87ef93815d514424baa28f9a8ca6d6d6.ppt
- Количество слайдов: 41
 Ne. Mo. Finder: Dissecting genomewide protein-protein intractions with meso-scale network motifs Mike Yuan
Ne. Mo. Finder: Dissecting genomewide protein-protein intractions with meso-scale network motifs Mike Yuan
 Outline of this presentation • • Introduction to PPI Introduction to Graph Mining Related work Problem statement Details of the Ne. Mo. Finder algorithm Summary References
Outline of this presentation • • Introduction to PPI Introduction to Graph Mining Related work Problem statement Details of the Ne. Mo. Finder algorithm Summary References
 Protein Interactions A Protein may interact with: – Other proteins – Nucleic Acids – Small molecules
Protein Interactions A Protein may interact with: – Other proteins – Nucleic Acids – Small molecules
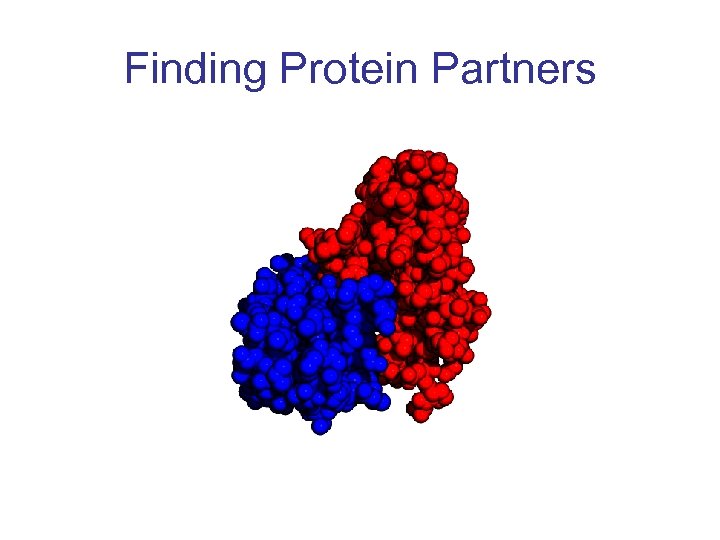 Finding Protein Partners
Finding Protein Partners
 Motivation • Important for biological functions • To understand the function of a protein, we need to find its interacting partners
Motivation • Important for biological functions • To understand the function of a protein, we need to find its interacting partners
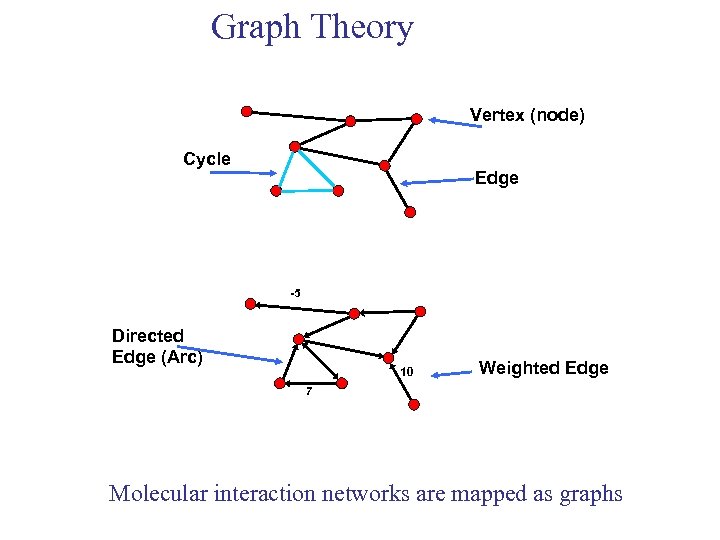 Graph Theory Vertex (node) Cycle Edge -5 Directed Edge (Arc) 10 Weighted Edge 7 Molecular interaction networks are mapped as graphs
Graph Theory Vertex (node) Cycle Edge -5 Directed Edge (Arc) 10 Weighted Edge 7 Molecular interaction networks are mapped as graphs
 The protein interaction network…
The protein interaction network…
 Graph mining • Methods for Mining Frequent Subgraphs • Mining Variant and Constrained Substructure Patterns • Applications: – Graph Indexing – Similarity Search – Classification and Clustering
Graph mining • Methods for Mining Frequent Subgraphs • Mining Variant and Constrained Substructure Patterns • Applications: – Graph Indexing – Similarity Search – Classification and Clustering
 Why Graph Mining? • Graphs are ubiquitous – Chemical compounds (Cheminformatics) – Protein structures, biological pathways/networks (Bioinformactics) – Program control flow, traffic flow, and workflow analysis – XML databases, Web, and social network analysis • Graph is a general model – Trees, lattices, sequences, and items are degenerated graphs • Complexity of algorithms: many problems are of high complexity
Why Graph Mining? • Graphs are ubiquitous – Chemical compounds (Cheminformatics) – Protein structures, biological pathways/networks (Bioinformactics) – Program control flow, traffic flow, and workflow analysis – XML databases, Web, and social network analysis • Graph is a general model – Trees, lattices, sequences, and items are degenerated graphs • Complexity of algorithms: many problems are of high complexity
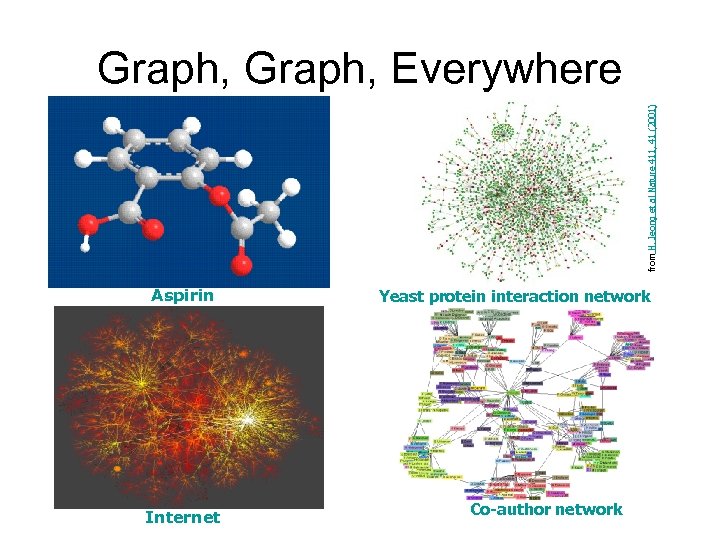 from H. Jeong et al Nature 411, 41 (2001) Graph, Everywhere Aspirin Internet Yeast protein interaction network Co-author network
from H. Jeong et al Nature 411, 41 (2001) Graph, Everywhere Aspirin Internet Yeast protein interaction network Co-author network
 Graph Pattern Mining • Frequent subgraphs – A (sub)graph is frequent if its support (occurrence frequency) in a given dataset is no less than a minimum support threshold • Applications of graph pattern mining – Mining biochemical structures – Program control flow analysis – Mining XML structures or Web communities – Building blocks for graph classification, clustering, compression, comparison, and correlation analysis
Graph Pattern Mining • Frequent subgraphs – A (sub)graph is frequent if its support (occurrence frequency) in a given dataset is no less than a minimum support threshold • Applications of graph pattern mining – Mining biochemical structures – Program control flow analysis – Mining XML structures or Web communities – Building blocks for graph classification, clustering, compression, comparison, and correlation analysis
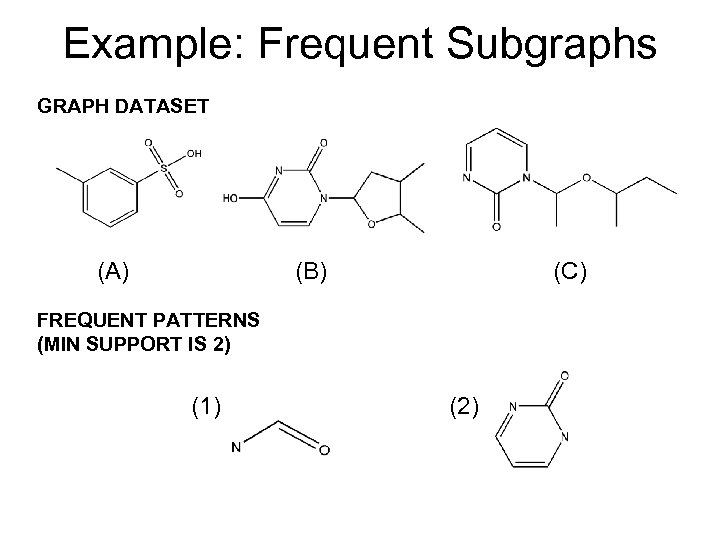 Example: Frequent Subgraphs GRAPH DATASET (A) (B) (C) FREQUENT PATTERNS (MIN SUPPORT IS 2) (1) (2)
