27fc5f746276c7cbabb5fc571c4589fb.ppt
- Количество слайдов: 54
 NIGMS and the NIH Roadmap for Medical Research Jeremy M. Berg National Institute of General Medical Sciences April 30, 2004
NIGMS and the NIH Roadmap for Medical Research Jeremy M. Berg National Institute of General Medical Sciences April 30, 2004
 Challenges for NIH n n n Revolutionary and rapid changes in science Increasing breadth of mission and growth Complex organization with many units (27 institutes and centers, multiple program offices, e. g. , OWHR, OAR, ORD, . . . ) Structured by disease, organ, life stage, disciplines Rapid convergence of science
Challenges for NIH n n n Revolutionary and rapid changes in science Increasing breadth of mission and growth Complex organization with many units (27 institutes and centers, multiple program offices, e. g. , OWHR, OAR, ORD, . . . ) Structured by disease, organ, life stage, disciplines Rapid convergence of science
 U. S. Health Expenditures (Percentage of GDP) 18 Actual Projected Percent 16 14 12 10 1985 1987 1989 1991 1993 1995 1997 1999 2001 2003 2005 2007 2009 2011 Year
U. S. Health Expenditures (Percentage of GDP) 18 Actual Projected Percent 16 14 12 10 1985 1987 1989 1991 1993 1995 1997 1999 2001 2003 2005 2007 2009 2011 Year
 Imperatives for NIH n n Accelerate pace of discoveries in life sciences Translate research more rapidly from laboratories to patients and back Explore novel approaches orders of magnitude more effective than current Develop new strategies: NIH Roadmap
Imperatives for NIH n n Accelerate pace of discoveries in life sciences Translate research more rapidly from laboratories to patients and back Explore novel approaches orders of magnitude more effective than current Develop new strategies: NIH Roadmap
 How was the Roadmap developed? n Extensive consultations with stakeholders, scientists, health care providers q q q What are today’s scientific challenges? What are the roadblocks to progress? What do we need to do to overcome roadblocks?
How was the Roadmap developed? n Extensive consultations with stakeholders, scientists, health care providers q q q What are today’s scientific challenges? What are the roadblocks to progress? What do we need to do to overcome roadblocks?
 What is the NIH Roadmap? n n n A framework of priorities the NIH as a whole must address in order to optimize its entire research portfolio. A vision for a more efficient, innovative and productive system of biomedical and behavioral research. A set of initiatives that are central to extending the quality of healthy life for people in this country and around the world.
What is the NIH Roadmap? n n n A framework of priorities the NIH as a whole must address in order to optimize its entire research portfolio. A vision for a more efficient, innovative and productive system of biomedical and behavioral research. A set of initiatives that are central to extending the quality of healthy life for people in this country and around the world.
 NIH Roadmap for Medical Research New Pathways to Discovery NIH Research Teams of the Future Re-engineering the Clinical Research Enterprise
NIH Roadmap for Medical Research New Pathways to Discovery NIH Research Teams of the Future Re-engineering the Clinical Research Enterprise
 The Biological Data of the Future n n n n Destructive Qualitative Uni-dimensional Low temporal resolution Low data density Variable standards Non cumulative n n n n Non-destructive Quantitative Multi-dimensional and spatially resolved High Temporal resolution High data density Stricter standards Cumulative
The Biological Data of the Future n n n n Destructive Qualitative Uni-dimensional Low temporal resolution Low data density Variable standards Non cumulative n n n n Non-destructive Quantitative Multi-dimensional and spatially resolved High Temporal resolution High data density Stricter standards Cumulative
 Multi- and Interdisciplinary Research will be Required to Solve the “Puzzle” of Complex Diseases and Conditions Genes Behavior Diet/Nutrition Infectious agents Environment Society ? ? ?
Multi- and Interdisciplinary Research will be Required to Solve the “Puzzle” of Complex Diseases and Conditions Genes Behavior Diet/Nutrition Infectious agents Environment Society ? ? ?
