8308095d65c54aee311c945fab2d86ee.ppt
- Количество слайдов: 74
NCBI Molecular Biology Resources A Field Guide NCBI Nov. 6, 2001

NCBI Resources n About NCBI n NCBI Sequence Databases • Primary Database – Gen. Bank • Derivative Databases - Ref. Seq Entrez Databases and Text Searching n BLAST Services n Genomic Resources NCBI n
The National Center for Biotechnology Information (NCBI) n Created as a part of the National Library of Medicine in 1988 • • Establish public databases Research in computational biology Develop software tools for sequence analysis Disseminate biomedical information Tools: BLAST(1990), Entrez (1992) n Gen. Bank (1992) n Free MEDLINE (Pub. Med, 1997) n Other databases: db. EST, db. GSS, db. STS, MMDB, OMIM, Uni. Gene, Gene. Map, Taxonomy, CGAP, SAGE, Locus. Link, Ref. Seq NCBI n
Molecular Databases n Primary Databases • • n Original submissions by experimentalists Database staff organize but don’t additional information • Human curated • Example: Gen. Bank Derivative Databases • • • Example: SWISS-PROT, NCBI Ref. Seq m. RNA Computationally Derived Example: Uni. Gene Combinations • Example: NCBI Genome Assembly NCBI • • compilation and correction of data
What is Gen. Bank? NCBI’s Primary Sequence Database n Nucleotide only sequence database n Archival in nature n Gen. Bank Data • • • n Direct submissions individual records (Bank. It, Sequin) Batch submissions via email (EST, GSS, STS) ftp accounts sequencing centers Data shared nightly among three collaborating databases Gen. Bank DNA Database of Japan (DDBJ). European Molecular Biology Laboratory Database (EMBL) at EBI. NCBI • • •
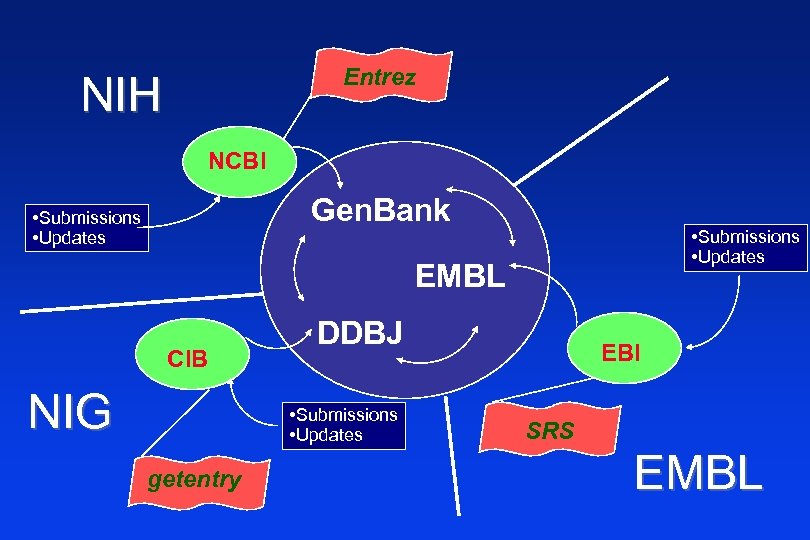
Entrez NIH NCBI Gen. Bank • Submissions • Updates EMBL CIB NIG DDBJ • Submissions • Updates getentry EBI SRS EMBL
NCBI
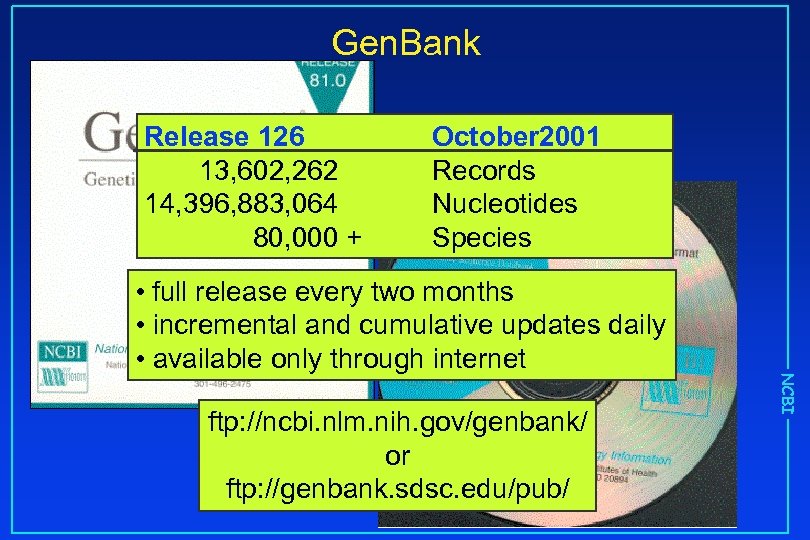
Gen. Bank Release 126 13, 602, 262 14, 396, 883, 064 80, 000 + October 2001 Records Nucleotides Species ftp: //ncbi. nlm. nih. gov/genbank/ or ftp: //genbank. sdsc. edu/pub/ NCBI • full release every two months • incremental and cumulative updates daily • available only through internet
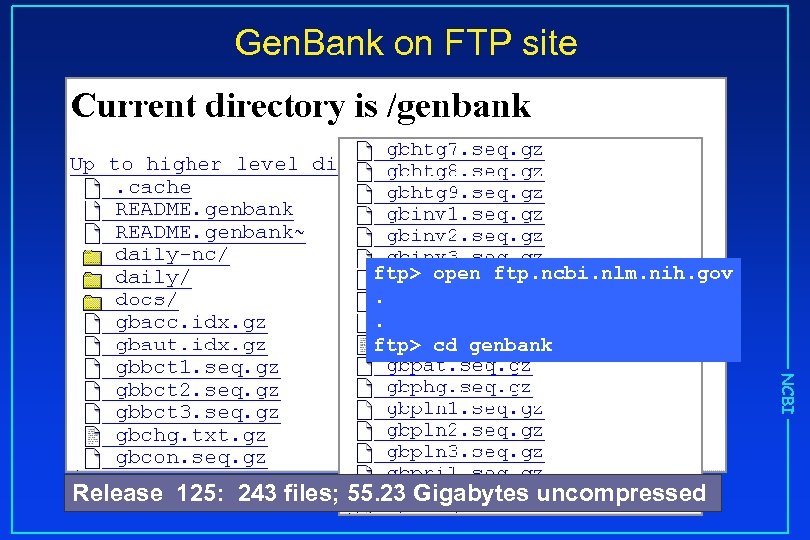
Gen. Bank on FTP site ftp> open ftp. ncbi. nlm. nih. gov. . ftp> cd genbank NCBI Release 125: 243 files; 55. 23 Gigabytes uncompressed