Example: Frequent Subgraphs GRAPH DATASET (A) (B) (C) FREQUENT PATTERNS (MIN SUPPORT IS 2) (1) (2)
 Frequent Subgraph Mining Approaches • Apriori-based approach: if a graph is frequent, all of its subgraphs are frequent ─ the Apriori property – AGM/Ac. GM: Inokuchi, et al. (PKDD’ 00) – FSG: Kuramochi and Karypis (ICDM’ 01) – PATH#: Vanetik and Gudes (ICDM’ 02, ICDM’ 04) – FFSM: Huan, et al. (ICDM’ 03) • Pattern growth approach – Mo. Fa, Borgelt and Berthold (ICDM’ 02) – g. Span: Yan and Han (ICDM’ 02) – Gaston: Nijssen and Kok (KDD’ 04)
Frequent Subgraph Mining Approaches • Apriori-based approach: if a graph is frequent, all of its subgraphs are frequent ─ the Apriori property – AGM/Ac. GM: Inokuchi, et al. (PKDD’ 00) – FSG: Kuramochi and Karypis (ICDM’ 01) – PATH#: Vanetik and Gudes (ICDM’ 02, ICDM’ 04) – FFSM: Huan, et al. (ICDM’ 03) • Pattern growth approach – Mo. Fa, Borgelt and Berthold (ICDM’ 02) – g. Span: Yan and Han (ICDM’ 02) – Gaston: Nijssen and Kok (KDD’ 04)
 Problem Statement • PPI network G=(V, E) _ each vertex represents a unique protein _ each edge between v. A and v. B indicates there is an interaction between A and B • Network motif _frequently occurring subgraph pattern in a network • fg is the number of occurrences of a subgraph g, g is repeated if fg>F. • fg_randi is the frequency of g in a randomized network Grandi, for 1 ≤ i ≤ N, N is the number of the randomized networks. sg is the number of times fg ≥ fg_randi, g is unique if its sg > S. • Network motif discovery algorithm
Problem Statement • PPI network G=(V, E) _ each vertex represents a unique protein _ each edge between v. A and v. B indicates there is an interaction between A and B • Network motif _frequently occurring subgraph pattern in a network • fg is the number of occurrences of a subgraph g, g is repeated if fg>F. • fg_randi is the frequency of g in a randomized network Grandi, for 1 ≤ i ≤ N, N is the number of the randomized networks. sg is the number of times fg ≥ fg_randi, g is unique if its sg > S. • Network motif discovery algorithm
 Problem Statement (cont) • • Motivation of Ne. Mo. Finder- existing research has following limitations: _Number of network motifs candidates increases exponentially _Interesting network motifs are repeated and unique and Apirori algorithms are not applicable _The graph isomorphism problem is an NP problem Ne. Mo. Finder _ a network motif discovery algorithm to discover repeated and unique meso-scale network motifs in a large PPI network
Problem Statement (cont) • • Motivation of Ne. Mo. Finder- existing research has following limitations: _Number of network motifs candidates increases exponentially _Interesting network motifs are repeated and unique and Apirori algorithms are not applicable _The graph isomorphism problem is an NP problem Ne. Mo. Finder _ a network motif discovery algorithm to discover repeated and unique meso-scale network motifs in a large PPI network
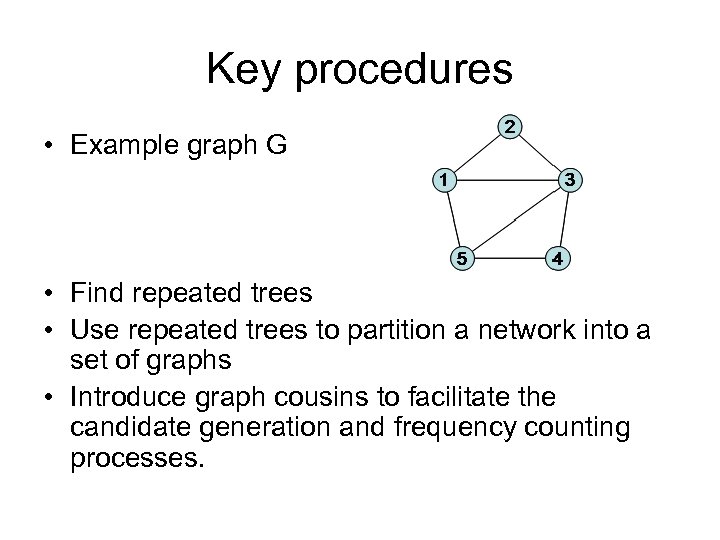 Key procedures • Example graph G • Find repeated trees • Use repeated trees to partition a network into a set of graphs • Introduce graph cousins to facilitate the candidate generation and frequency counting processes.
Key procedures • Example graph G • Find repeated trees • Use repeated trees to partition a network into a set of graphs • Introduce graph cousins to facilitate the candidate generation and frequency counting processes.
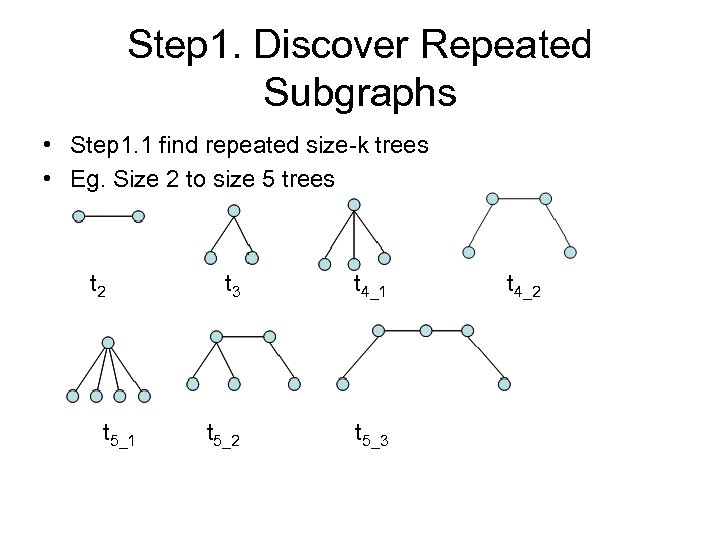 Step 1. Discover Repeated Subgraphs • Step 1. 1 find repeated size-k trees • Eg. Size 2 to size 5 trees t 2 t 5_1 t 3 t 4_1 t 5_2 t 5_3 t 4_2
Step 1. Discover Repeated Subgraphs • Step 1. 1 find repeated size-k trees • Eg. Size 2 to size 5 trees t 2 t 5_1 t 3 t 4_1 t 5_2 t 5_3 t 4_2
 Step 1. discover repeated subgraphs (cont) • ft 2 = 7, ft 3 = 13, ft 4_1 = 6, ft 4_2 =17, ft 5_1=1, ft 5_2 = 5, ft 5_3 = 7. • T 2 = {t 2}, T 3 = {t 3}, T 4 ={t 4_1, t 4_2} and T 5 = {t 5_2, t 5_3}.