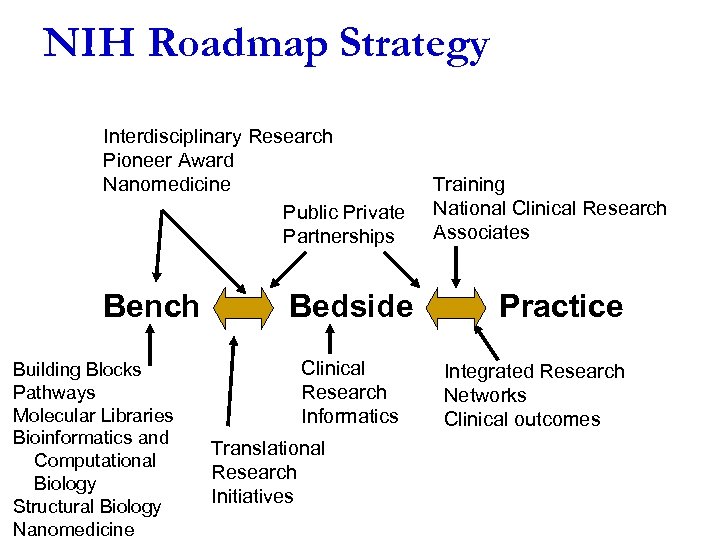 NIH Roadmap Strategy Interdisciplinary Research Pioneer Award Nanomedicine Public Private Partnerships Bench Building Blocks Pathways Molecular Libraries Bioinformatics and Computational Biology Structural Biology Nanomedicine Bedside Clinical Research Informatics Translational Research Initiatives Training National Clinical Research Associates Practice Integrated Research Networks Clinical outcomes
NIH Roadmap Strategy Interdisciplinary Research Pioneer Award Nanomedicine Public Private Partnerships Bench Building Blocks Pathways Molecular Libraries Bioinformatics and Computational Biology Structural Biology Nanomedicine Bedside Clinical Research Informatics Translational Research Initiatives Training National Clinical Research Associates Practice Integrated Research Networks Clinical outcomes
 Re-engineering the Clinical Research Enterprise Molecular Libraries and Imaging Clinical Enterprise Public-Private Partnerships Implementation Groups Structural Biology Bioinformatics and Computational Biology High-risk Research Interdisciplinary Research Teams Building Blocks, Biological Pathways and Networks Nanomedicine New Pathways to Discovery
Re-engineering the Clinical Research Enterprise Molecular Libraries and Imaging Clinical Enterprise Public-Private Partnerships Implementation Groups Structural Biology Bioinformatics and Computational Biology High-risk Research Interdisciplinary Research Teams Building Blocks, Biological Pathways and Networks Nanomedicine New Pathways to Discovery
 Key elements of Roadmap funding and management n All Institutes: q q q Participate with their scientific community in defining all components of the Roadmap Contribute equally and proportionately Participate directly in decision making and have a direct liaison to the Roadmap n All Roadmap initiatives are offered for competition to researchers from all fields n All research communities can compete for all initiatives The peer-review process will ensure appropriate expertise n
Key elements of Roadmap funding and management n All Institutes: q q q Participate with their scientific community in defining all components of the Roadmap Contribute equally and proportionately Participate directly in decision making and have a direct liaison to the Roadmap n All Roadmap initiatives are offered for competition to researchers from all fields n All research communities can compete for all initiatives The peer-review process will ensure appropriate expertise n
 Roadmap Funding dollars in millions FY 2004 Funding = $128. 3 (dollars in millions) New Pathways to Discovery $64. 1 NIH $26. 6 Research Teams of the Future $37. 6 Re-engineering the Clinical Research Enterprise
Roadmap Funding dollars in millions FY 2004 Funding = $128. 3 (dollars in millions) New Pathways to Discovery $64. 1 NIH $26. 6 Research Teams of the Future $37. 6 Re-engineering the Clinical Research Enterprise
 Roadmap Funding dollars in millions FY 04 FY 05 FY 06 FY 07 FY 08 FY 09 Total Pathways to Discovery 64 137 169 182 209 188 948 Research Teams 27 39 44 92 96 93 390 Clinical Research 38 61 120 174 214 227 833 Total 128 237 332 448 520 507 2, 172 0. 34% 0. 63% To be competed for in a common pool of initiatives by all researchers from every discipline ~0. 9%
Roadmap Funding dollars in millions FY 04 FY 05 FY 06 FY 07 FY 08 FY 09 Total Pathways to Discovery 64 137 169 182 209 188 948 Research Teams 27 39 44 92 96 93 390 Clinical Research 38 61 120 174 214 227 833 Total 128 237 332 448 520 507 2, 172 0. 34% 0. 63% To be competed for in a common pool of initiatives by all researchers from every discipline ~0. 9%