Gen. Bank Divisions Bulk Sequence Divisions PAT EST STS GSS HTG HTC CON Patent Expressed Sequence Tags (133 files) Sequence Tagged Site Genome Survey Sequence (41 files) High Throughput Genome (25 files) High Throughput c. DNA Contig Traditional Divisions BCT INV MAM PHG PLN PRI ROD SYN UNA VRL VRT
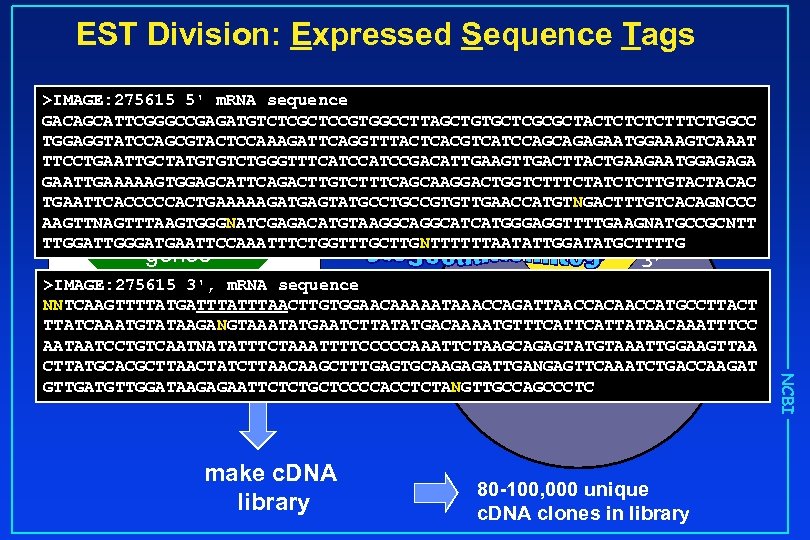
EST Division: Expressed Sequence Tags >IMAGE: 275615 5' m. RNA sequence GACAGCATTCGGGCCGAGATGTCTCGCTCCGTGGCCTTAGCTGTGCTCGCGCTACTCTTTCTGGCC TGGAGGTATCCAGCGTACTCCAAAGATTCAGGTTTACTCACGTCATCCAGCAGAGAATGGAAAGTCAAAT TTCCTGAATTGCTATGTGTCTGGGTTTCATCCGACATTGAAGTTGACTTACTGAAGAATGGAGAGA GAATTGAAAAAGTGGAGCATTCAGACTTGTCTTTCAGCAAGGACTGGTCTTTCTATCTCTTGTACTACAC TGAATTCACCCCCACTGAAAAAGATGAGTATGCCGTGTTGAACCATGTNGACTTTGTCACAGNCCC 5’ AAGTTNAGTTTAAGTGGGNATCGAGACATGTAAGGCATCATGGGAGGTTTTGAAGNATGCCGCNTT 30, 000 TTGGATTGGGATGAATTCCAAATTTCTGGTTTGCTTGNTTTTTTAATATTGGATATGCTTTTG nucleus genes 3’ make c. DNA library 80 -100, 000 unique c. DNA clones in library NCBI >IMAGE: 275615 3', m. RNA sequence - isolate unique clones NNTCAAGTTTTATGATTTAACTTGTGGAACAAAAATAAACCAGATTAACCACAACCATGCCTTACT -sequence once TTATCAAATGTATAAGANGTAAATATGAATCTTATATGACAAAATGTTTCATTATAACAAATTTCC 80 -100, 000 RNA AATAATCCTGTCAATNATATTTCTAAATTTTCCCCCAAATTCTAAGCAGAGTATGTAAATTGGAAGTTAA from each end gene products CTTATGCACGCTTAACTATCTTAACAAGCTTTGAGTGCAAGAGATTGANGAGTTCAAATCTGACCAAGAT GTTGATGTTGGATAAGAGAATTCTCTGCTCCCCACCTCTANGTTGCCAGCCCTC
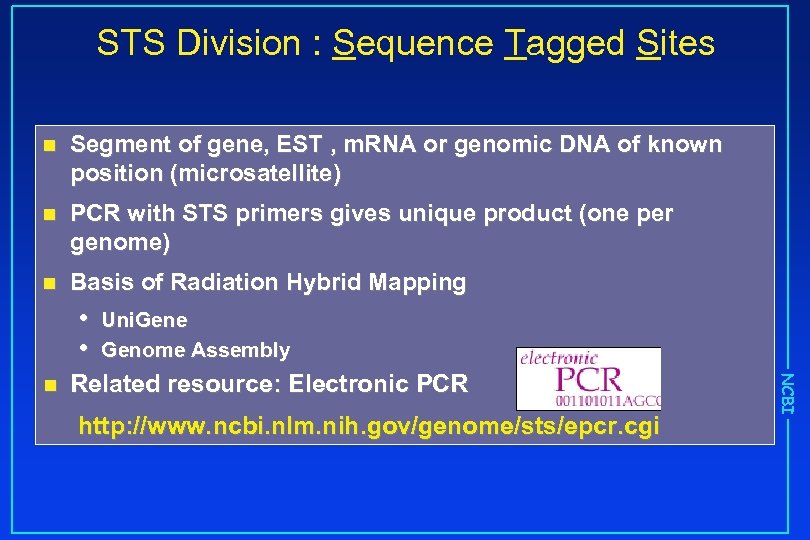
STS Division : Sequence Tagged Sites n Segment of gene, EST , m. RNA or genomic DNA of known position (microsatellite) n PCR with STS primers gives unique product (one per genome) n Basis of Radiation Hybrid Mapping • • Related resource: Electronic PCR http: //www. ncbi. nlm. nih. gov/genome/sts/epcr. cgi NCBI n Uni. Gene Genome Assembly
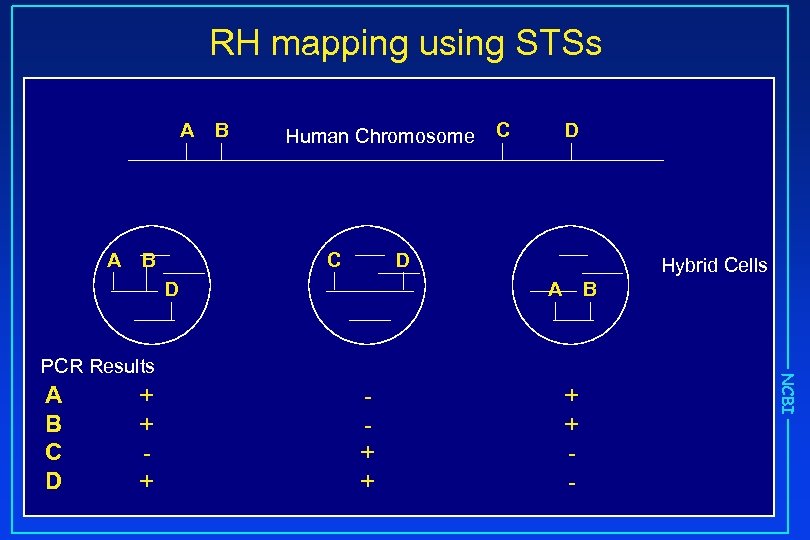
RH mapping using STSs A A B B Human Chromosome C C D D D Hybrid Cells A B C D + + + + - NCBI PCR Results
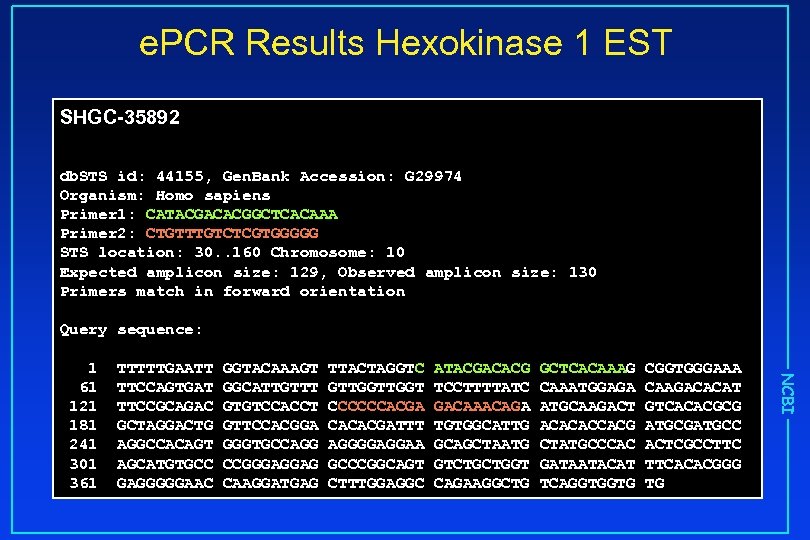
e. PCR Results Hexokinase 1 EST SHGC-35892 db. STS id: 44155, Gen. Bank Accession: G 29974 Organism: Homo sapiens Primer 1: CATACGACACGGCTCACAAA Primer 2: CTGTTTGTCTCGTGGGGG STS location: 30. . 