60f2a5bb05295e48c3350b8d0733cacc.ppt
- Количество слайдов: 42
 Next generation sequencing Platforms, chemistries, and applications
Next generation sequencing Platforms, chemistries, and applications
 Outline • Sanger sequencing – Chain termination with modified d. NTPs • “Next generation sequencing” (NGS) – “Sequencing by synthesis” systems – Pyrosequencing refers to Roche GS FLX (formerly “ 454”) • 3 rd generation sequencing (discussed by Kristen) – e. g. , Nanostring
Outline • Sanger sequencing – Chain termination with modified d. NTPs • “Next generation sequencing” (NGS) – “Sequencing by synthesis” systems – Pyrosequencing refers to Roche GS FLX (formerly “ 454”) • 3 rd generation sequencing (discussed by Kristen) – e. g. , Nanostring
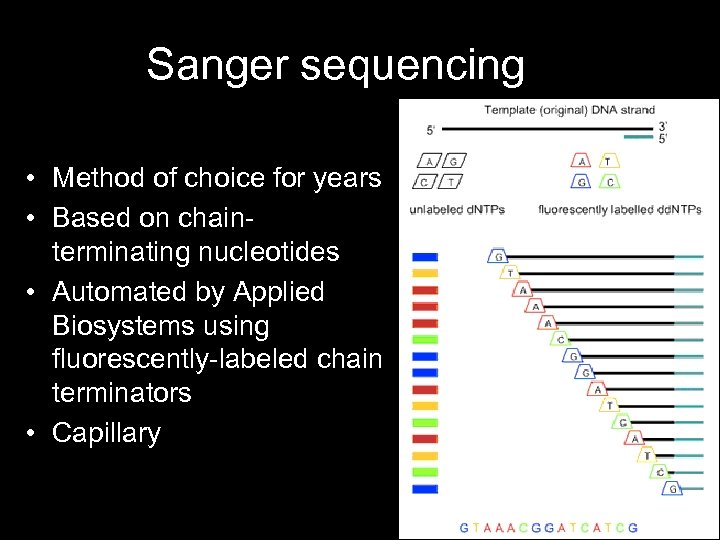 Sanger sequencing • Method of choice for years • Based on chainterminating nucleotides • Automated by Applied Biosystems using fluorescently-labeled chain terminators • Capillary
Sanger sequencing • Method of choice for years • Based on chainterminating nucleotides • Automated by Applied Biosystems using fluorescently-labeled chain terminators • Capillary
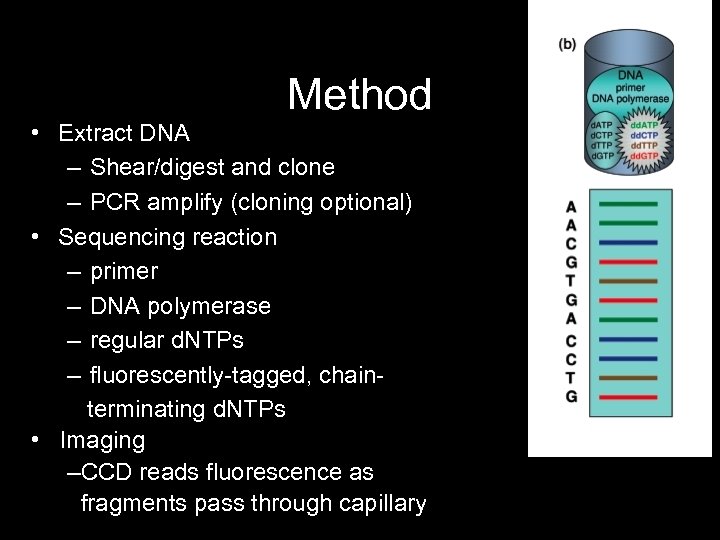 Method • Extract DNA – Shear/digest and clone – PCR amplify (cloning optional) • Sequencing reaction – primer – DNA polymerase – regular d. NTPs – fluorescently-tagged, chainterminating d. NTPs • Imaging –CCD reads fluorescence as fragments pass through capillary
Method • Extract DNA – Shear/digest and clone – PCR amplify (cloning optional) • Sequencing reaction – primer – DNA polymerase – regular d. NTPs – fluorescently-tagged, chainterminating d. NTPs • Imaging –CCD reads fluorescence as fragments pass through capillary