 NEW PATHWAYS TO DISCOVERY Working Group and Co-Chairs n n n Molecular Library and Imaging Francis Collins, NHGRI Tom Insel, NIMH Rod Pettigrew, NIBIB Building Blocks and Pathways Francis Collins, NHGRI Richard Hodes, NIA T-K Li, NIAAA Allen Spiegel, NIDDK Structural Biology Jeremy Berg, NIGMS Paul Sieving, NEI Bioinformatics and Computational Biology Jeremy Berg, NIGMS Don Lindberg, NLM Nanomedicine Jeffery Schloss, NHGRI Paul Sieving, NEI
NEW PATHWAYS TO DISCOVERY Working Group and Co-Chairs n n n Molecular Library and Imaging Francis Collins, NHGRI Tom Insel, NIMH Rod Pettigrew, NIBIB Building Blocks and Pathways Francis Collins, NHGRI Richard Hodes, NIA T-K Li, NIAAA Allen Spiegel, NIDDK Structural Biology Jeremy Berg, NIGMS Paul Sieving, NEI Bioinformatics and Computational Biology Jeremy Berg, NIGMS Don Lindberg, NLM Nanomedicine Jeffery Schloss, NHGRI Paul Sieving, NEI
 New Pathways to Discovery § Molecular Libraries and Imaging § Building Blocks, Biological Pathways and Networks § Structural Biology § Bioinformatics and Computational Biology § Nanomedicine
New Pathways to Discovery § Molecular Libraries and Imaging § Building Blocks, Biological Pathways and Networks § Structural Biology § Bioinformatics and Computational Biology § Nanomedicine
 Three recent developments make small molecule/chemical genomics initiatives feasible Human Genome Project Modern Synthetic Chemistry Availability of targets Compound Collections Availability of compounds Public sector screening and chemistry initiative Robotic Technology Availability of screening
Three recent developments make small molecule/chemical genomics initiatives feasible Human Genome Project Modern Synthetic Chemistry Availability of targets Compound Collections Availability of compounds Public sector screening and chemistry initiative Robotic Technology Availability of screening
 Molecular Libraries: Putting Chemistry to Work for Medicine n n n Six national screening centers for small molecules Public database for “chemical genomics” Technology advances in combinatorial chemistry, robotics, virtual screening
Molecular Libraries: Putting Chemistry to Work for Medicine n n n Six national screening centers for small molecules Public database for “chemical genomics” Technology advances in combinatorial chemistry, robotics, virtual screening
 Collaborative Pipeline of a NIH Chemical Genomics Center Peer review Assay Customized Assay Compound Repository Investigator Screen Limited Med. Cheminformatics, Pub. Chem (NCBI) Probe List Probe picking, confirmation, secondary screens
Collaborative Pipeline of a NIH Chemical Genomics Center Peer review Assay Customized Assay Compound Repository Investigator Screen Limited Med. Cheminformatics, Pub. Chem (NCBI) Probe List Probe picking, confirmation, secondary screens
 Molecular Imaging Roadmap Components n n n Development of high resolution probes for cellular imaging Ø RFA issued in 2004 Ø http: //grants. nih. gov/grants/guide/rfa-files/RFA-RM 04 -001. html Development of an imaging probe database Ø In process, with links to Pub. Chem Core synthesis facility to produce imaging probes Ø Efforts to establish an intramural facility are underway
Molecular Imaging Roadmap Components n n n Development of high resolution probes for cellular imaging Ø RFA issued in 2004 Ø http: //grants. nih. gov/grants/guide/rfa-files/RFA-RM 04 -001. html Development of an imaging probe database Ø In process, with links to Pub. Chem Core synthesis facility to produce imaging probes Ø Efforts to establish an intramural facility are underway
 New Pathways to Discovery § Molecular Libraries and Imaging § Building Blocks, Biological Pathways and Networks § Structural Biology § Bioinformatics and Computational Biology § Nanomedicine
New Pathways to Discovery § Molecular Libraries and Imaging § Building Blocks, Biological Pathways and Networks § Structural Biology § Bioinformatics and Computational Biology § Nanomedicine
 Structural Biology n n n Initiative: Centers for Innovation in Membrane Protein Production Applications due March 11, 2004 $5 M FY 2004 Roadmap funding (~2 Centers, P 50 Mechanism)