160 Chromosome: 10 Expected amplicon size: 129, Observed amplicon size: 130 Primers match in forward orientation Query sequence: TTTTTGAATT TTCCAGTGAT TTCCGCAGAC GCTAGGACTG AGGCCACAGT AGCATGTGCC GAGGGGGAAC GGTACAAAGT GGCATTGTTT GTGTCCACCT GTTCCACGGA GGGTGCCAGG CCGGGAGGAG CAAGGATGAG TTACTAGGTC GTTGGT CCCCCCACGA CACACGATTT AGGGGAGGAA GCCCGGCAGT CTTTGGAGGC ATACGACACG TCCTTTTATC GACAAACAGA TGTGGCATTG GCAGCTAATG GTCTGCTGGT CAGAAGGCTG GCTCACAAAG CAAATGGAGA ATGCAAGACT ACACACCACG CTATGCCCAC GATAATACAT TCAGGTGGTG CGGTGGGAAA CAAGACACAT GTCACACGCG ATGCGATGCC ACTCGCCTTC TTCACACGGG TG NCBI 1 61 121 181 241 301 361
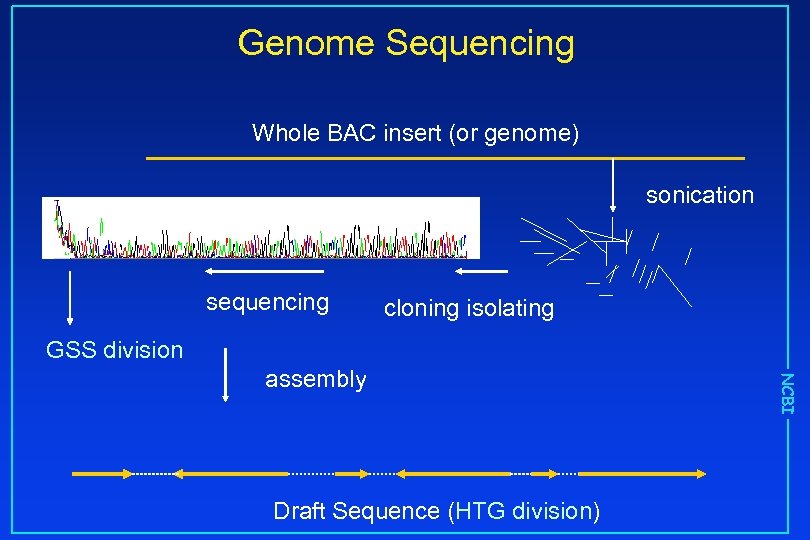
Genome Sequencing Whole BAC insert (or genome) sonication sequencing cloning isolating GSS division Draft Sequence (HTG division) NCBI assembly
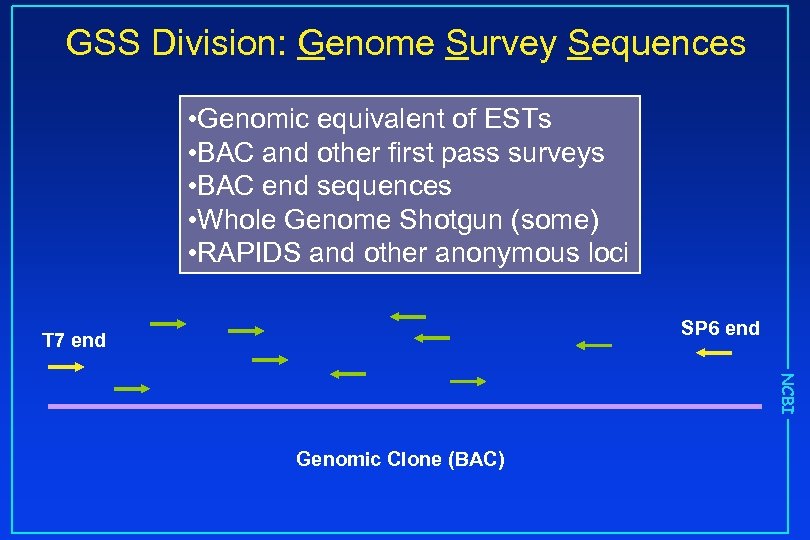
GSS Division: Genome Survey Sequences • Genomic equivalent of ESTs • BAC and other first pass surveys • BAC end sequences • Whole Genome Shotgun (some) • RAPIDS and other anonymous loci SP 6 end T 7 end NCBI Genomic Clone (BAC)
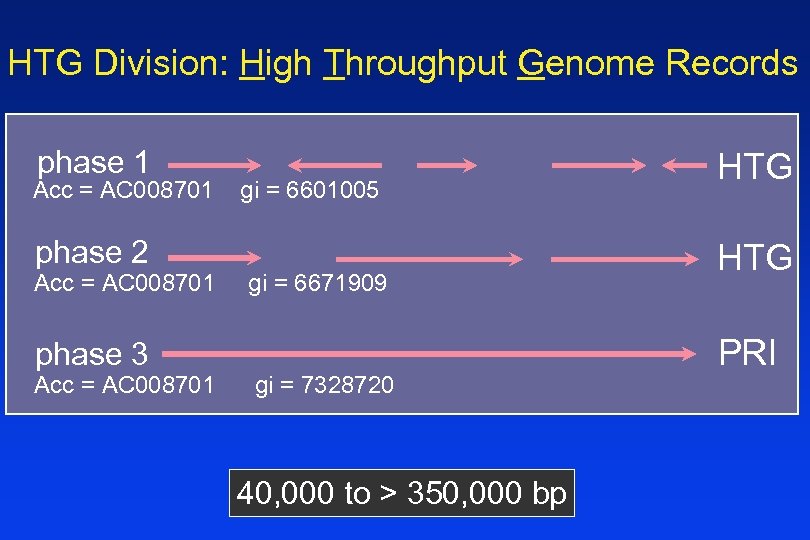
HTG Division: High Throughput Genome Records phase 1 Acc = AC 008701 phase 2 Acc = AC 008701 phase 3 Acc = AC 008701 gi = 6601005 gi = 6671909 gi = 7328720 40, 000 to > 350, 000 bp HTG PRI

The Gen. Bank Record NCBI

A Simple Gen. Bank Record LOCUS DEFINITION ACCESSION VERSION KEYWORDS SOURCE ORGANISM REFERENCE AUTHORS TITLE JOURNAL REMARK COMMENT AF 062069 3808 bp m. RNA INV 02 -MAR-2000 Limulus polyphemus myosin III m. RNA, complete cds. AF 062069. 2 GI: 7144484. Atlantic horseshoe crab. Limulus polyphemus Eukaryota; Metazoa; Arthropoda; Chelicerata; Merostomata; Xiphosura; Limulidae; Limulus. 1 (bases 1 to 3808) Battelle, B. -A. , Andrews, A. W. , Calman, B. G. , Sellers, J. R. , Greenberg, R. M. and Smith, W. C. A myosin III from Limulus eyes is a clock-regulated phosphoprotein J. Neurosci. (1998) In press 2 (bases 1 to 3808) Battelle, B. -A. , Andrews, A. W. , Calman, B. G. , Sellers, J. R. , Greenberg, R. M. and Smith, W. C. Direct Submission Submitted (29 -APR-1998) Whitney Laboratory, University of Florida, 9505 Ocean Shore Blvd. , St. Augustine, FL 32086, USA 3 (bases 1 to 3808) Battelle, B. -A. , Andrews, A. W. , Calman, B. G. , Sellers, J. R. , Greenberg, R. M. and Smith, W. C. Direct Submission Submitted (02 -MAR-2000) Whitney Laboratory, University of Florida, 9505 Ocean Shore Blvd. , St. Augustine, FL 32086, USA Sequence update by submitter On Mar 2, 2000 this sequence version replaced gi: 3132700.