 Sanger sequencing: pros & cons • Pros – Long read lengths: up to ~700 bp – Most flexible in throughput: from 1 to 1, 000 s of samples – Convenient: found in many facilities • Cons – Expensive: ~$3/sequence – Requires PCR or bacterial -mediated preamplification – Cannot quantify genome copies or transcripts from DNA/c. DNA libraries* *Unless doing SAGE
Sanger sequencing: pros & cons • Pros – Long read lengths: up to ~700 bp – Most flexible in throughput: from 1 to 1, 000 s of samples – Convenient: found in many facilities • Cons – Expensive: ~$3/sequence – Requires PCR or bacterial -mediated preamplification – Cannot quantify genome copies or transcripts from DNA/c. DNA libraries* *Unless doing SAGE
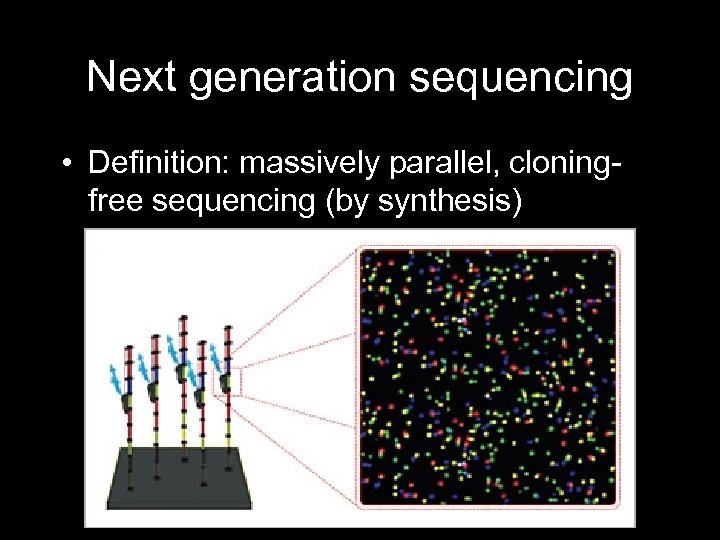 Next generation sequencing • Definition: massively parallel, cloningfree sequencing (by synthesis) – Roche GS FLX (pyrosequencing) – Illumina (Solexa sequencing) – Applied Biosystems (SOLi. D)
Next generation sequencing • Definition: massively parallel, cloningfree sequencing (by synthesis) – Roche GS FLX (pyrosequencing) – Illumina (Solexa sequencing) – Applied Biosystems (SOLi. D)
 Roche GS FLX (“ 454”) • The original “pyrosequencer” • Pyrosequencing is not new (Nyren 1996) • Was converted into high-throughput system in 2005 (Margulis et al. Nature)
Roche GS FLX (“ 454”) • The original “pyrosequencer” • Pyrosequencing is not new (Nyren 1996) • Was converted into high-throughput system in 2005 (Margulis et al. Nature)
 GS FLX library preparation • Shear DNA/c. DNA and ligate to adaptors – Amount of shearing is dependent on desired read length – New reagents “claim” reads up to 500 bp – How much variation does this lead to?