Step 1. discover repeated subgraphs (cont) • ft 2 = 7, ft 3 = 13, ft 4_1 = 6, ft 4_2 =17, ft 5_1=1, ft 5_2 = 5, ft 5_3 = 7. • T 2 = {t 2}, T 3 = {t 3}, T 4 ={t 4_1, t 4_2} and T 5 = {t 5_2, t 5_3}.
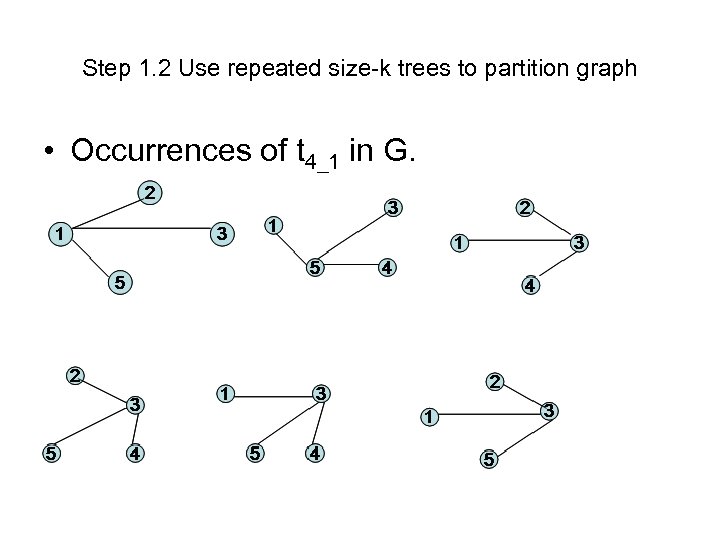 Step 1. 2 Use repeated size-k trees to partition graph • Occurrences of t 4_1 in G.
Step 1. 2 Use repeated size-k trees to partition graph • Occurrences of t 4_1 in G.
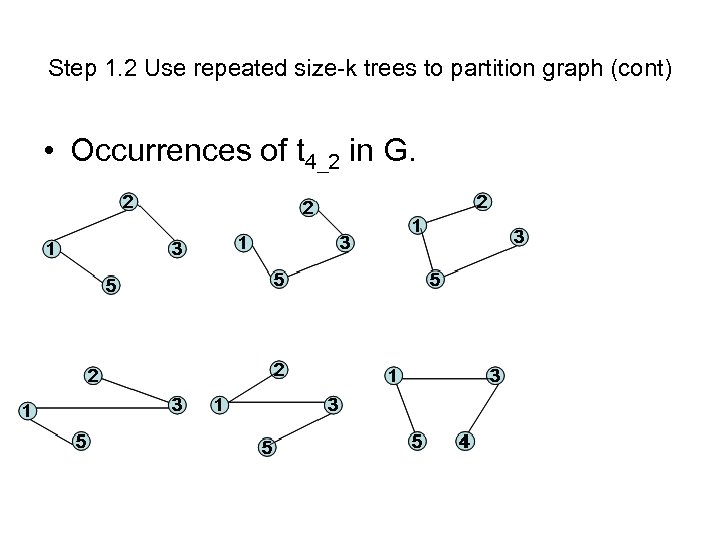 Step 1. 2 Use repeated size-k trees to partition graph (cont) • Occurrences of t 4_2 in G.
Step 1. 2 Use repeated size-k trees to partition graph (cont) • Occurrences of t 4_2 in G.
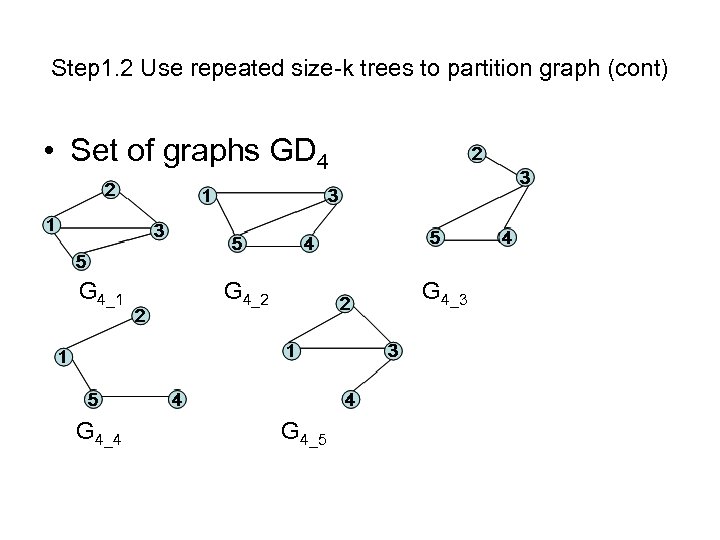 Step 1. 2 Use repeated size-k trees to partition graph (cont) • Set of graphs GD 4 G 4_1 G 4_4 G 4_2 G 4_3 G 4_5
Step 1. 2 Use repeated size-k trees to partition graph (cont) • Set of graphs GD 4 G 4_1 G 4_4 G 4_2 G 4_3 G 4_5
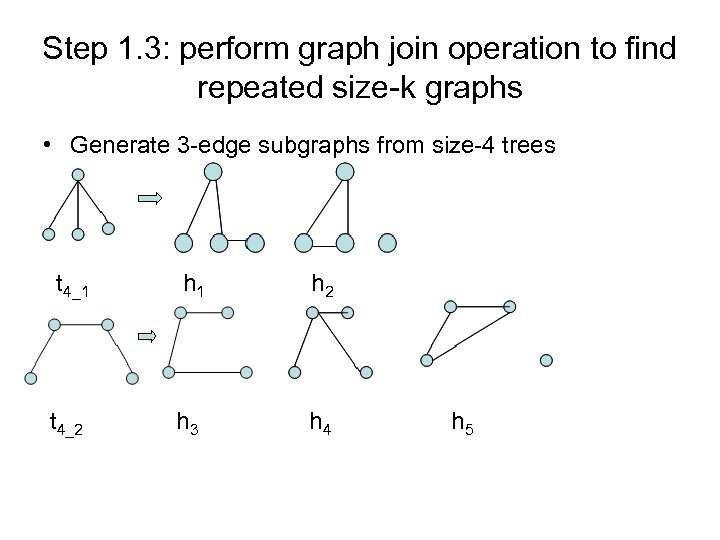 Step 1. 3: perform graph join operation to find repeated size-k graphs • Generate 3 -edge subgraphs from size-4 trees t 4_1 h 2 t 4_2 h 3 h 4 h 5
Step 1. 3: perform graph join operation to find repeated size-k graphs • Generate 3 -edge subgraphs from size-4 trees t 4_1 h 2 t 4_2 h 3 h 4 h 5
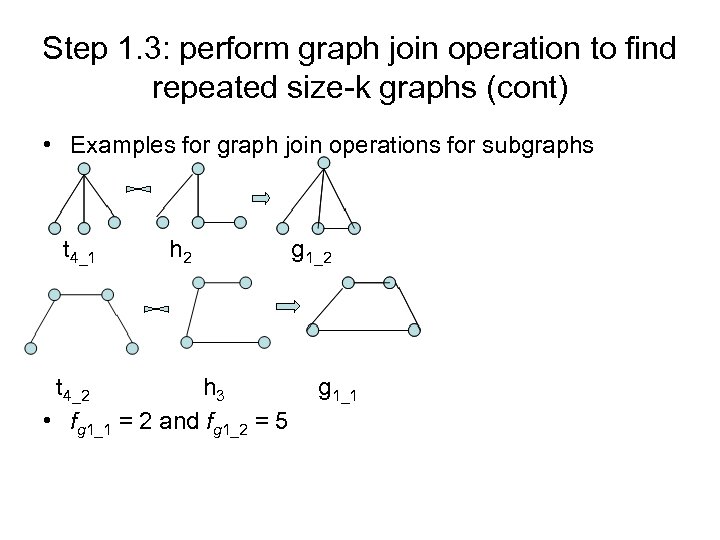 Step 1. 3: perform graph join operation to find repeated size-k graphs (cont) • Examples for graph join operations for subgraphs t 4_1 h 2 t 4_2 h 3 • fg 1_1 = 2 and fg 1_2 = 5 g 1_2 g 1_1
Step 1. 3: perform graph join operation to find repeated size-k graphs (cont) • Examples for graph join operations for subgraphs t 4_1 h 2 t 4_2 h 3 • fg 1_1 = 2 and fg 1_2 = 5 g 1_2 g 1_1
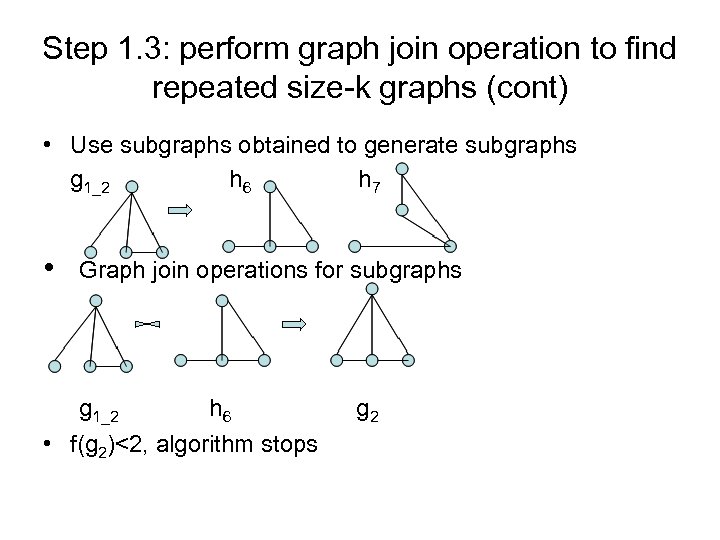 Step 1. 3: perform graph join operation to find repeated size-k graphs (cont) • Use subgraphs obtained to generate subgraphs g 1_2 h 6 h 7 • Graph join operations for subgraphs g 1_2 h 6 • f(g 2)<2, algorithm stops g 2
Step 1. 3: perform graph join operation to find repeated size-k graphs (cont) • Use subgraphs obtained to generate subgraphs g 1_2 h 6 h 7 • Graph join operations for subgraphs g 1_2 h 6 • f(g 2)<2, algorithm stops g 2
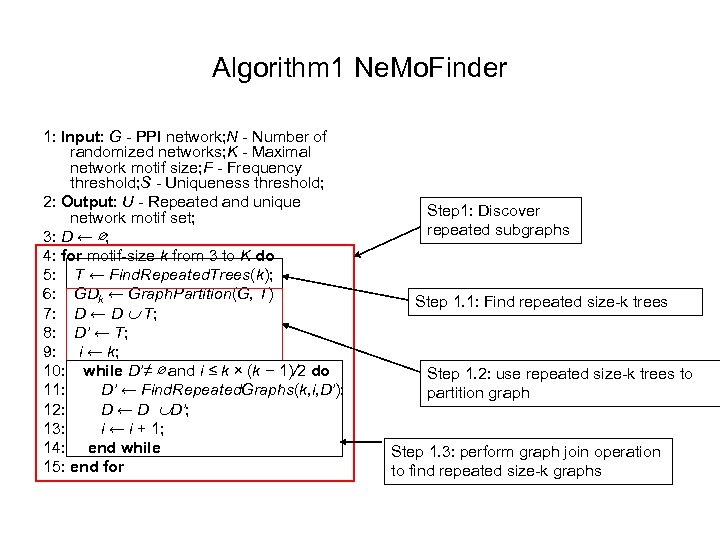 Algorithm 1 Ne. Mo. Finder 1: Input: G - PPI network; N - Number of randomized networks; K - Maximal network motif size; F - Frequency threshold; S - Uniqueness threshold; 2: Output: U - Repeated and unique network motif set; 3: D ← ∅; 4: for motif-size k from 3 to K do 5: T ← Find. Repeated. Trees(k); 6: GDk ← Graph. Partition(G, T) 7: D ← D T; 8: D’ ← T; 9: i ← k; 10: while D’≠ ∅ and i ≤ k × (k − 1)/2 do 11: D’ ← Find. Repeated. Graphs(k, i, D’); 12: D ← D D’; 13: i ← i + 1; 14: end while 15: end for Step 1: Discover repeated subgraphs Step 1. 1: Find repeated size-k trees Step 1. 2: use repeated size-k trees to partition graph Step 1. 3: perform graph join operation to find repeated size-k graphs