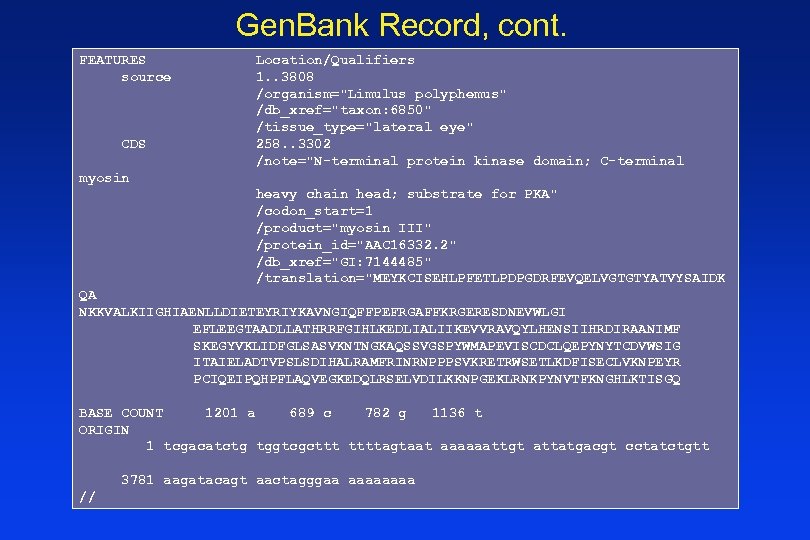
Gen. Bank Record, cont. FEATURES source CDS Location/Qualifiers 1. . 3808 /organism="Limulus polyphemus" /db_xref="taxon: 6850" /tissue_type="lateral eye" 258. . 3302 /note="N-terminal protein kinase domain; C-terminal myosin heavy chain head; substrate for PKA" /codon_start=1 /product="myosin III" /protein_id="AAC 16332. 2" /db_xref="GI: 7144485" /translation="MEYKCISEHLPFETLPDPGDRFEVQELVGTGTYATVYSAIDK QA NKKVALKIIGHIAENLLDIETEYRIYKAVNGIQFFPEFRGAFFKRGERESDNEVWLGI EFLEEGTAADLLATHRRFGIHLKEDLIALIIKEVVRAVQYLHENSIIHRDIRAANIMF SKEGYVKLIDFGLSASVKNTNGKAQSSVGSPYWMAPEVISCDCLQEPYNYTCDVWSIG ITAIELADTVPSLSDIHALRAMFRINRNPPPSVKRETRWSETLKDFISECLVKNPEYR PCIQEIPQHPFLAQVEGKEDQLRSELVDILKKNPGEKLRNKPYNVTFKNGHLKTISGQ BASE COUNT 1201 a 689 c 782 g 1136 t ORIGIN 1 tcgacatctg tggtcgcttt ttttagtaat aaaaaattgt attatgacgt cctatctgtt 3781 aagatacagt aactagggaa aaaa //
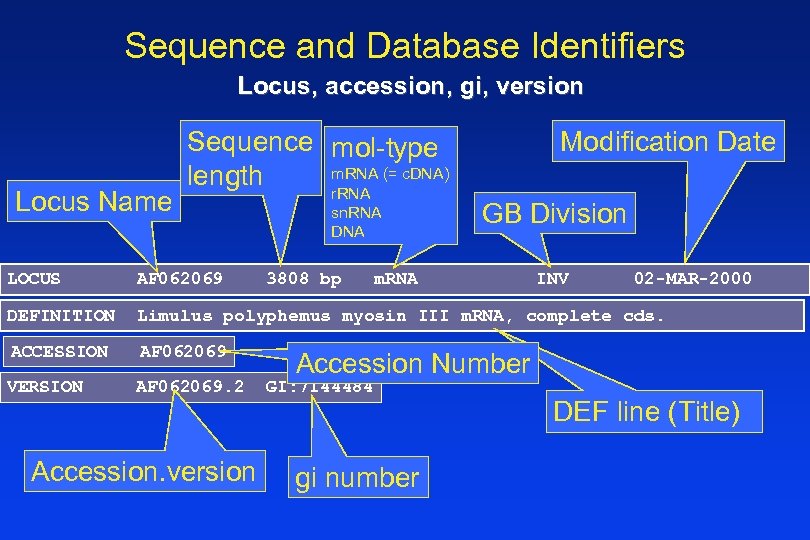
Sequence and Database Identifiers Locus, accession, gi, version Locus Name Sequence mol-type m. RNA (= c. DNA) length r. RNA sn. RNA DNA GB Division LOCUS AF 062069 DEFINITION Limulus polyphemus myosin III m. RNA, complete cds. ACCESSION AF 062069 VERSION AF 062069. 2 Accession. version 3808 bp Modification Date m. RNA INV 02 -MAR-2000 Accession Number GI: 7144484 gi number DEF line (Title)

Keywords, Source-organism Legacy field exception • EST • GSS • HTG KEYWORDS SOURCE ORGANISM Accepted common name . Scientific name Atlantic horseshoe crab. Limulus polyphemus Eukaryota; Metazoa; Arthropoda; Chelicerata; Merostomata; Xiphosura; Limulidae; Limulus. NCBI Taxonomic lineage according to Gen. Bank
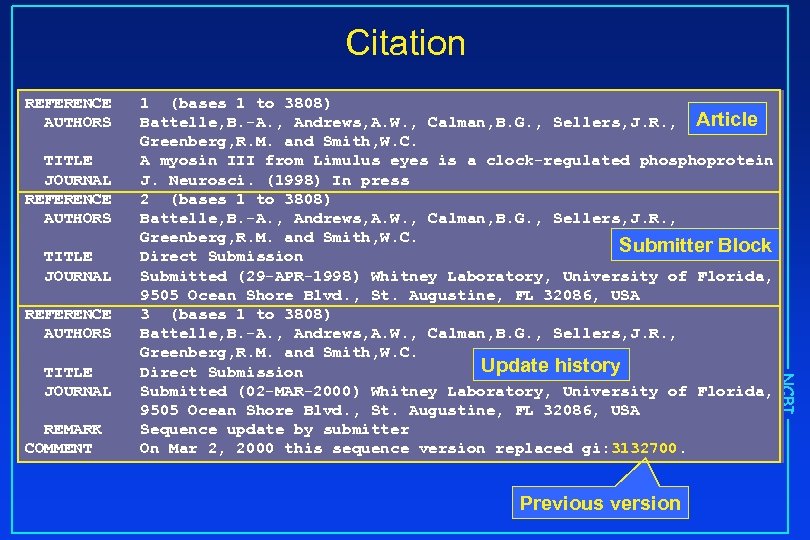
Citation REFERENCE AUTHORS TITLE JOURNAL REFERENCE AUTHORS REMARK COMMENT Previous version NCBI TITLE JOURNAL 1 (bases 1 to 3808) Battelle, B. -A. , Andrews, A. W. , Calman, B. G. , Sellers, J. R. , Article Greenberg, R. M. and Smith, W. C. A myosin III from Limulus eyes is a clock-regulated phosphoprotein J. Neurosci. (1998) In press 2 (bases 1 to 3808) Battelle, B. -A. , Andrews, A. W. , Calman, B. G. , Sellers, J. R. , Greenberg, R. M. and Smith, W. C. Submitter Block Direct Submission Submitted (29 -APR-1998) Whitney Laboratory, University of Florida, 9505 Ocean Shore Blvd. , St. Augustine, FL 32086, USA 3 (bases 1 to 3808) Battelle, B. -A. , Andrews, A. W. , Calman, B. G. , Sellers, J. R. , Greenberg, R. M. and Smith, W. C. Update history Direct Submission Submitted (02 -MAR-2000) Whitney Laboratory, University of Florida, 9505 Ocean Shore Blvd. , St. Augustine, FL 32086, USA Sequence update by submitter On Mar 2, 2000 this sequence version replaced gi: 3132700.