Structural Biology n n n Initiative: Centers for Innovation in Membrane Protein Production Applications due March 11, 2004 $5 M FY 2004 Roadmap funding (~2 Centers, P 50 Mechanism)
 Centers for Innovation in Membrane Protein Production n n Many physiologically and pharmaceutically important proteins are membrane proteins Few membrane proteins structures known All eukaryotic membrane protein structures determined to date have been from proteins derived from naturally rich sources Detergents and other agents required for solubilization and crystallization Development of methods for the production of structurally and functionally intact membrane proteins for subsequent structural studies
Centers for Innovation in Membrane Protein Production n n Many physiologically and pharmaceutically important proteins are membrane proteins Few membrane proteins structures known All eukaryotic membrane protein structures determined to date have been from proteins derived from naturally rich sources Detergents and other agents required for solubilization and crystallization Development of methods for the production of structurally and functionally intact membrane proteins for subsequent structural studies
 B. W. Matthews Ann. Rev. Phys. Chem. 27, 493 (1976) http: //www. mpibp-frankfurt. pg. de/ michel/public/memprotstruct. html progress in membrane protein structure determinations parallels that of water-soluble proteins with a ~25 year offset Courtesy of Doug Rees, Caltech
B. W. Matthews Ann. Rev. Phys. Chem. 27, 493 (1976) http: //www. mpibp-frankfurt. pg. de/ michel/public/memprotstruct. html progress in membrane protein structure determinations parallels that of water-soluble proteins with a ~25 year offset Courtesy of Doug Rees, Caltech
 Structural Biology Roadmap Plans n n n Wide range of structural biology programs throughout NIH (intramural and extramural) Synchrotron sources supported by DOE, NIH (NCRR, NCI, NIGMS), and others NMR instrumentation supported (NCRR, NIGMS) Protein Structure Initiative-Network of Centers devoted to structural genomics Roadmap initiatives will be used to provide integration of these programs
Structural Biology Roadmap Plans n n n Wide range of structural biology programs throughout NIH (intramural and extramural) Synchrotron sources supported by DOE, NIH (NCRR, NCI, NIGMS), and others NMR instrumentation supported (NCRR, NIGMS) Protein Structure Initiative-Network of Centers devoted to structural genomics Roadmap initiatives will be used to provide integration of these programs
 Protein Structure Initiative
Protein Structure Initiative
 Protein Structure Initiative (PSI) PSI Pilot phase n Nine research centers funded 2000 -2001 q Pilots to examine the best strategies q Methodology and technology development q Construction of structural genomics pipeline and automation of all steps q Increases in efficiency and success rates and lower costs q Production of unique protein structures
Protein Structure Initiative (PSI) PSI Pilot phase n Nine research centers funded 2000 -2001 q Pilots to examine the best strategies q Methodology and technology development q Construction of structural genomics pipeline and automation of all steps q Increases in efficiency and success rates and lower costs q Production of unique protein structures
 PSI Pilot Research Centers p UK UK, Japan, Israel
PSI Pilot Research Centers p UK UK, Japan, Israel
 PSI Goals n n n To make three-dimensional atomic level structures of most proteins easily available from knowledge of their corresponding DNA sequences Information on function Value of comparisons of protein structures Key biochemical and biophysical problems q Protein folding, prediction, folds, evolution Other benefits to biologists q Methodology and technology developments q Structural biology facilities q Availability of reagents and materials q Experimental outcome data on protein production and crystallization