Algorithm 1 Ne. Mo. Finder 1: Input: G - PPI network; N - Number of randomized networks; K - Maximal network motif size; F - Frequency threshold; S - Uniqueness threshold; 2: Output: U - Repeated and unique network motif set; 3: D ← ∅; 4: for motif-size k from 3 to K do 5: T ← Find. Repeated. Trees(k); 6: GDk ← Graph. Partition(G, T) 7: D ← D T; 8: D’ ← T; 9: i ← k; 10: while D’≠ ∅ and i ≤ k × (k − 1)/2 do 11: D’ ← Find. Repeated. Graphs(k, i, D’); 12: D ← D D’; 13: i ← i + 1; 14: end while 15: end for Step 1: Discover repeated subgraphs Step 1. 1: Find repeated size-k trees Step 1. 2: use repeated size-k trees to partition graph Step 1. 3: perform graph join operation to find repeated size-k graphs
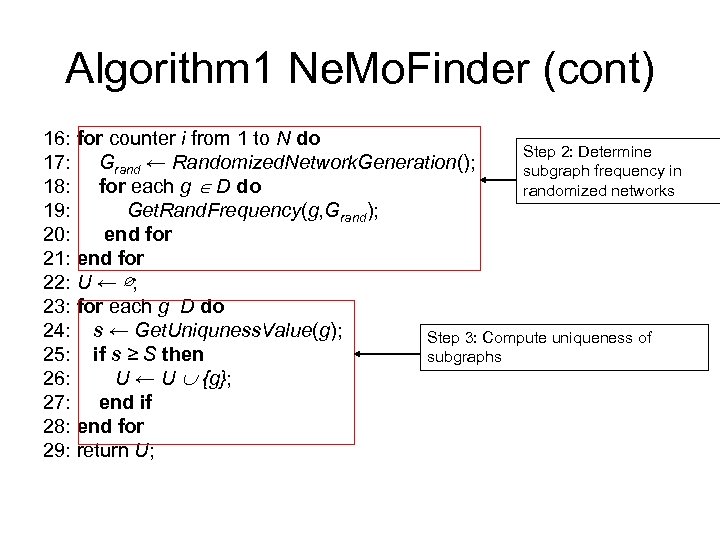 Algorithm 1 Ne. Mo. Finder (cont) 16: for counter i from 1 to N do Step 2: Determine 17: Grand ← Randomized. Network. Generation(); subgraph frequency in 18: for each g D do randomized networks 19: Get. Rand. Frequency(g, Grand); 20: end for 21: end for 22: U ← ∅; 23: for each g D do 24: s ← Get. Uniquness. Value(g); Step 3: Compute uniqueness of 25: if s ≥ S then subgraphs 26: U ← U {g}; 27: end if 28: end for 29: return U;
Algorithm 1 Ne. Mo. Finder (cont) 16: for counter i from 1 to N do Step 2: Determine 17: Grand ← Randomized. Network. Generation(); subgraph frequency in 18: for each g D do randomized networks 19: Get. Rand. Frequency(g, Grand); 20: end for 21: end for 22: U ← ∅; 23: for each g D do 24: s ← Get. Uniquness. Value(g); Step 3: Compute uniqueness of 25: if s ≥ S then subgraphs 26: U ← U {g}; 27: end if 28: end for 29: return U;
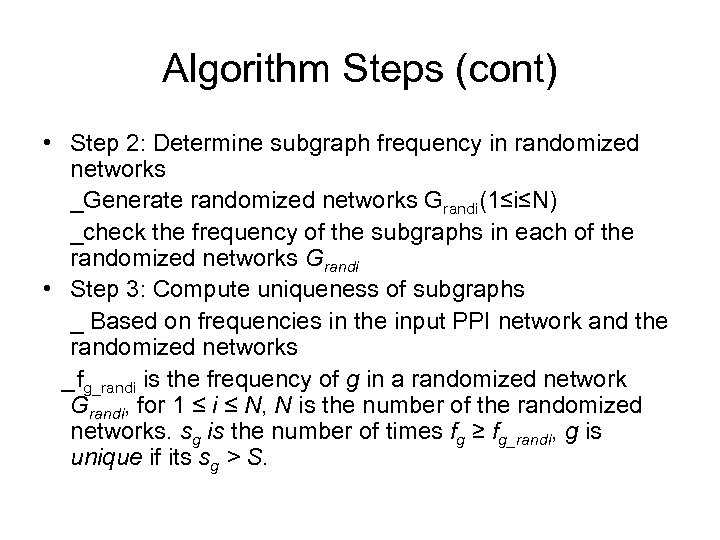 Algorithm Steps (cont) • Step 2: Determine subgraph frequency in randomized networks _Generate randomized networks Grandi(1≤i≤N) _check the frequency of the subgraphs in each of the randomized networks Grandi • Step 3: Compute uniqueness of subgraphs _ Based on frequencies in the input PPI network and the randomized networks _fg_randi is the frequency of g in a randomized network Grandi, for 1 ≤ i ≤ N, N is the number of the randomized networks. sg is the number of times fg ≥ fg_randi, g is unique if its sg > S.