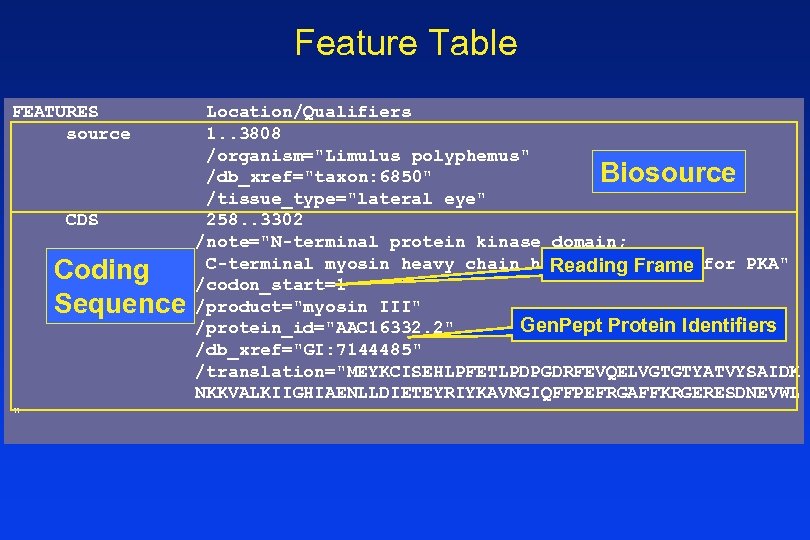
Feature Table FEATURES source CDS Coding Sequence " Location/Qualifiers 1. . 3808 /organism="Limulus polyphemus" Biosource /db_xref="taxon: 6850" /tissue_type="lateral eye" 258. . 3302 /note="N-terminal protein kinase domain; C-terminal myosin heavy chain head; substrate for PKA" Reading Frame /codon_start=1 /product="myosin III" Gen. Pept Protein Identifiers /protein_id="AAC 16332. 2" /db_xref="GI: 7144485" /translation="MEYKCISEHLPFETLPDPGDRFEVQELVGTGTYATVYSAIDK NKKVALKIIGHIAENLLDIETEYRIYKAVNGIQFFPEFRGAFFKRGERESDNEVWL

Sequence Indicates beginning of sequence data BASE COUNT 1201 a 689 c 782 g 1136 t ORIGIN 1 tcgacatctg tggtcgcttt ttttagtaat aaaaaattgt attatgacgt cctatctgtt <sequence omitted> 3721 accaatgtta taatatgaaataaag cagtcatggt agcagtggct gtttgaaata 3781 aagatacagt aactagggaa aaaa // End of record
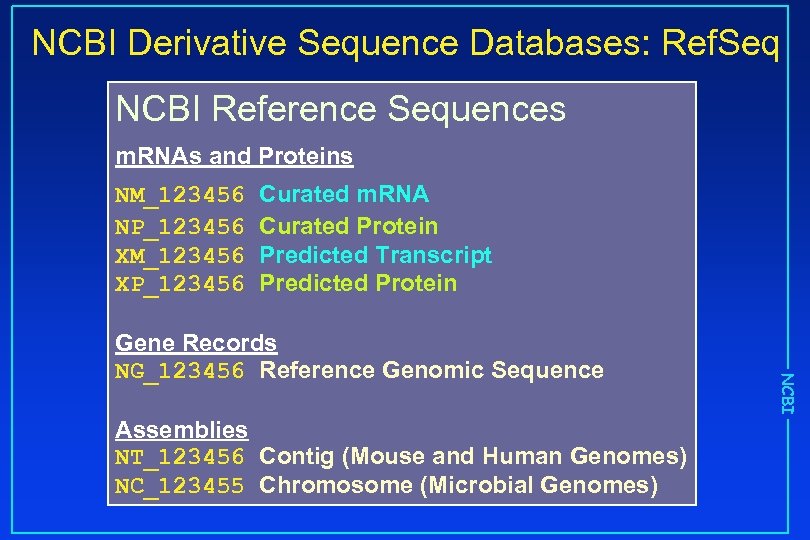
NCBI Derivative Sequence Databases: Ref. Seq NCBI Reference Sequences m. RNAs and Proteins NM_123456 NP_123456 XM_123456 XP_123456 Curated m. RNA Curated Protein Predicted Transcript Predicted Protein Assemblies NT_123456 Contig (Mouse and Human Genomes) NC_123455 Chromosome (Microbial Genomes) NCBI Gene Records NG_123456 Reference Genomic Sequence

Curated Ref. Seq Records: NM_, NP_ LOCUS NM_000492 6159 bp m. RNA PRI 26 -JUL-1999 DEFINITION Homo sapiens cystic fibrosis transmembrane conductance regulator(CFTR) m. RNA. REFSEQ: This reference sequence was derived from M 28668. 1, Ref. Seq Nucleotide ACCESSION M 55131. 1. NM_000492 On Feb 17, 2000 this sequence version replaced gi: 4502784. Summary: Cystic fibrosis transmembrane conductance regulator is LOCUS 1480 cassete sub-family C. PRI protein 26 -JUL-1999 member 7 NP_000483 of the ATP-bindingaa The DEFINITION cystic chloride channel and controls the regulation of functions as a fibrosis transmembrane conductance regulator. ACCESSION transport pathways. Mutations in this gene cause the NP_000483 other PID autosomalg 4502785 Ref. Seq Protein recessive disorder, cystic fibrosis (CF) and congenital VERSION bilateral. NP_000483. 1 the vas deferens (CBAVD). Alternative splice aplasia of GI: 4502785 DBSOURCE variants REFSEQ: accession NM_000492. 1 which result from mutations have been described, many of in the CFTR gene. COMPLETENESS: full length. COMMENT REFSEQ: This reference sequence was derived from M 55131. PROVISIONAL Ref. Seq: This is a provisional reference sequence record that has not yet been subject to human review. The final curated reference sequence record may be somewhat different from this one. Reviewed