GS FLX library preparation • Shear DNA/c. DNA and ligate to adaptors – Amount of shearing is dependent on desired read length – New reagents “claim” reads up to 500 bp – How much variation does this lead to?
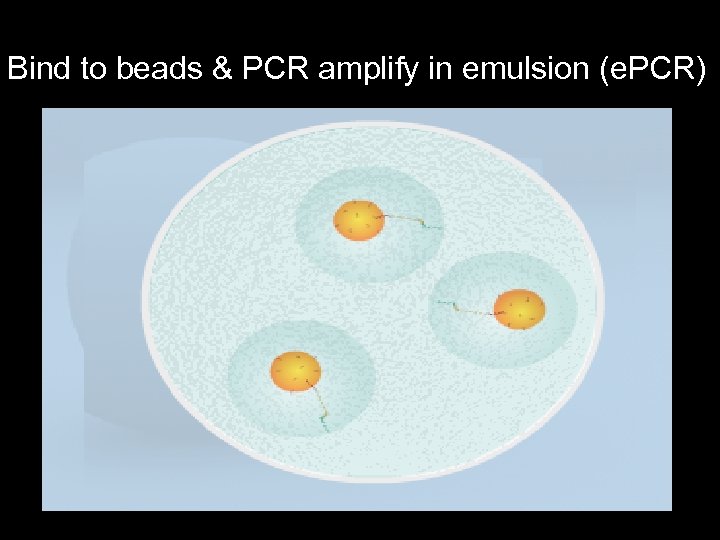 Bind to beads & PCR amplify in emulsion (e. PCR)
Bind to beads & PCR amplify in emulsion (e. PCR)
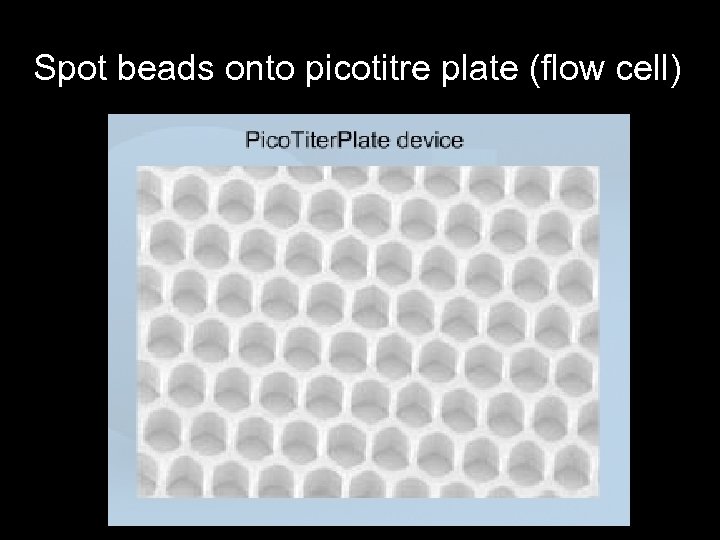 Spot beads onto picotitre plate (flow cell)
Spot beads onto picotitre plate (flow cell)
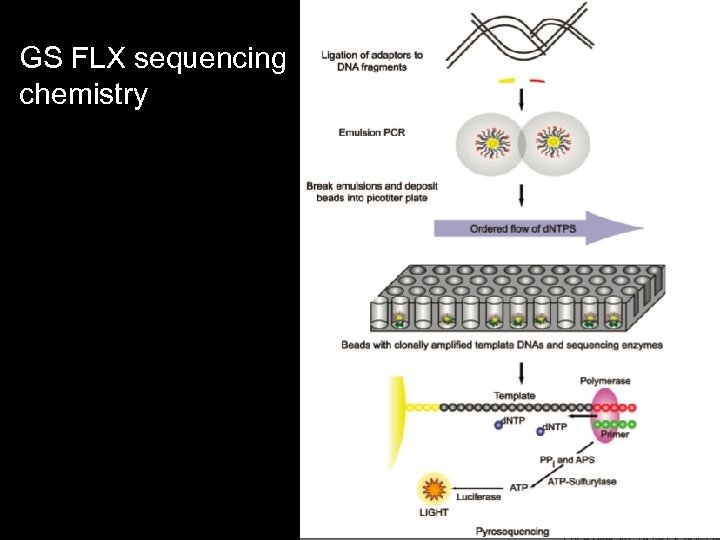 GS FLX sequencing chemistry
GS FLX sequencing chemistry
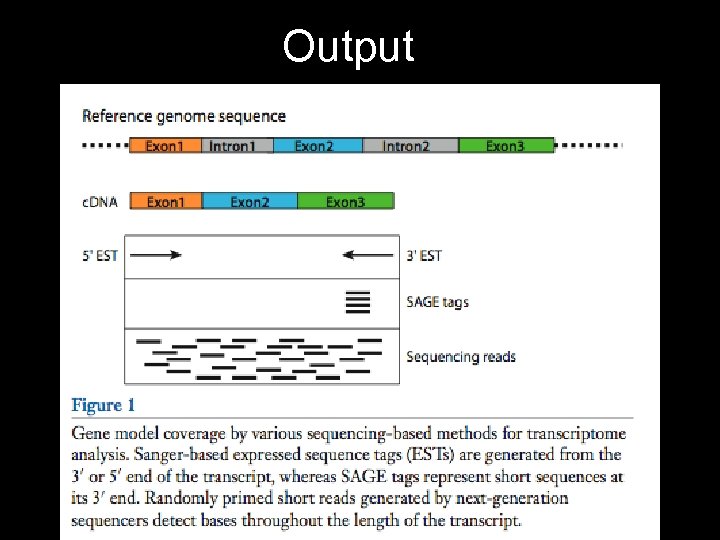 Output • Creates an image for every read • ~13 Mbp/hr, ~400 -500 bp/read • Best instrument for de novo work
Output • Creates an image for every read • ~13 Mbp/hr, ~400 -500 bp/read • Best instrument for de novo work
 GS FLX pros & cons vs. Sanger • Pros – Cloning-free – Generates Mbp of DNA sequence – Massively parallel: all sequencing done simultaneously – Quantitative: # reads => # molecules in sample – Cheaper than Sanger at $/bp • Cons – Shorter read lengths: 200 -400 bp – Low biological replication (n = 8 for $10 k run) – Low flexibility in throughput: must do high throughput
GS FLX pros & cons vs. Sanger • Pros – Cloning-free – Generates Mbp of DNA sequence – Massively parallel: all sequencing done simultaneously – Quantitative: # reads => # molecules in sample – Cheaper than Sanger at $/bp • Cons – Shorter read lengths: 200 -400 bp – Low biological replication (n = 8 for $10 k run) – Low flexibility in throughput: must do high throughput