PSI Goals n n n To make three-dimensional atomic level structures of most proteins easily available from knowledge of their corresponding DNA sequences Information on function Value of comparisons of protein structures Key biochemical and biophysical problems q Protein folding, prediction, folds, evolution Other benefits to biologists q Methodology and technology developments q Structural biology facilities q Availability of reagents and materials q Experimental outcome data on protein production and crystallization
 PSI Policies n n n n Deposition and release of coordinates in PDB upon completion Public listing of targets and progress Results on PSI webpage and all center websites Technical workshops: protein production and crystallization; data management; target selection; comparative modeling; structural determination Repository for materials -- clones, reagents, samples Databases: PDB, Target. DB, Pepc. DB Administrative supplements to R 01 s for functional studies of PSI structures
PSI Policies n n n n Deposition and release of coordinates in PDB upon completion Public listing of targets and progress Results on PSI webpage and all center websites Technical workshops: protein production and crystallization; data management; target selection; comparative modeling; structural determination Repository for materials -- clones, reagents, samples Databases: PDB, Target. DB, Pepc. DB Administrative supplements to R 01 s for functional studies of PSI structures
 PSI technology and methodology n n Robotic systems for cloning, expression, purification, characterization, crystallization, data collection, sample changers Automated structure determination LIMS Developments: Solubility engineering, capillary crystallization, auto-inducing media, cell-free protein production, domain parsing, protein-pair discovery, expression vectors, disorder predictions and methods, direct crystallography
PSI technology and methodology n n Robotic systems for cloning, expression, purification, characterization, crystallization, data collection, sample changers Automated structure determination LIMS Developments: Solubility engineering, capillary crystallization, auto-inducing media, cell-free protein production, domain parsing, protein-pair discovery, expression vectors, disorder predictions and methods, direct crystallography
 Research Centers n n Structures determined: 403 in first three years (doubling each year) Unique structures: 70% for PSI (10% for PDB) New folds: 12% for PSI (3% for PDB) Average costs per structure – decreasing significantly (<$240 K)
Research Centers n n Structures determined: 403 in first three years (doubling each year) Unique structures: 70% for PSI (10% for PDB) New folds: 12% for PSI (3% for PDB) Average costs per structure – decreasing significantly (<$240 K)
 Andrzej Joachimiak, P 50 GM 062414
Andrzej Joachimiak, P 50 GM 062414
 Ian A. Wilson, Scripps Research Institute, P 50 GM 062411 Technology Status – Gene to Structure PROTEIN PRODUCTION 4 th Generation System In use since Dec, 2000 NANOVOLUME CRYSTALLIZATION Established, May 1998 PROTEIN PURIFICATION CRYSTALLIZATION Generation System In use since March, 2002 2 nd Generation System In use since Feb. , 2001 IMAGING HT Data Collection 1 st Generation Hardware 6 th Generation Software 1 st Generation System 3 rd Generation Software 3 rd
Ian A. Wilson, Scripps Research Institute, P 50 GM 062411 Technology Status – Gene to Structure PROTEIN PRODUCTION 4 th Generation System In use since Dec, 2000 NANOVOLUME CRYSTALLIZATION Established, May 1998 PROTEIN PURIFICATION CRYSTALLIZATION Generation System In use since March, 2002 2 nd Generation System In use since Feb. , 2001 IMAGING HT Data Collection 1 st Generation Hardware 6 th Generation Software 1 st Generation System 3 rd Generation Software 3 rd