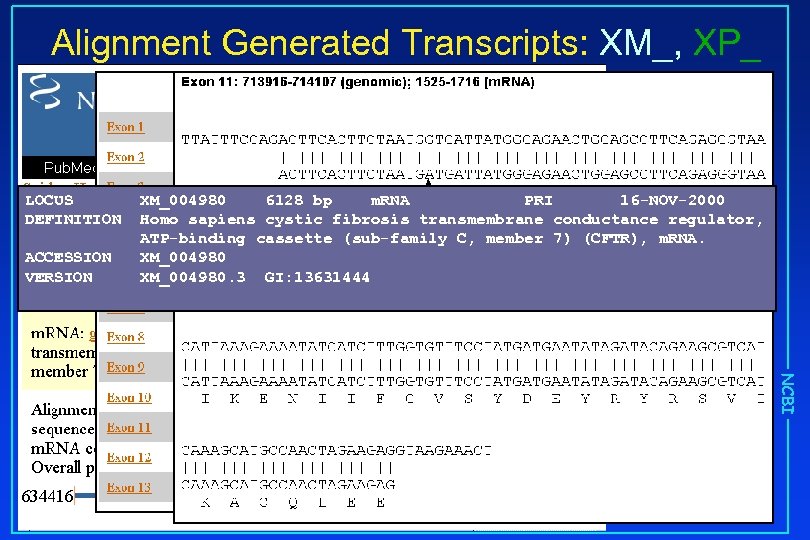
Alignment Generated Transcripts: XM_, XP_ LOCUS DEFINITION ACCESSION VERSION XM_004980 6128 bp m. RNA PRI 16 -NOV-2000 Homo sapiens cystic fibrosis transmembrane conductance regulator, ATP-binding cassette (sub-family C, member 7) (CFTR), m. RNA. mismatch XM_004980. 3 GI: 13631444 NCBI
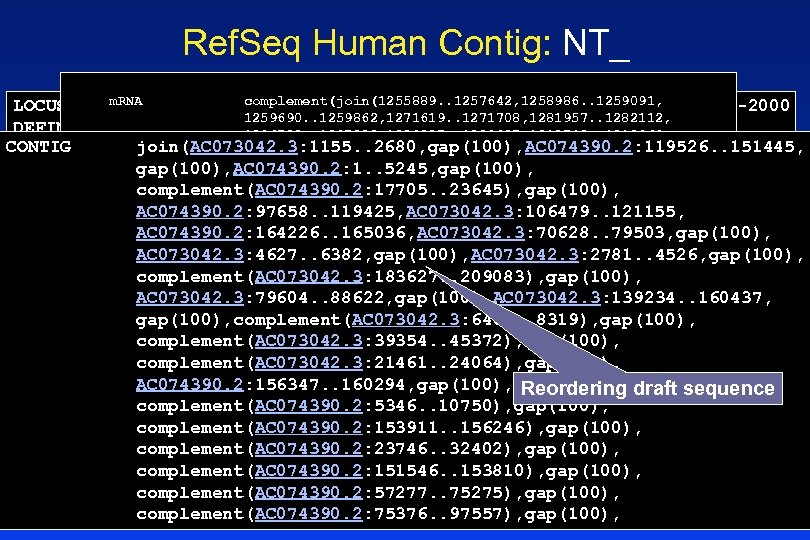
Ref. Seq Human Contig: NT_ m. RNA complement(join(1255889. . 1257642, 1258986. . 1259091, 16 -NOV-2000 LOCUS NT_007935 1888399 bp DNA CON 1259690. . 1259862, 1271619. . 1271708, 1281957. . 1282112, DEFINITION Homo sapiens chromosome 7 working draft sequence segment, 1296780. . 1297028, 1309837. . 1309937, 1312742. . 1312969, CONTIG join(AC 073042. 3: 1155. . 2680, gap(100), AC 074390. 2: 119526. . 151445, complete sequence. 1313881. . 1314031, 1317797. . 1317876, 1320768. . 1321018, gap(100), AC 074390. 2: 1. . 5245, gap(100), ACCESSION NT_007935 1321687. . 1321724, 1329492. . 1329620, 1331893. . 1332616, 1334111. . 1334197, 1336717. . 1336811, 1364895. . 1365086, complement(AC 074390. 2: 17705. . 23645), gap(100), VERSION NT_007935. 1 GI: 11422165 1375727. . 1375909, 1382442. . 1382534, 1384204. . 1384450, AC 074390. 2: 97658. . 119425, AC 073042. 3: 106479. . 121155, KEYWORDS HTG. 1387877. . 1388002, 1389139. . 1389302, 1390185. . 1390274, AC 074390. 2: 164226. . 165036, AC 073042. 3: 70628. . 79503, gap(100), 1393436. . 1393651, 1415408. . 1415516, 1420187. . 1420297, SOURCE human. 1444403. . 1444587)) AC 073042. 3: 4627. . 6382, gap(100), AC 073042. 3: 2781. . 4526, gap(100), ORGANISM Homo sapiens /partial complement(AC 073042. 3: 183627. . 209083), gap(100), Eukaryota; /gene="CFTR" Chordata; Craniata; Vertebrata; Metazoa; AC 073042. 3: 79604. . 88622, gap(100), AC 073042. 3: 139234. . 160437, Euteleostomi; Mammalia; Eutheria; Primates; Catarrhini; /product="cystic fibrosis transmembrane conductance gap(100), complement(AC 073042. 3: 6483. . 8319), gap(100), Hominidae; regulator, ATP-binding cassette (sub-family C, member 7)" Homo. /transcript_id="XM_004980. 1" complement(AC 073042. 3: 39354. . 45372), gap(100), REFERENCE 1 (bases 1 to 1888399) /db_xref="Locus. ID: 1080" complement(AC 073042. 3: 21461. . 24064), gap(100), AUTHORS International Human Genome Project collaborators. /db_xref="MIM: 602421" AC 074390. 