Algorithm Steps (cont) • Step 2: Determine subgraph frequency in randomized networks _Generate randomized networks Grandi(1≤i≤N) _check the frequency of the subgraphs in each of the randomized networks Grandi • Step 3: Compute uniqueness of subgraphs _ Based on frequencies in the input PPI network and the randomized networks _fg_randi is the frequency of g in a randomized network Grandi, for 1 ≤ i ≤ N, N is the number of the randomized networks. sg is the number of times fg ≥ fg_randi, g is unique if its sg > S.
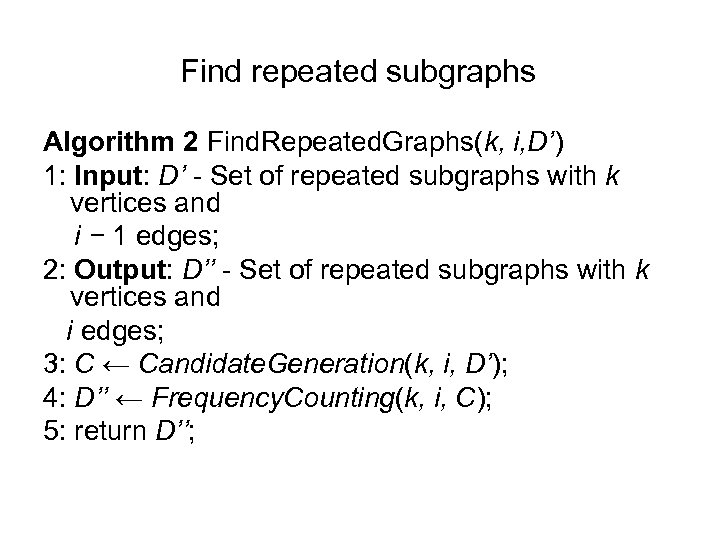 Find repeated subgraphs Algorithm 2 Find. Repeated. Graphs(k, i, D’) 1: Input: D’ - Set of repeated subgraphs with k vertices and i − 1 edges; 2: Output: D’’ - Set of repeated subgraphs with k vertices and i edges; 3: C ← Candidate. Generation(k, i, D’); 4: D’’ ← Frequency. Counting(k, i, C); 5: return D’’;
Find repeated subgraphs Algorithm 2 Find. Repeated. Graphs(k, i, D’) 1: Input: D’ - Set of repeated subgraphs with k vertices and i − 1 edges; 2: Output: D’’ - Set of repeated subgraphs with k vertices and i edges; 3: C ← Candidate. Generation(k, i, D’); 4: D’’ ← Frequency. Counting(k, i, C); 5: return D’’;
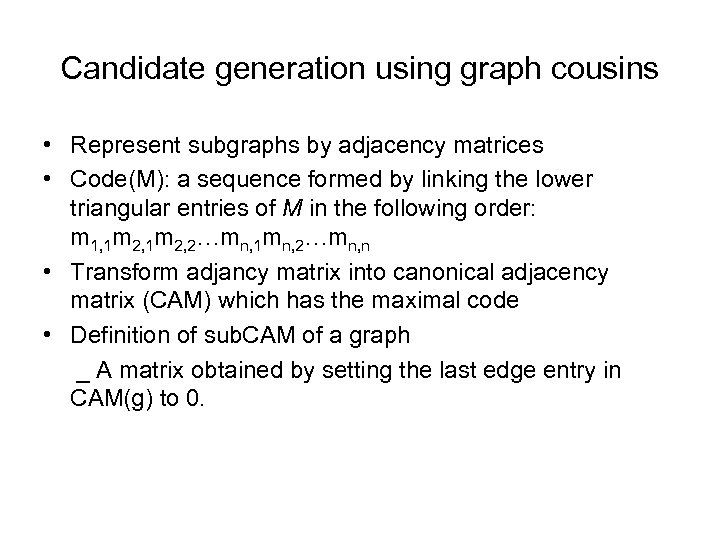 Candidate generation using graph cousins • Represent subgraphs by adjacency matrices • Code(M): a sequence formed by linking the lower triangular entries of M in the following order: m 1, 1 m 2, 2…mn, 1 mn, 2…mn, n • Transform adjancy matrix into canonical adjacency matrix (CAM) which has the maximal code • Definition of sub. CAM of a graph _ A matrix obtained by setting the last edge entry in CAM(g) to 0.
Candidate generation using graph cousins • Represent subgraphs by adjacency matrices • Code(M): a sequence formed by linking the lower triangular entries of M in the following order: m 1, 1 m 2, 2…mn, 1 mn, 2…mn, n • Transform adjancy matrix into canonical adjacency matrix (CAM) which has the maximal code • Definition of sub. CAM of a graph _ A matrix obtained by setting the last edge entry in CAM(g) to 0.
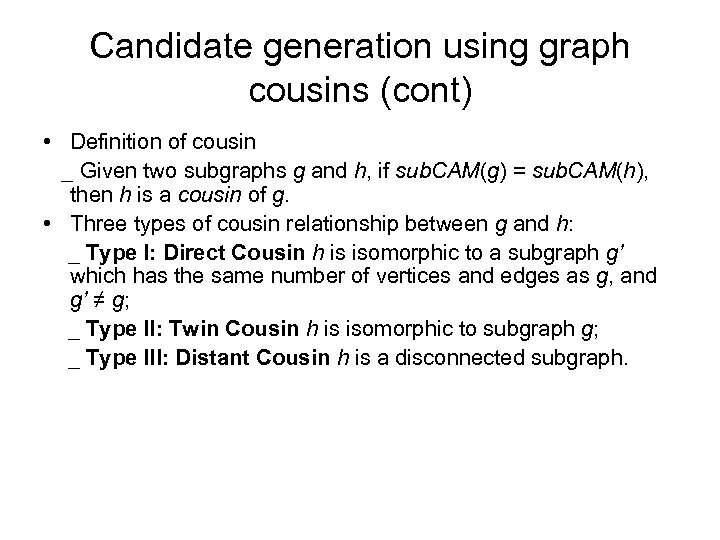 Candidate generation using graph cousins (cont) • Definition of cousin _ Given two subgraphs g and h, if sub. CAM(g) = sub. CAM(h), then h is a cousin of g. • Three types of cousin relationship between g and h: _ Type I: Direct Cousin h is isomorphic to a subgraph g’ which has the same number of vertices and edges as g, and g’ ≠ g; _ Type II: Twin Cousin h is isomorphic to subgraph g; _ Type III: Distant Cousin h is a disconnected subgraph.