 Illumina (formerly Solexa) • Polymerasebased sequencing by synthesis
Illumina (formerly Solexa) • Polymerasebased sequencing by synthesis
 Protocol • Shear DNA/c. DNA and link to adaptors • Adaptors bind to probes on flow cell • Adaptor “lawn” (similar to a probe array)
Protocol • Shear DNA/c. DNA and link to adaptors • Adaptors bind to probes on flow cell • Adaptor “lawn” (similar to a probe array)
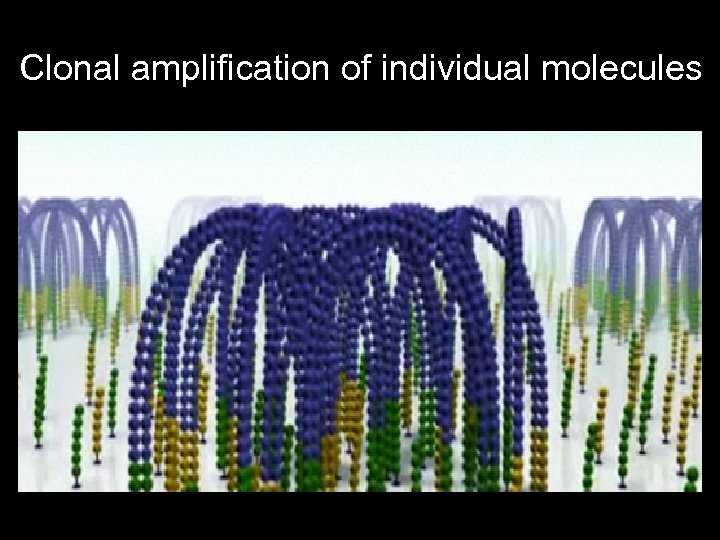 Clonal amplification of individual molecules
Clonal amplification of individual molecules
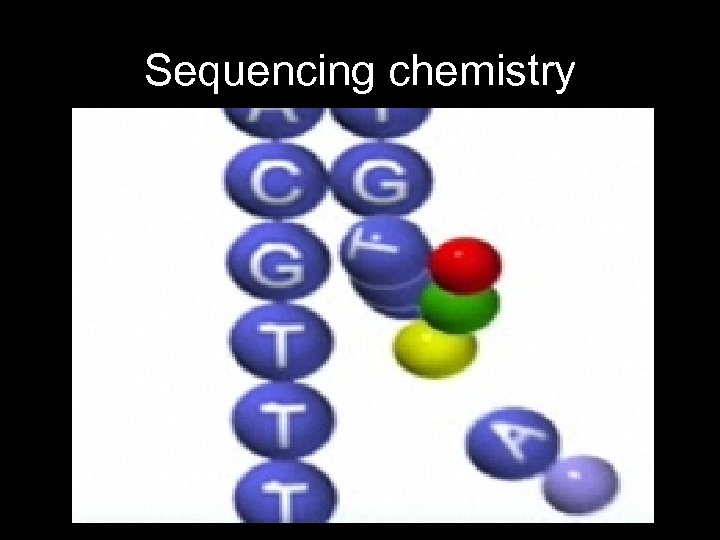 Sequencing chemistry – Fluorescently labeled bases • Initially blocked to prevent polymerization • Laser reads fluorescence • Unblocked so that next base can be added
Sequencing chemistry – Fluorescently labeled bases • Initially blocked to prevent polymerization • Laser reads fluorescence • Unblocked so that next base can be added
 Output • Superimposed image of 4 colors • RNA-seq application (Kristen)
Output • Superimposed image of 4 colors • RNA-seq application (Kristen)
 Illumina : pros & cons vs. Sanger • Pros – Cloning-free – Generates Gbp of DNA sequence – Massively parallel: all sequencing done simultaneously – Quantitative: # reads => # molecules in sample – Cheaper at $/bp • Cons – Short read lengths: 20100 bp – Low biological replication (n = 8 for $10 k run) – Low flexibility in throughput: must do high throughput – Run lasts from 1 -3 days
Illumina : pros & cons vs. Sanger • Pros – Cloning-free – Generates Gbp of DNA sequence – Massively parallel: all sequencing done simultaneously – Quantitative: # reads => # molecules in sample – Cheaper at $/bp • Cons – Short read lengths: 20100 bp – Low biological replication (n = 8 for $10 k run) – Low flexibility in throughput: must do high throughput – Run lasts from 1 -3 days