2: 156347. . 160294, gap(100), human genome using TITLE Toward the /note="derived by automated the Reordering draft sequence complete sequence of computational analysis gene prediction method: Acembly. Supporting complement(AC 074390. 2: 5346. . 10750), gap(100), evidence JOURNAL Unpublished includes similarity to: 9 proteins, 1 m. RNAs See complement(AC 074390. 2: 153911. . 156246), gap(100), details in COMMENT GENOME ANNOTATION REFSEQ: NCBI contigs are derived from Ace. View" complement(AC 074390. 2: 23746. . 32402), gap(100), assembled genomic sequence data. They may include both gene complement(1255889. . 1444587) /gene="CFTR" complement(AC 074390. 2: 151546. . 153810), gap(100), draft and finished sequence. /note="CF; MRP 7; ABC 35; complement(AC 074390. 2: 57277. . 75275), gap(100), COMPLETENESS: not full length. ABCC 7" /db_xref="Locus. ID: 1080" complement(AC 074390. 2: 75376. . 97557), gap(100),
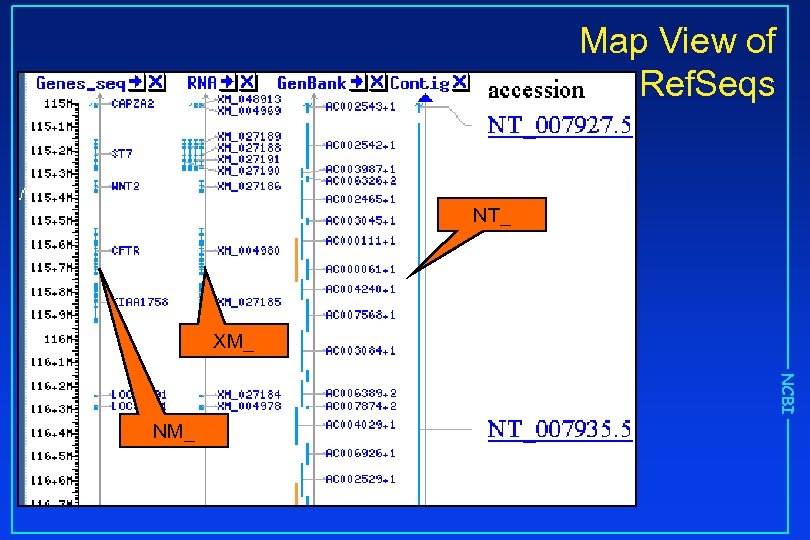
Map View of Ref. Seqs NT_ XM_ NCBI NM_

Ref. Seq Genome Records: NG_ NCBI
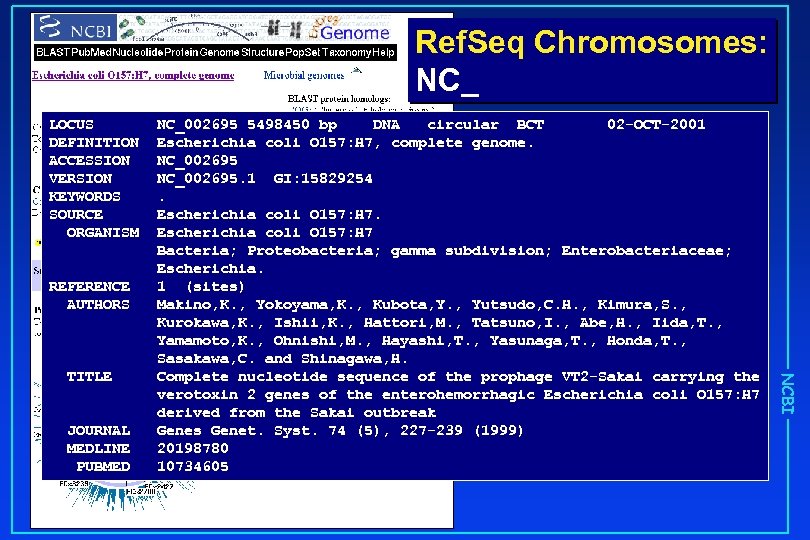
Ref. Seq Chromosomes: NC_ LOCUS DEFINITION ACCESSION VERSION KEYWORDS SOURCE ORGANISM REFERENCE AUTHORS JOURNAL MEDLINE PUBMED NCBI TITLE NC_002695 5498450 bp DNA circular BCT 02 -OCT-2001 Escherichia coli O 157: H 7, complete genome. NC_002695. 1 GI: 15829254. Escherichia coli O 157: H 7 Bacteria; Proteobacteria; gamma subdivision; Enterobacteriaceae; Escherichia. 1 (sites) Makino, K. , Yokoyama, K. , Kubota, Y. , Yutsudo, C. H. , Kimura, S. , Kurokawa, K. , Ishii, K. , Hattori, M. , Tatsuno, I. , Abe, H. , Iida, T. , Yamamoto, K. , Ohnishi, M. , Hayashi, T. , Yasunaga, T. , Honda, T. , Sasakawa, C. and Shinagawa, H. Complete nucleotide sequence of the prophage VT 2 -Sakai carrying the verotoxin 2 genes of the enterohemorrhagic Escherichia coli O 157: H 7 derived from the Sakai outbreak Genes Genet. Syst. 74 (5), 227 -239 (1999) 20198780 10734605
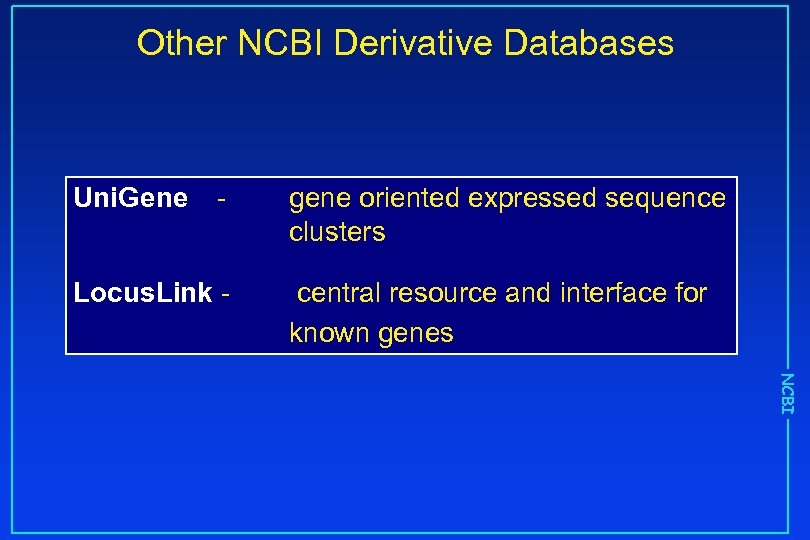
Other NCBI Derivative Databases Uni. Gene - Locus. Link - gene oriented expressed sequence clusters central resource and interface for known genes NCBI

NCBI Homepage NCBI

Mendelian Inheritance in Man Entrez NCBI Similarity Searching NCBI Homepage

Using Entrez An integrated database search and retrieval system NCBI
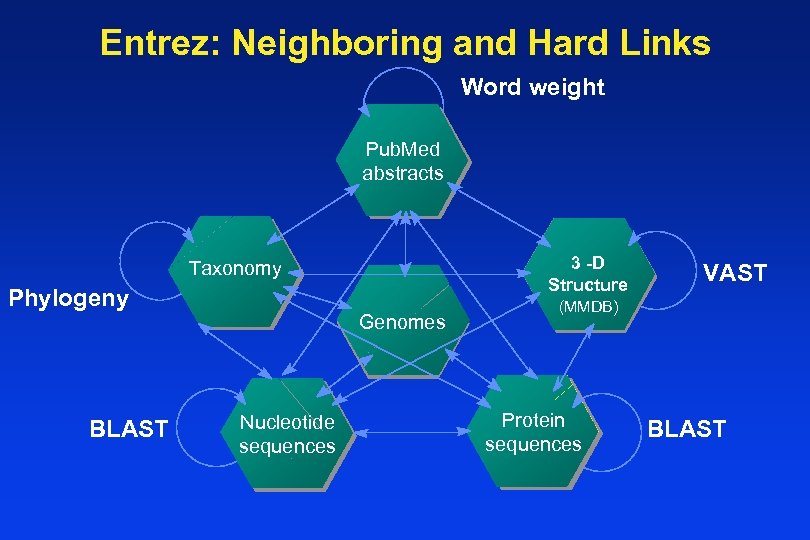
Entrez: Neighboring and Hard Links Word weight Pub. Med abstracts 3 -D 3 -D Structure Taxonomy Phylogeny BLAST Genomes Nucleotide sequences VAST (MMDB) Protein sequences BLAST