 IR 24 ER 14 Gaetano T. Montelione, P 50 GM 062413 FGF-2 Structures analyzed with automated NMR analysis software developed by NESG IL 13 WR 41 JR 19 ZR 18 ER 115 WR 64 OP 3 WR 33 MMP-1 N-Tm. Zip Z-domain C-Tm. Zip LC 8 ZR 31 WR 90 ER 75
IR 24 ER 14 Gaetano T. Montelione, P 50 GM 062413 FGF-2 Structures analyzed with automated NMR analysis software developed by NESG IL 13 WR 41 JR 19 ZR 18 ER 115 WR 64 OP 3 WR 33 MMP-1 N-Tm. Zip Z-domain C-Tm. Zip LC 8 ZR 31 WR 90 ER 75
 Sung-Hou Kim, P 50 GM 062412 Samples of Structure-based discovery of function (BSGC) MJ 0882 TM 841 Sequence inference: No molecular or cellular function Structural inference: Methyl transferase Biochemical assay: Methyl transferease Sequence inference: No molecular or cellular function Structural inference: Fatty acid binding protein MJ 0577 Sequence inference: No molecular or cellular function Weak Ham 1 homology Structural inference: Nucleotide binding protein (weak) Biochem. and complementation assay: Nucleotide housekeeping Sequence inference: No molecular or cellular function Structural inference: ATPase or Molecular switch Biochemical assay: Molecular switch
Sung-Hou Kim, P 50 GM 062412 Samples of Structure-based discovery of function (BSGC) MJ 0882 TM 841 Sequence inference: No molecular or cellular function Structural inference: Methyl transferase Biochemical assay: Methyl transferease Sequence inference: No molecular or cellular function Structural inference: Fatty acid binding protein MJ 0577 Sequence inference: No molecular or cellular function Weak Ham 1 homology Structural inference: Nucleotide binding protein (weak) Biochem. and complementation assay: Nucleotide housekeeping Sequence inference: No molecular or cellular function Structural inference: ATPase or Molecular switch Biochemical assay: Molecular switch
 PSI Pilot Phase -- Lessons Learned 1. 2. 3. 4. 5. 6. Structural genomics pipelines can be constructed and scaled-up High throughput operation works for many proteins Genomic approach works for structures Bottlenecks remain for some proteins A coordinated, 5 -year target selection policy must be developed Homology modeling methods need improvement
PSI Pilot Phase -- Lessons Learned 1. 2. 3. 4. 5. 6. Structural genomics pipelines can be constructed and scaled-up High throughput operation works for many proteins Genomic approach works for structures Bottlenecks remain for some proteins A coordinated, 5 -year target selection policy must be developed Homology modeling methods need improvement
 PSI-2 Large-scale Centers Goals n n Increase the number of sequence families that have at least one experimental structure Increase the number of sequenced genes for which homology models can be built Increase the biomedical significance of the structures Requires 4 -6, 000 unique experimental structures
PSI-2 Large-scale Centers Goals n n Increase the number of sequence families that have at least one experimental structure Increase the number of sequenced genes for which homology models can be built Increase the biomedical significance of the structures Requires 4 -6, 000 unique experimental structures
 PSI-2 Production Phase (2005) n n n Interacting network with three or four components q Large-scale centers q Specialized centers for technology development for challenging proteins q Disease-targeted structural genomics centers (pending) q Knowledge Base (future) Cooperative agreements Affiliated with the NIH Structural Biology Roadmap
PSI-2 Production Phase (2005) n n n Interacting network with three or four components q Large-scale centers q Specialized centers for technology development for challenging proteins q Disease-targeted structural genomics centers (pending) q Knowledge Base (future) Cooperative agreements Affiliated with the NIH Structural Biology Roadmap