 Applied Biosystems SOLi. D • Supported oligonucleotide ligation and detection system • Similar to FLX but uses DNA ligase • e. PCR beads coated onto slide
Applied Biosystems SOLi. D • Supported oligonucleotide ligation and detection system • Similar to FLX but uses DNA ligase • e. PCR beads coated onto slide
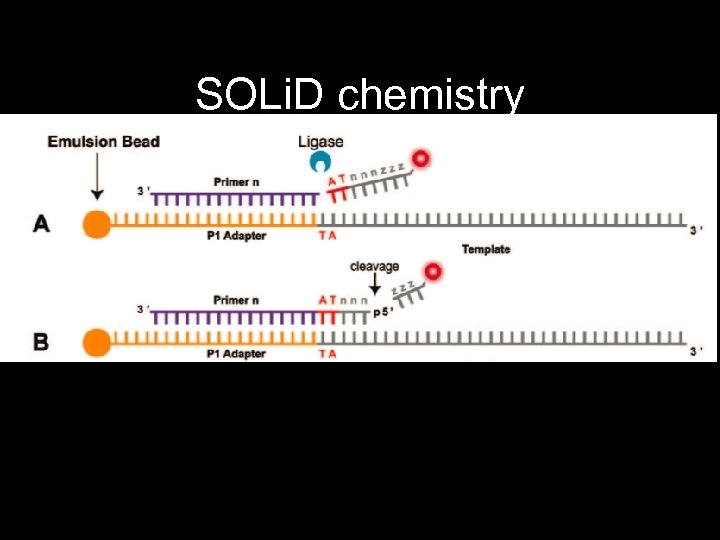 SOLi. D chemistry
SOLi. D chemistry
 Coverage: 20 X
Coverage: 20 X
 SOLi. D : pros & cons vs. Sanger • Pros – Cloning-free – Generates Gbp of DNA sequence – Massively parallel: all sequencing done simultaneously – Quantitative: # reads => # molecules in sample – Cheaper at $/bp • Cons – Short read lengths: 2550 bp – Low biological replication (n = 8 for $12 k run) – Low flexibility in throughput: must do high throughput – Run lasts from 3 -6 days
SOLi. D : pros & cons vs. Sanger • Pros – Cloning-free – Generates Gbp of DNA sequence – Massively parallel: all sequencing done simultaneously – Quantitative: # reads => # molecules in sample – Cheaper at $/bp • Cons – Short read lengths: 2550 bp – Low biological replication (n = 8 for $12 k run) – Low flexibility in throughput: must do high throughput – Run lasts from 3 -6 days
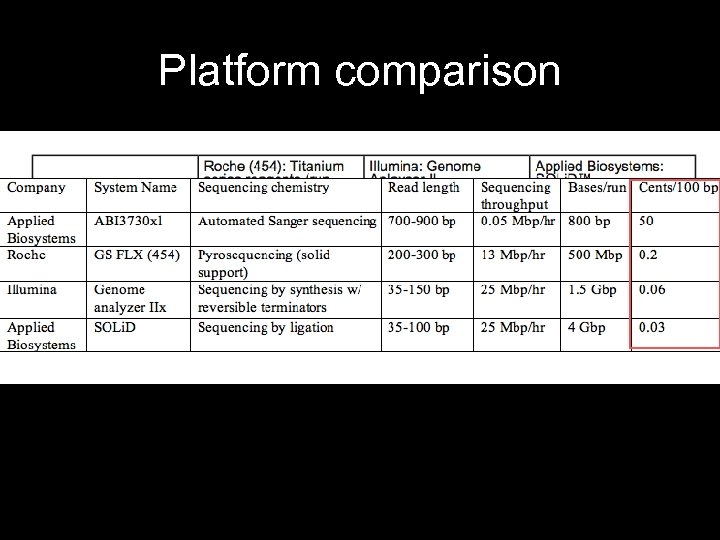 Platform comparison
Platform comparison
 Applications • • Genome sequencing Resequencing Transcriptome characterization Comparative transcriptomics mi. RNA profiling Epigenetics CHi. P sequencing
Applications • • Genome sequencing Resequencing Transcriptome characterization Comparative transcriptomics mi. RNA profiling Epigenetics CHi. P sequencing