Candidate generation using graph cousins (cont) • Definition of cousin _ Given two subgraphs g and h, if sub. CAM(g) = sub. CAM(h), then h is a cousin of g. • Three types of cousin relationship between g and h: _ Type I: Direct Cousin h is isomorphic to a subgraph g’ which has the same number of vertices and edges as g, and g’ ≠ g; _ Type II: Twin Cousin h is isomorphic to subgraph g; _ Type III: Distant Cousin h is a disconnected subgraph.
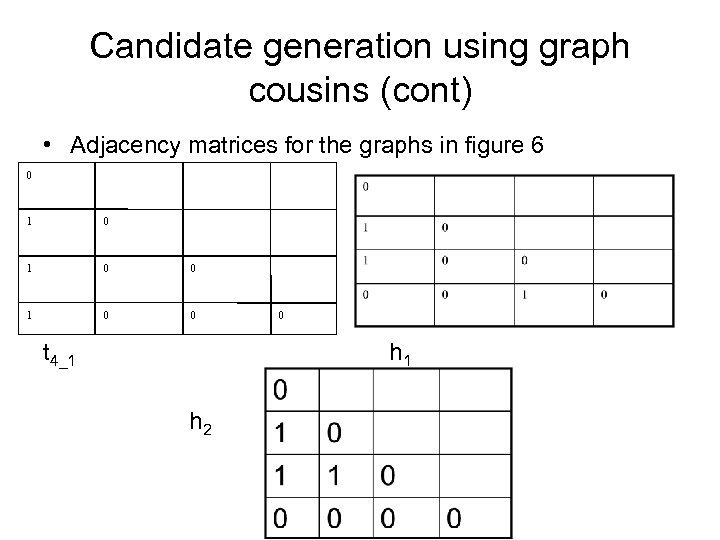 Candidate generation using graph cousins (cont) • Adjacency matrices for the graphs in figure 6 0 1 0 0 t 4_1 0 h 1 h 2
Candidate generation using graph cousins (cont) • Adjacency matrices for the graphs in figure 6 0 1 0 0 t 4_1 0 h 1 h 2
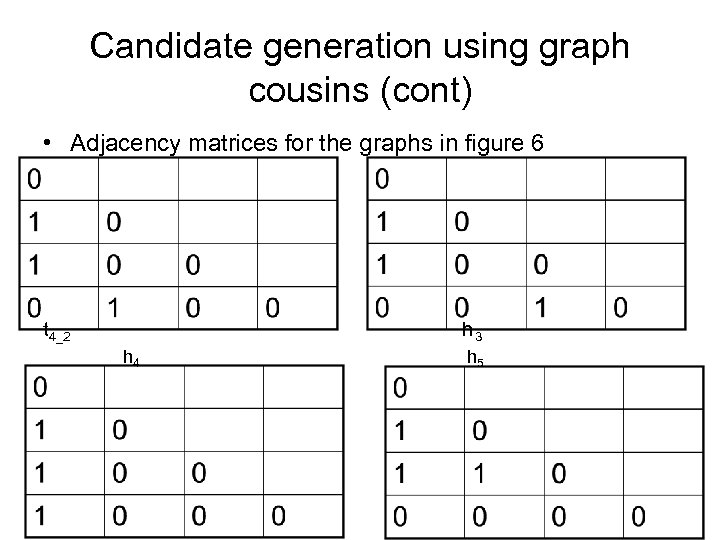 Candidate generation using graph cousins (cont) • Adjacency matrices for the graphs in figure 6 t 4_2 h 3 h 4 h 5
Candidate generation using graph cousins (cont) • Adjacency matrices for the graphs in figure 6 t 4_2 h 3 h 4 h 5
 Candidate generation using graph cousins (cont) • Observations of above example _h 1 is a type 1 direct cousin of t 4_1 _h 2 is a type 3 distant cousin of t 4_1 _h 3 is a type 2 twin cousin of t 4_2 _h 4 is a type 1 direct cousin of t 4_2 _h 5 is a type 3 distant cousin of t 4_2
Candidate generation using graph cousins (cont) • Observations of above example _h 1 is a type 1 direct cousin of t 4_1 _h 2 is a type 3 distant cousin of t 4_1 _h 3 is a type 2 twin cousin of t 4_2 _h 4 is a type 1 direct cousin of t 4_2 _h 5 is a type 3 distant cousin of t 4_2
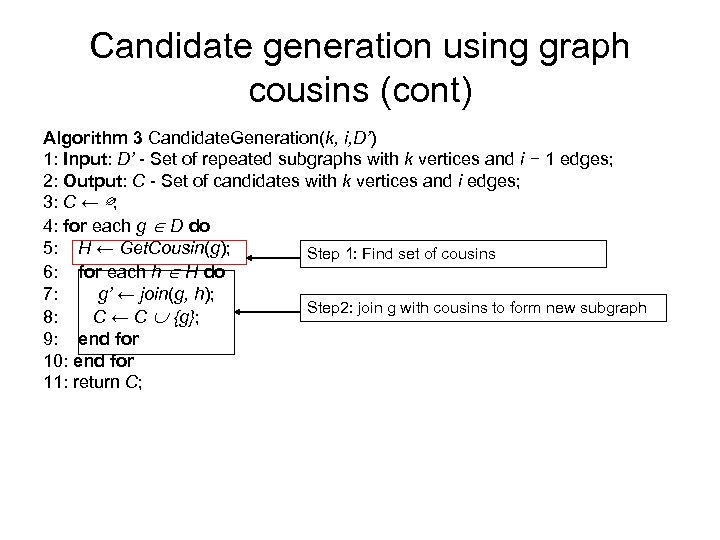 Candidate generation using graph cousins (cont) Algorithm 3 Candidate. Generation(k, i, D’) 1: Input: D’ - Set of repeated subgraphs with k vertices and i − 1 edges; 2: Output: C - Set of candidates with k vertices and i edges; 3: C ← ∅; 4: for each g D do 5: H ← Get. Cousin(g); Step 1: Find set of cousins 6: for each h H do 7: g’ ← join(g, h); Step 2: join g with cousins to form new subgraph 8: C ← C {g}; 9: end for 10: end for 11: return C;
Candidate generation using graph cousins (cont) Algorithm 3 Candidate. Generation(k, i, D’) 1: Input: D’ - Set of repeated subgraphs with k vertices and i − 1 edges; 2: Output: C - Set of candidates with k vertices and i edges; 3: C ← ∅; 4: for each g D do 5: H ← Get. Cousin(g); Step 1: Find set of cousins 6: for each h H do 7: g’ ← join(g, h); Step 2: join g with cousins to form new subgraph 8: C ← C {g}; 9: end for 10: end for 11: return C;
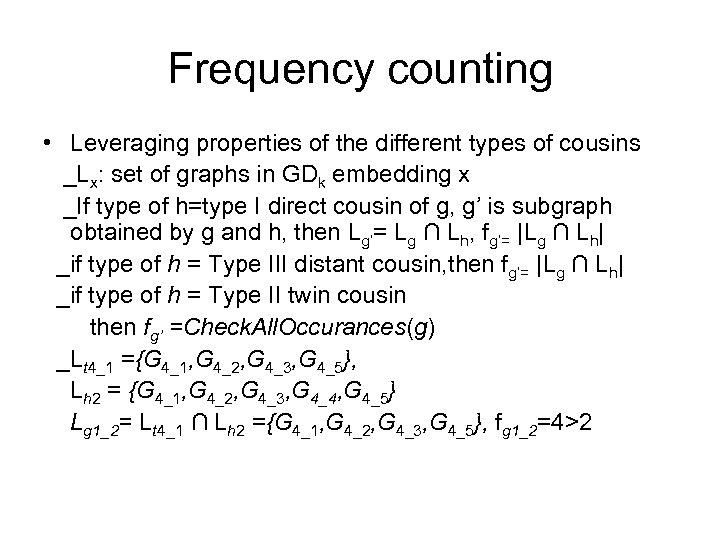 Frequency counting • Leveraging properties of the different types of cousins _Lx: set of graphs in GDk embedding x _If type of h=type I direct cousin of g, g’ is subgraph obtained by g and h, then Lg’= Lg ∩ Lh, fg’= |Lg ∩ Lh| _if type of h = Type III distant cousin, then fg’= |Lg ∩ Lh| _if type of h = Type II twin cousin then fg’ =Check. All. Occurances(g) _Lt 4_1 ={G 4_1, G 4_2, G 4_3, G 4_5}, Lh 2 = {G 4_1, G 4_2, G 4_3, G 4_4, G 4_5} Lg 1_2= Lt 4_1 ∩ Lh 2 ={G 4_1, G 4_2, G 4_3, G 4_5}, fg 1_2=4>2
Frequency counting • Leveraging properties of the different types of cousins _Lx: set of graphs in GDk embedding x _If type of h=type I direct cousin of g, g’ is subgraph obtained by g and h, then Lg’= Lg ∩ Lh, fg’= |Lg ∩ Lh| _if type of h = Type III distant cousin, then fg’= |Lg ∩ Lh| _if type of h = Type II twin cousin then fg’ =Check. All. Occurances(g) _Lt 4_1 ={G 4_1, G 4_2, G 4_3, G 4_5}, Lh 2 = {G 4_1, G 4_2, G 4_3, G 4_4, G 4_5} Lg 1_2= Lt 4_1 ∩ Lh 2 ={G 4_1, G 4_2, G 4_3, G 4_5}, fg 1_2=4>2
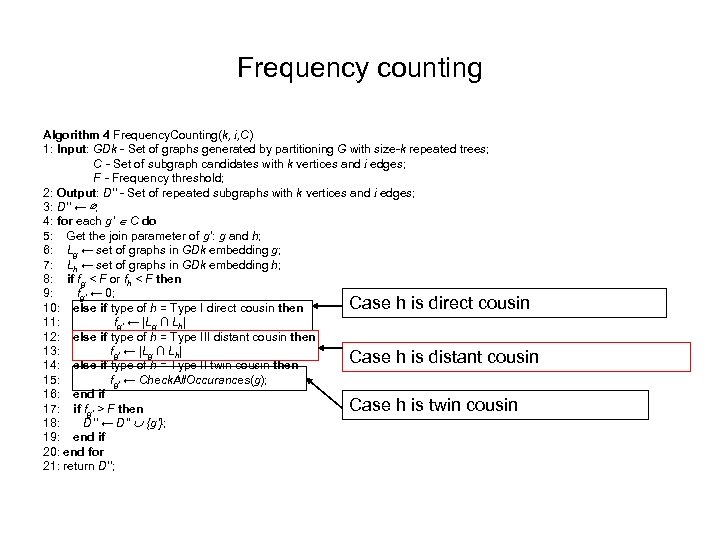 Frequency counting Algorithm 4 Frequency. Counting(k, i, C) 1: Input: GDk - Set of graphs generated by partitioning G with size-k repeated trees; C - Set of subgraph candidates with k vertices and i edges; F - Frequency threshold; 2: Output: D’’ - Set of repeated subgraphs with k vertices and i edges; 3: D’’ ← ∅; 4: for each g’ C do 5: Get the join parameter of g’: g and h; 6: Lg ← set of graphs in GDk embedding g; 7: Lh ← set of graphs in GDk embedding h; 8: if fg < F or fh < F then 9: fg’ ← 0; Case h is direct cousin 10: else if type of h = Type I direct cousin then 11: fg’ ← |Lg ∩ Lh| 12: else if type of h = Type III distant cousin then 13: fg’ ← |Lg ∩ Lh| Case h is distant cousin 14: else if type of h = Type II twin cousin then 15: fg’ ← Check. All. Occurances(g); 16: end if Case h is twin cousin 17: if fg’ > F then 18: D’’ ← D’’ {g’}; 19: end if 20: end for 21: return D’’;