 PSI-2 Large-scale Centers n n n High throughput structure output Continued technology and methodology development High throughput operation of all pipeline tasks Provisions for sharing facilities with the scientific community GM-05 -001
PSI-2 Large-scale Centers n n n High throughput structure output Continued technology and methodology development High throughput operation of all pipeline tasks Provisions for sharing facilities with the scientific community GM-05 -001
 PSI-2 Specialized Centers n Methodology and technology development for challenging proteins q Membrane proteins Higher eukaryote proteins, especially human q Small protein complexes q Other major bottlenecks to high throughput Major impact and applicability to PSI goals Leading toward high throughput operation GM-05 -002 q n n n
PSI-2 Specialized Centers n Methodology and technology development for challenging proteins q Membrane proteins Higher eukaryote proteins, especially human q Small protein complexes q Other major bottlenecks to high throughput Major impact and applicability to PSI goals Leading toward high throughput operation GM-05 -002 q n n n
 PSI-2 Disease-targeted Structural Genomics Centers (pending) n n n Protein structures from pathogens and from tissues and organ systems related to disease Member of the PSI network Under consideration by the NIH Structural Biology Roadmap
PSI-2 Disease-targeted Structural Genomics Centers (pending) n n n Protein structures from pathogens and from tissues and organ systems related to disease Member of the PSI network Under consideration by the NIH Structural Biology Roadmap
 http: //www. nigms. nih. gov/psi. html/
http: //www. nigms. nih. gov/psi. html/
 New Pathways to Discovery § Molecular Libraries and Imaging § Building Blocks, Biological Pathways and Networks § Structural Biology § Bioinformatics and Computational Biology § Nanomedicine
New Pathways to Discovery § Molecular Libraries and Imaging § Building Blocks, Biological Pathways and Networks § Structural Biology § Bioinformatics and Computational Biology § Nanomedicine
 Bioinformatics and Computational Biology n n n Initiative: National Centers for Biomedical Computing Applications received January 23, 2004 $12 M FY 2004 Roadmap funding (~4 Centers, U 54 Mechanism)
Bioinformatics and Computational Biology n n n Initiative: National Centers for Biomedical Computing Applications received January 23, 2004 $12 M FY 2004 Roadmap funding (~4 Centers, U 54 Mechanism)
 National Centers for Biomedical Computing n Partnerships of: Ø Ø Ø n n Computer scientists Biomedical computational scientists Experimental and clinical biomedical and behavioral researchers Focused on software rather than hardware Each National Center to have Driving Biological Projects Open source requirement Programs in preparation for partnerships between individual investigators and National Centers
National Centers for Biomedical Computing n Partnerships of: Ø Ø Ø n n Computer scientists Biomedical computational scientists Experimental and clinical biomedical and behavioral researchers Focused on software rather than hardware Each National Center to have Driving Biological Projects Open source requirement Programs in preparation for partnerships between individual investigators and National Centers
 RESEARCH TEAMS OF THE FUTURE Working Groups and Co-Chairs § Interdisciplinary Research Patricia Grady, NINR Ken Olden, NIEHS Larry Tabak, NIDCR § High-risk Research Ellie Ehrenfeld, NIAID Stephen Straus, NCCAM § Public-Private Partnerships Andy von Eschenbach, NCI Richard Hodes, NIA
RESEARCH TEAMS OF THE FUTURE Working Groups and Co-Chairs § Interdisciplinary Research Patricia Grady, NINR Ken Olden, NIEHS Larry Tabak, NIDCR § High-risk Research Ellie Ehrenfeld, NIAID Stephen Straus, NCCAM § Public-Private Partnerships Andy von Eschenbach, NCI Richard Hodes, NIA