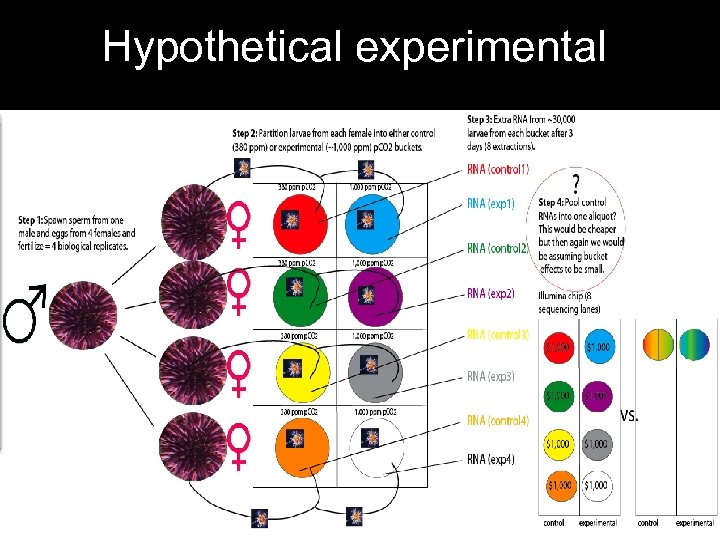 Hypothetical experimental
Hypothetical experimental
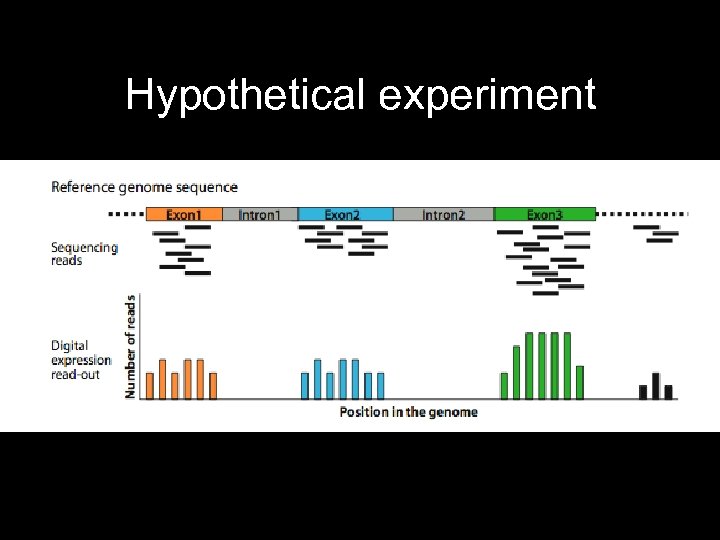 Hypothetical experiment • Sequence c. DNA libraries from each bucket and/or treatment • Count reads for each transcript • Compare transcript abundances between treatments • BLAST against reference genome
Hypothetical experiment • Sequence c. DNA libraries from each bucket and/or treatment • Count reads for each transcript • Compare transcript abundances between treatments • BLAST against reference genome
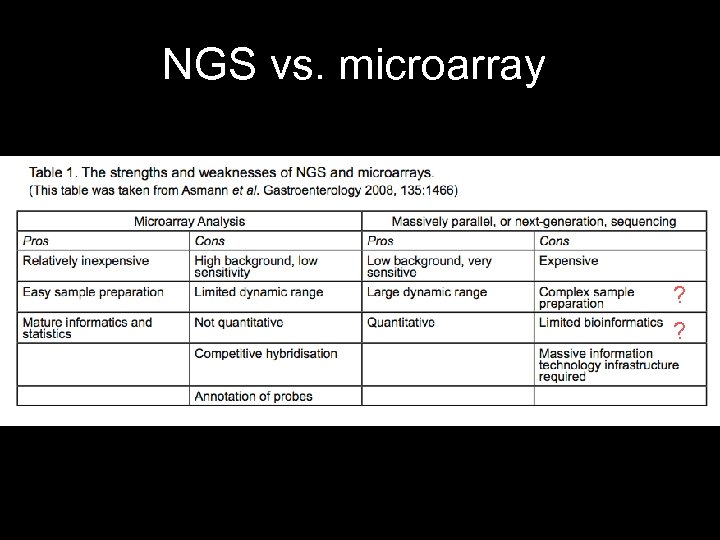 NGS vs. microarray • With microarray: must have sequences in hand to design probes. • With NGS: there is no such bias. – Sequence everything. – # of reads is proportional to # of transcripts. – Also no bias to particular gene region. ? ?
NGS vs. microarray • With microarray: must have sequences in hand to design probes. • With NGS: there is no such bias. – Sequence everything. – # of reads is proportional to # of transcripts. – Also no bias to particular gene region. ? ?
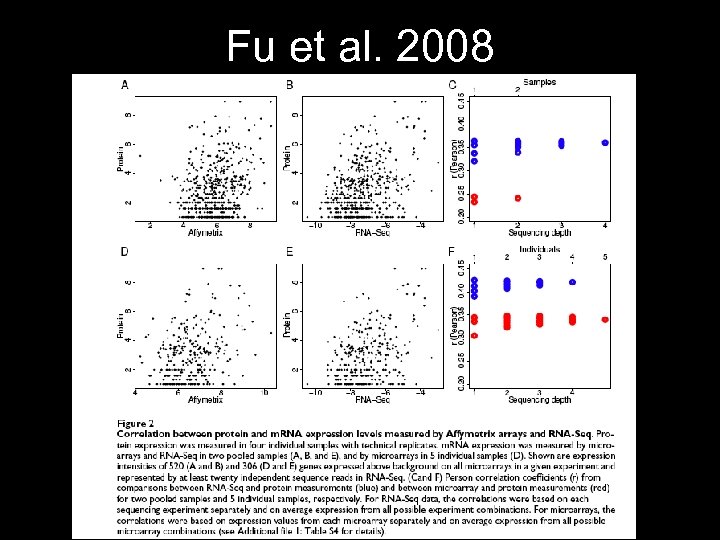 Fu et al. 2008
Fu et al. 2008
 Microarrays: a dying technology? • • • Must generate sequences first Difficulty in interpreting data Probe hybridization issues Can only resolve large differences NGS shows higher correlation w/ protein But NGS is a bioinformatics nightmare!!
Microarrays: a dying technology? • • • Must generate sequences first Difficulty in interpreting data Probe hybridization issues Can only resolve large differences NGS shows higher correlation w/ protein But NGS is a bioinformatics nightmare!!