Frequency counting Algorithm 4 Frequency. Counting(k, i, C) 1: Input: GDk - Set of graphs generated by partitioning G with size-k repeated trees; C - Set of subgraph candidates with k vertices and i edges; F - Frequency threshold; 2: Output: D’’ - Set of repeated subgraphs with k vertices and i edges; 3: D’’ ← ∅; 4: for each g’ C do 5: Get the join parameter of g’: g and h; 6: Lg ← set of graphs in GDk embedding g; 7: Lh ← set of graphs in GDk embedding h; 8: if fg < F or fh < F then 9: fg’ ← 0; Case h is direct cousin 10: else if type of h = Type I direct cousin then 11: fg’ ← |Lg ∩ Lh| 12: else if type of h = Type III distant cousin then 13: fg’ ← |Lg ∩ Lh| Case h is distant cousin 14: else if type of h = Type II twin cousin then 15: fg’ ← Check. All. Occurances(g); 16: end if Case h is twin cousin 17: if fg’ > F then 18: D’’ ← D’’ {g’}; 19: end if 20: end for 21: return D’’;
 Summary • Nemo. Finder-an efficient network motif discovery algorithm to discover largersized repeated and unique network motifs in PPI networks. • Use repeated trees to partition network into graphs • Graph cousins for candidate generation and frequency counting
Summary • Nemo. Finder-an efficient network motif discovery algorithm to discover largersized repeated and unique network motifs in PPI networks. • Use repeated trees to partition network into graphs • Graph cousins for candidate generation and frequency counting
 References (1) • T. Asai, et al. “Efficient substructure discovery from large semi-structured data”, SDM'02 • C. Borgelt and M. R. Berthold, “Mining molecular fragments: Finding relevant substructures of molecules”, ICDM'02 • D. Cai, Z. Shao, X. He, X. Yan, and J. Han, “Community Mining from Multi-Relational Networks”, PKDD'05. • J. Chen, W. Hsu, M. Lee, Ne. Mo. Finder: Dissecting genome wide protein-protein interactions with repeated and unique network motifs, Seekiong Ng, SIGKDD 2006 • M. Deshpande, M. Kuramochi, and G. Karypis, “Frequent Sub-structure Based Approaches for Classifying Chemical Compounds”, ICDM 2003 • M. Deshpande, M. Kuramochi, and G. Karypis. “Automated approaches for classifying structures”, BIOKDD'02 • C. Faloutsos, K. Mc. Curley, and A. Tomkins, “Fast Discovery of 'Connection Subgraphs”, KDD'04 • H. Fröhlich, J. Wegner, F. Sieker, and A. Zell, “Optimal Assignment Kernels For Attributed Molecular Graphs”, ICML’ 05
References (1) • T. Asai, et al. “Efficient substructure discovery from large semi-structured data”, SDM'02 • C. Borgelt and M. R. Berthold, “Mining molecular fragments: Finding relevant substructures of molecules”, ICDM'02 • D. Cai, Z. Shao, X. He, X. Yan, and J. Han, “Community Mining from Multi-Relational Networks”, PKDD'05. • J. Chen, W. Hsu, M. Lee, Ne. Mo. Finder: Dissecting genome wide protein-protein interactions with repeated and unique network motifs, Seekiong Ng, SIGKDD 2006 • M. Deshpande, M. Kuramochi, and G. Karypis, “Frequent Sub-structure Based Approaches for Classifying Chemical Compounds”, ICDM 2003 • M. Deshpande, M. Kuramochi, and G. Karypis. “Automated approaches for classifying structures”, BIOKDD'02 • C. Faloutsos, K. Mc. Curley, and A. Tomkins, “Fast Discovery of 'Connection Subgraphs”, KDD'04 • H. Fröhlich, J. Wegner, F. Sieker, and A. Zell, “Optimal Assignment Kernels For Attributed Molecular Graphs”, ICML’ 05
 References (2) • L. Holder, D. Cook, and S. Djoko. “Substructure discovery in the subdue system”, KDD'94 • J. Huan, W. Wang, D. Bandyopadhyay, J. Snoeyink, J. Prins, and A. Tropsha. “Mining spatial motifs from protein structure graphs”, RECOMB’ 04 • J. Huan, W. Wang, and J. Prins. “Efficient mining of frequent subgraph in the presence of isomorphism”, ICDM'03 • H. Hu, X. Yan, Yu, J. Han and X. J. Zhou, “Mining Coherent Dense Subgraphs across Massive Biological Networks for Functional Discovery”, ISMB'05 • A. Inokuchi, T. Washio, and H. Motoda. “An apriori-based algorithm for mining frequent substructures from graph data”, PKDD'00 • C. James, D. Weininger, and J. Delany. “Daylight Theory Manual Daylight Version 4. 82”. Daylight Chemical Information Systems, Inc. , 2003. • G. Jeh, and J. Widom, “Mining the Space of Graph Properties”, KDD'04 • H. Kashima, K. Tsuda, and A. Inokuchi, “Marginalized Kernels Between Labeled Graphs”, ICML’ 03