 Multi- and Interdisciplinary Research A B A Work on Multidisciplinary common problem Interaction forges new discipline B C Interdisciplinary
Multi- and Interdisciplinary Research A B A Work on Multidisciplinary common problem Interaction forges new discipline B C Interdisciplinary
 Challenges to Interdisciplinary Research § § § The current system of academic advancement favors the independent investigator Most institutions house scientists in discrete departments Interdisciplinary science requires interdisciplinary peer-review Project management and oversight is currently performed by discrete ICs Interdisciplinary research teams take time to assemble and require unique resources
Challenges to Interdisciplinary Research § § § The current system of academic advancement favors the independent investigator Most institutions house scientists in discrete departments Interdisciplinary science requires interdisciplinary peer-review Project management and oversight is currently performed by discrete ICs Interdisciplinary research teams take time to assemble and require unique resources
 NIH Director’s Pioneer Award • New program to support individuals with untested, potentially groundbreaking ideas! • Encourages innovation, risk-taking • Totally new application and peer review process • Expected to be highly competitive • Expanded eligibility – (not only traditional biomedical investigators) • Provides $500, 000/year for 5 years
NIH Director’s Pioneer Award • New program to support individuals with untested, potentially groundbreaking ideas! • Encourages innovation, risk-taking • Totally new application and peer review process • Expected to be highly competitive • Expanded eligibility – (not only traditional biomedical investigators) • Provides $500, 000/year for 5 years
 RE-ENGINEERING THE CLINICAL RESEARCH ENTERPRISE Co-Chairs Working Groups and Co-Chairs Stephen Katz, NIAMS Stephen E. Straus, NCCAM Subgroups n n n Harmonization of Clinical Research Regulatory Processes Amy Patterson, OSP Integration of Clinical Research Networks, including NECTAR Larry Friedman, NHLBI Stephen Katz, NIAMS Enhance Clinical Research Workforce Training Duane Alexander, NICHD Rob Star, NIDDK Enabling Technologies for Improved Assessment of Clinical Outcomes Deborah Ader, NIAMS Larry Fine, OBSSR Stephen Katz, NIAMS Regional Translational Research Centers Stephen E. Straus, NCCAM Steve Zalcman, NIMH Translational Research Service Cores Josephine Briggs, NIDDK Stephen E. Straus, NCCAM
RE-ENGINEERING THE CLINICAL RESEARCH ENTERPRISE Co-Chairs Working Groups and Co-Chairs Stephen Katz, NIAMS Stephen E. Straus, NCCAM Subgroups n n n Harmonization of Clinical Research Regulatory Processes Amy Patterson, OSP Integration of Clinical Research Networks, including NECTAR Larry Friedman, NHLBI Stephen Katz, NIAMS Enhance Clinical Research Workforce Training Duane Alexander, NICHD Rob Star, NIDDK Enabling Technologies for Improved Assessment of Clinical Outcomes Deborah Ader, NIAMS Larry Fine, OBSSR Stephen Katz, NIAMS Regional Translational Research Centers Stephen E. Straus, NCCAM Steve Zalcman, NIMH Translational Research Service Cores Josephine Briggs, NIDDK Stephen E. Straus, NCCAM
 Clinical Research: Navigating the Roadway § Clinical research impeded by multiple and variable requirements to address fundamentally the same oversight concerns § Variability among and within agencies § Creates uncertainty about how to comply § Hampers efficiency and effectiveness
Clinical Research: Navigating the Roadway § Clinical research impeded by multiple and variable requirements to address fundamentally the same oversight concerns § Variability among and within agencies § Creates uncertainty about how to comply § Hampers efficiency and effectiveness
 The NIH Roadmap: A Work in Progress
The NIH Roadmap: A Work in Progress
 www. nihroadmap. nih. gov
www. nihroadmap. nih. gov