 The beginning of the end of the microarray? • • Knowledge of sequences on array Cross-hyb problematic if seq are similar Difficult to detect low abundant species Reproducibility b/w labs and platforms
The beginning of the end of the microarray? • • Knowledge of sequences on array Cross-hyb problematic if seq are similar Difficult to detect low abundant species Reproducibility b/w labs and platforms
 RNA-Seq: a new tool for transcriptomics - “shotgun transcriptomic sequencing/short read” - more precise method of measuring expression • • • Illumina, Applied Biosystems SOLi. D, 454 Life Sciences Transcriptomics on non-model organisms Reveal SNPs Reveal connectivity b/w exons (long or paired reads) High accuracy, on par with q. PCR Quantitation • Spike-in RNA standards • No upper limit, 5 orders of magnitude • No extensive normalization required across treatments
RNA-Seq: a new tool for transcriptomics - “shotgun transcriptomic sequencing/short read” - more precise method of measuring expression • • • Illumina, Applied Biosystems SOLi. D, 454 Life Sciences Transcriptomics on non-model organisms Reveal SNPs Reveal connectivity b/w exons (long or paired reads) High accuracy, on par with q. PCR Quantitation • Spike-in RNA standards • No upper limit, 5 orders of magnitude • No extensive normalization required across treatments
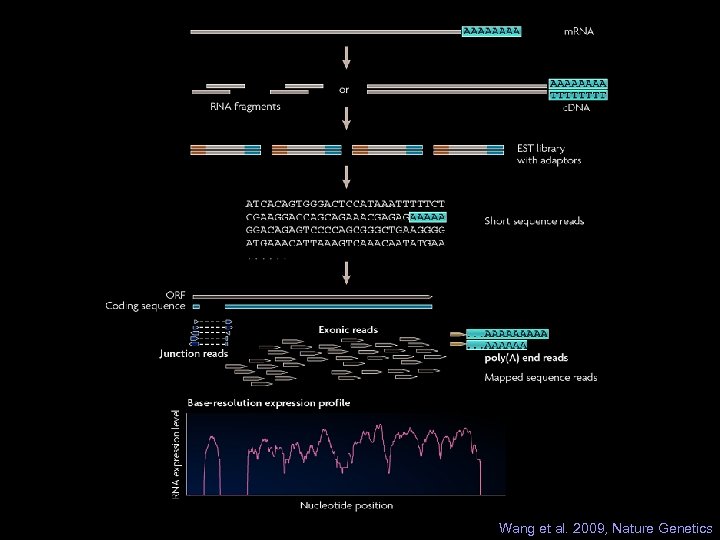 Wang et al. 2009, Nature Genetics
Wang et al. 2009, Nature Genetics
 -Illumina sequencing ~35 bp, single end reads, ~ 15 M reads Nagalakshmi et al. 2008, Science
-Illumina sequencing ~35 bp, single end reads, ~ 15 M reads Nagalakshmi et al. 2008, Science
