~40% of patients
 Conclusions § By using Real-time PCR and PAP assays in parallel, the technique was found to be: – – Reliable (sensitivity and specificity 100%) Easy to perform Low in cost Capable of providing a diagnosis within 24 hours § High rate of no results: – Majority of samples received as plasma aliquots from other laboratories § 1 aliquot per sample: no possibly of repeating if scored as a no result § Further work is being carried out to determine what gestation to offer testing from. – Currently validating samples from 7 -10 weeks gestation § Method to confirm the presence of ff. DNA where Y is absent
Conclusions § By using Real-time PCR and PAP assays in parallel, the technique was found to be: – – Reliable (sensitivity and specificity 100%) Easy to perform Low in cost Capable of providing a diagnosis within 24 hours § High rate of no results: – Majority of samples received as plasma aliquots from other laboratories § 1 aliquot per sample: no possibly of repeating if scored as a no result § Further work is being carried out to determine what gestation to offer testing from. – Currently validating samples from 7 -10 weeks gestation § Method to confirm the presence of ff. DNA where Y is absent
 Acknowledgements § West Midlands Regional Genetics Laboratory – – Joanne Dunlop Stephanie Allen Jennie Bell Fiona Macdonald § Manchester Regional Genetics Service – Helene Schlecht § International Blood Reference Laboratory § University College London
Acknowledgements § West Midlands Regional Genetics Laboratory – – Joanne Dunlop Stephanie Allen Jennie Bell Fiona Macdonald § Manchester Regional Genetics Service – Helene Schlecht § International Blood Reference Laboratory § University College London