References (2) • L. Holder, D. Cook, and S. Djoko. “Substructure discovery in the subdue system”, KDD'94 • J. Huan, W. Wang, D. Bandyopadhyay, J. Snoeyink, J. Prins, and A. Tropsha. “Mining spatial motifs from protein structure graphs”, RECOMB’ 04 • J. Huan, W. Wang, and J. Prins. “Efficient mining of frequent subgraph in the presence of isomorphism”, ICDM'03 • H. Hu, X. Yan, Yu, J. Han and X. J. Zhou, “Mining Coherent Dense Subgraphs across Massive Biological Networks for Functional Discovery”, ISMB'05 • A. Inokuchi, T. Washio, and H. Motoda. “An apriori-based algorithm for mining frequent substructures from graph data”, PKDD'00 • C. James, D. Weininger, and J. Delany. “Daylight Theory Manual Daylight Version 4. 82”. Daylight Chemical Information Systems, Inc. , 2003. • G. Jeh, and J. Widom, “Mining the Space of Graph Properties”, KDD'04 • H. Kashima, K. Tsuda, and A. Inokuchi, “Marginalized Kernels Between Labeled Graphs”, ICML’ 03
 References (3) • M. Koyuturk, A. Grama, and W. Szpankowski. “An efficient algorithm for detecting frequent subgraphs in biological networks”, Bioinformatics, 20: I 200 --I 207, 2004. • T. Kudo, E. Maeda, and Y. Matsumoto, “An Application of Boosting to Graph Classification”, NIPS’ 04 • M. Kuramochi and G. Karypis. “Frequent subgraph discovery”, ICDM'01 • M. Kuramochi and G. Karypis, “GREW: A Scalable Frequent Subgraph Discovery Algorithm”, ICDM’ 04 • C. Liu, X. Yan, H. Yu, J. Han, and P. S. Yu, “Mining Behavior Graphs for ‘Backtrace'' of Noncrashing Bugs’'', SDM'05 • P. Mahé, N. Ueda, T. Akutsu, J. Perret, and J. Vert, “Extensions of Marginalized Graph Kernels”, ICML’ 04 • S. Nijssen and J. Kok. A quickstart in frequent structure mining can make a difference. KDD'04 • J. Prins, J. Yang, J. Huan, and W. Wang. “Spin: Mining maximal frequent subgraphs from graph databases”. KDD'04
References (3) • M. Koyuturk, A. Grama, and W. Szpankowski. “An efficient algorithm for detecting frequent subgraphs in biological networks”, Bioinformatics, 20: I 200 --I 207, 2004. • T. Kudo, E. Maeda, and Y. Matsumoto, “An Application of Boosting to Graph Classification”, NIPS’ 04 • M. Kuramochi and G. Karypis. “Frequent subgraph discovery”, ICDM'01 • M. Kuramochi and G. Karypis, “GREW: A Scalable Frequent Subgraph Discovery Algorithm”, ICDM’ 04 • C. Liu, X. Yan, H. Yu, J. Han, and P. S. Yu, “Mining Behavior Graphs for ‘Backtrace'' of Noncrashing Bugs’'', SDM'05 • P. Mahé, N. Ueda, T. Akutsu, J. Perret, and J. Vert, “Extensions of Marginalized Graph Kernels”, ICML’ 04 • S. Nijssen and J. Kok. A quickstart in frequent structure mining can make a difference. KDD'04 • J. Prins, J. Yang, J. Huan, and W. Wang. “Spin: Mining maximal frequent subgraphs from graph databases”. KDD'04
 References (4) • • • D. Shasha, J. T. -L. Wang, and R. Giugno. “Algorithmics and applications of tree and graph searching”, PODS'02 J. R. Ullmann. “An algorithm for subgraph isomorphism”, J. ACM, 23: 31 --42, 1976. N. Vanetik, E. Gudes, and S. E. Shimony. “Computing frequent graph patterns from semistructured data”, ICDM'02 C. Wang, W. Wang, J. Pei, Y. Zhu, and B. Shi. “Scalable mining of large disk-base graph databases”, KDD'04 T. Washio and H. Motoda, “State of the art of graph-based data mining”, SIGKDD Explorations, 5: 59 -68, 2003 X. Yan and J. Han, “g. Span: Graph-Based Substructure Pattern Mining”, ICDM'02 X. Yan and J. Han, “Close. Graph: Mining Closed Frequent Graph Patterns”, KDD'03 X. Yan, P. S. Yu, and J. Han, “Graph Indexing: A Frequent Structure-based Approach”, SIGMOD'04 X. Yan, X. J. Zhou, and J. Han, “Mining Closed Relational Graphs with Connectivity Constraints”, KDD'05 X. Yan, P. S. Yu, and J. Han, “Substructure Similarity Search in Graph Databases”, SIGMOD'05 X. Yan, F. Zhu, J. Han, and P. S. Yu, “Searching Substructures with Superimposed Distance”, ICDE'06 M. J. Zaki. “Efficiently mining frequent trees in a forest”, KDD'02
References (4) • • • D. Shasha, J. T. -L. Wang, and R. Giugno. “Algorithmics and applications of tree and graph searching”, PODS'02 J. R. Ullmann. “An algorithm for subgraph isomorphism”, J. ACM, 23: 31 --42, 1976. N. Vanetik, E. Gudes, and S. E. Shimony. “Computing frequent graph patterns from semistructured data”, ICDM'02 C. Wang, W. Wang, J. Pei, Y. Zhu, and B. Shi. “Scalable mining of large disk-base graph databases”, KDD'04 T. Washio and H. Motoda, “State of the art of graph-based data mining”, SIGKDD Explorations, 5: 59 -68, 2003 X. Yan and J. Han, “g. Span: Graph-Based Substructure Pattern Mining”, ICDM'02 X. Yan and J. Han, “Close. Graph: Mining Closed Frequent Graph Patterns”, KDD'03 X. Yan, P. S. Yu, and J. Han, “Graph Indexing: A Frequent Structure-based Approach”, SIGMOD'04 X. Yan, X. J. Zhou, and J. Han, “Mining Closed Relational Graphs with Connectivity Constraints”, KDD'05 X. Yan, P. S. Yu, and J. Han, “Substructure Similarity Search in Graph Databases”, SIGMOD'05 X. Yan, F. Zhu, J. Han, and P. S. Yu, “Searching Substructures with Superimposed Distance”, ICDE'06 M. J. Zaki. “Efficiently mining frequent trees in a forest”, KDD'02