 RNA-Seq pitfalls • Difficulty with the following: – – – Mapping short reads to the genome Appropriate assign. of ‘multi-mapping’ reads Identification of new splice junctions Sample comparison to ID diff. exp. genes Reads mapping outside annotated boundaries • Genomic DNA contamination • Pre-spliced heterogeneous nuclear RNA – Bioinformatic challenge Shendure 2008, Nature Methods
RNA-Seq pitfalls • Difficulty with the following: – – – Mapping short reads to the genome Appropriate assign. of ‘multi-mapping’ reads Identification of new splice junctions Sample comparison to ID diff. exp. genes Reads mapping outside annotated boundaries • Genomic DNA contamination • Pre-spliced heterogeneous nuclear RNA – Bioinformatic challenge Shendure 2008, Nature Methods

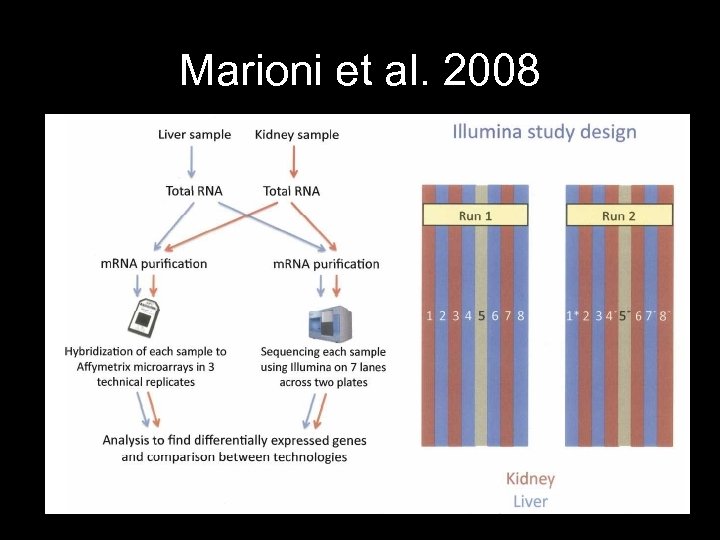 Marioni et al. 2008
Marioni et al. 2008
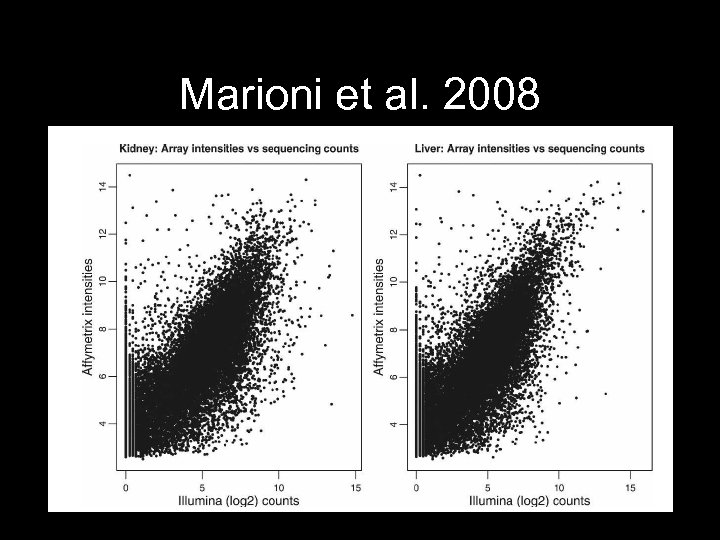 Marioni et al. 2008
Marioni et al. 2008
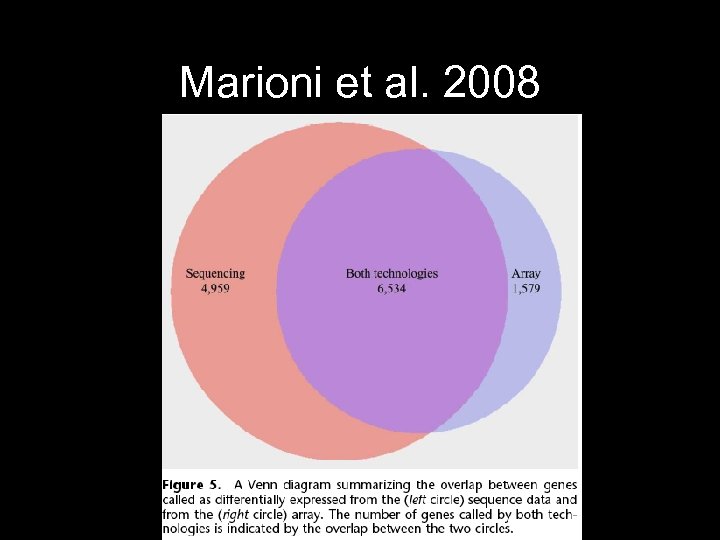 Marioni et al. 2008
Marioni et al. 2008
 Nano. String Technology -Minimal background signal -No amplification (induce bias) -Less sample needed -Improved detection of low exp. RNAs - single copy per cell Fortina and Surrey 2008, Nature Biotechnoloy
Nano. String Technology -Minimal background signal -No amplification (induce bias) -Less sample needed -Improved detection of low exp. RNAs - single copy per cell Fortina and Surrey 2008, Nature Biotechnoloy
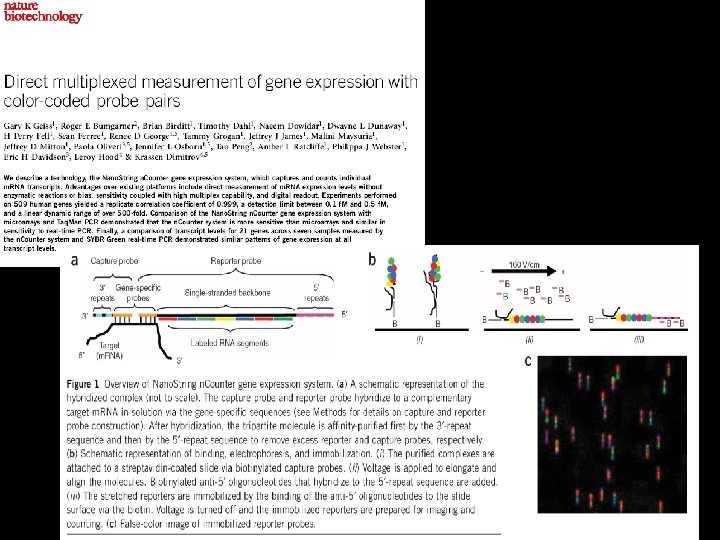
 Probe Design • 2 ss. DNA probes/ m. RNA (35 -50 bp oligo) • Overnight hybridization to m. RNA (solution-based) • Slide adhesion via biotin labeled capture probe • Reporter probe, 4 spectrally distinct dyes, 7 spaces • ‘Barcode’, 47 or 16, 384 barcodes
Probe Design • 2 ss. DNA probes/ m. RNA (35 -50 bp oligo) • Overnight hybridization to m. RNA (solution-based) • Slide adhesion via biotin labeled capture probe • Reporter probe, 4 spectrally distinct dyes, 7 spaces • ‘Barcode’, 47 or 16, 384 barcodes