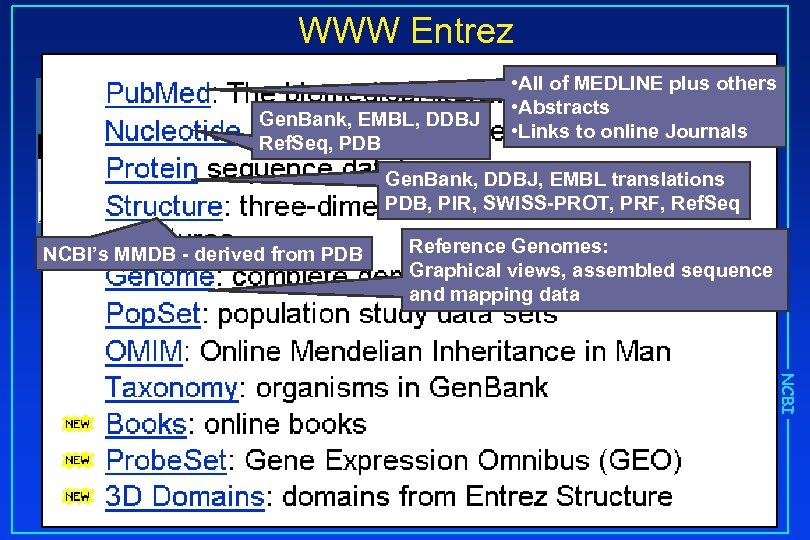
WWW Entrez Gen. Bank, EMBL, DDBJ Ref. Seq, PDB • All of MEDLINE plus others • Abstracts • Links to online Journals Gen. Bank, DDBJ, EMBL translations PDB, PIR, SWISS-PROT, PRF, Ref. Seq NCBI’s MMDB - derived from PDB Reference Genomes: Graphical views, assembled sequence and mapping data NCBI

Database Searching with Entrez Using limits and field restriction to find mouse GAPD u Linking and neighboring with mouse GAPD NCBI u
Entrez Nucleotides Mouse NCBI
![Document Summaries: Mouse[All Fields] 3 million records Chicken not mouse !? NCBI Document Summaries: Mouse[All Fields] 3 million records Chicken not mouse !? NCBI](https://present5.com/presentation/8308095d65c54aee311c945fab2d86ee/image-41.jpg)
Document Summaries: Mouse[All Fields] 3 million records Chicken not mouse !? NCBI
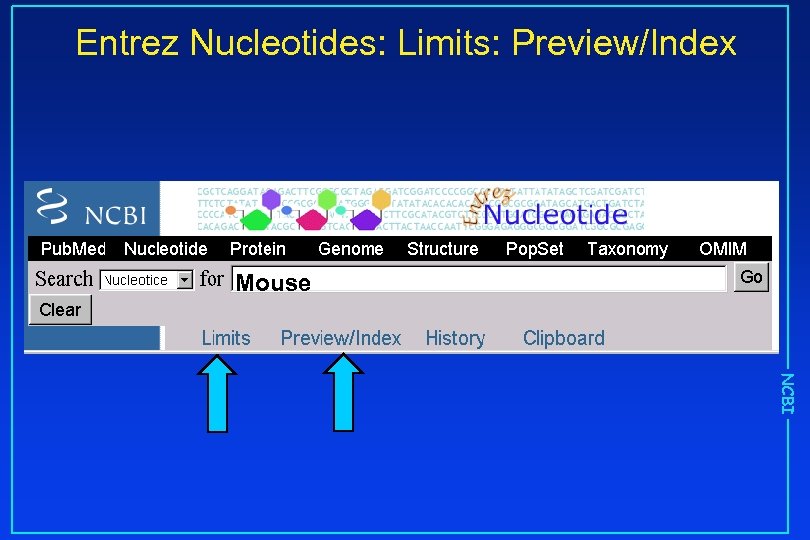
Entrez Nucleotides: Limits: Preview/Index Mouse NCBI
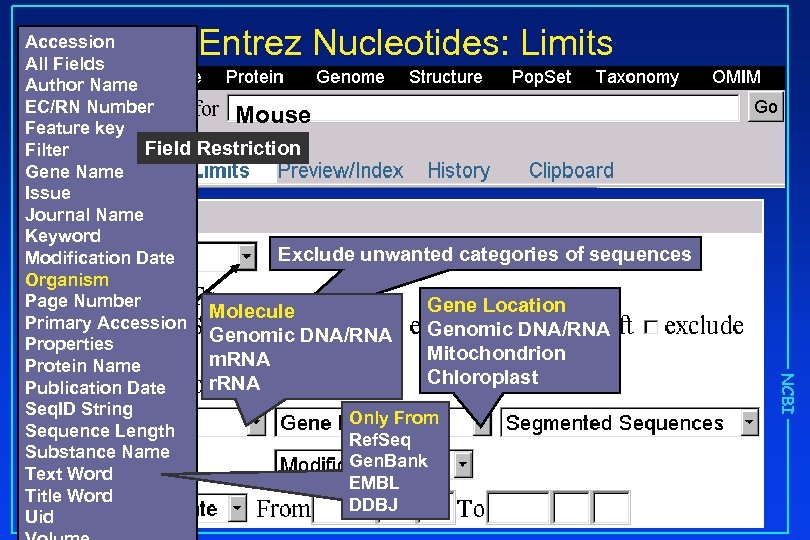
Entrez Nucleotides: Limits Mouse Restriction Exclude unwanted categories of sequences Molecule Genomic DNA/RNA m. RNA r. RNA Gene Location Genomic DNA/RNA Mitochondrion Chloroplast Only From Ref. Seq Gen. Bank EMBL DDBJ NCBI Accession All Fields Author Name EC/RN Number Feature key Field Filter Gene Name Issue Journal Name Keyword Modification Date Organism Page Number Primary Accession Properties Protein Name Publication Date Seq. ID String Sequence Length Substance Name Text Word Title Word Uid
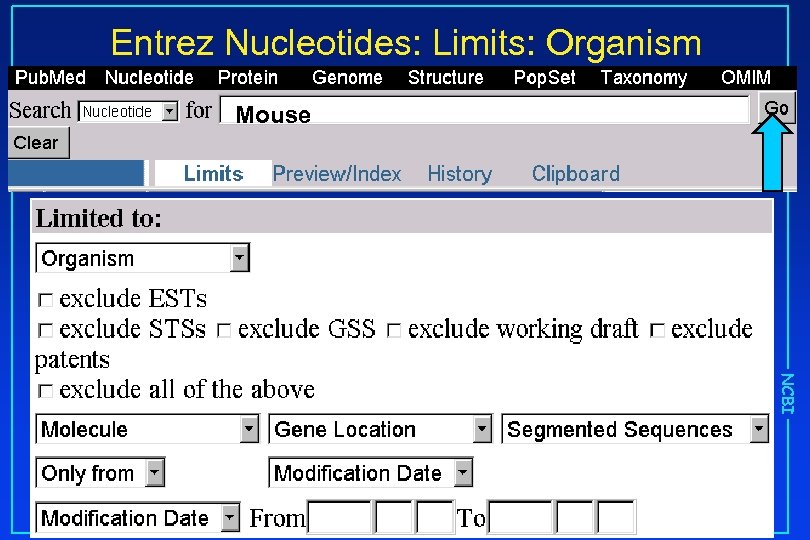
Entrez Nucleotides: Limits: Organism Mouse NCBI
![Document Summaries: Mouse[Organism] 2, 976, 070[All Fields] -2, 921, 009[Organism] 55, 061 NCBI Document Summaries: Mouse[Organism] 2, 976, 070[All Fields] -2, 921, 009[Organism] 55, 061 NCBI](https://present5.com/presentation/8308095d65c54aee311c945fab2d86ee/image-45.jpg)
Document Summaries: Mouse[Organism] 2, 976, 070[All Fields] -2, 921, 009[Organism] 55, 061 NCBI
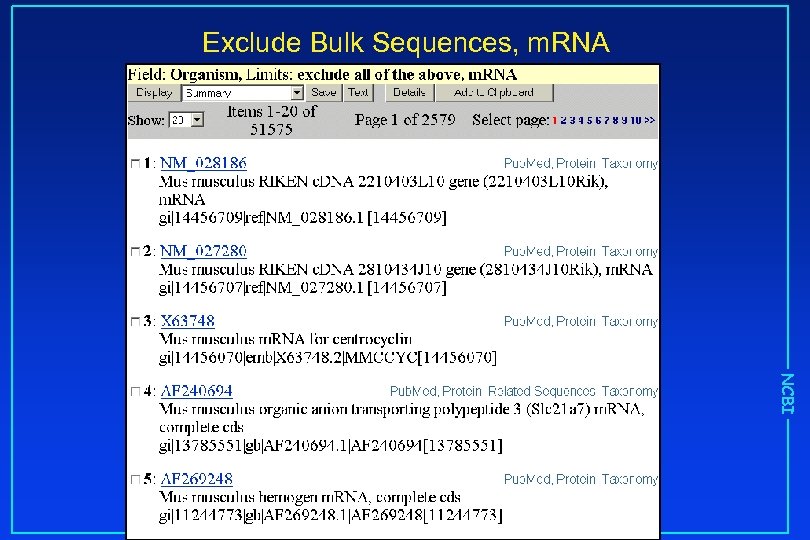
Exclude Bulk Sequences, m. RNA NCBI
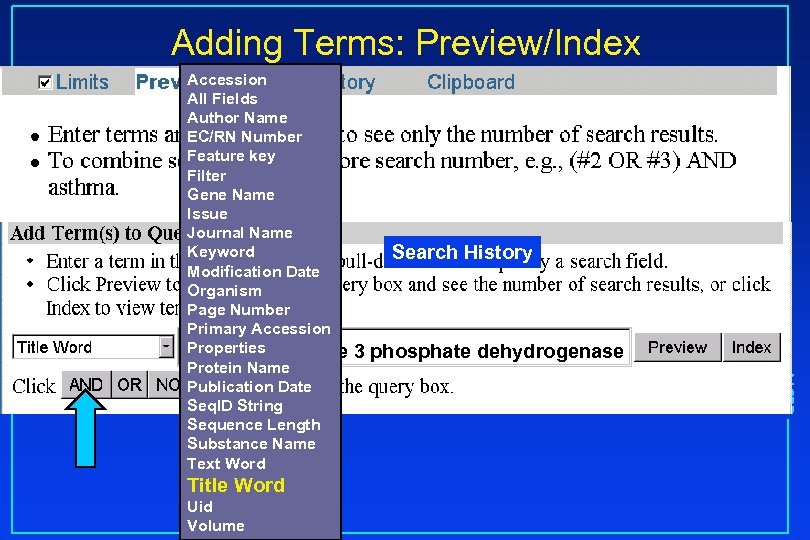
Adding Terms: Preview/Index Title Word Uid Volume Search History 3 phosphate dehydrogenase NCBI Accession All Fields Author Name EC/RN Number Feature key Filter Gene Name Issue Journal Name Keyword Modification Date Organism Page Number Primary Accession Properties glyceraldehyde Protein Name Publication Date Seq. ID String Sequence Length Substance Name Text Word
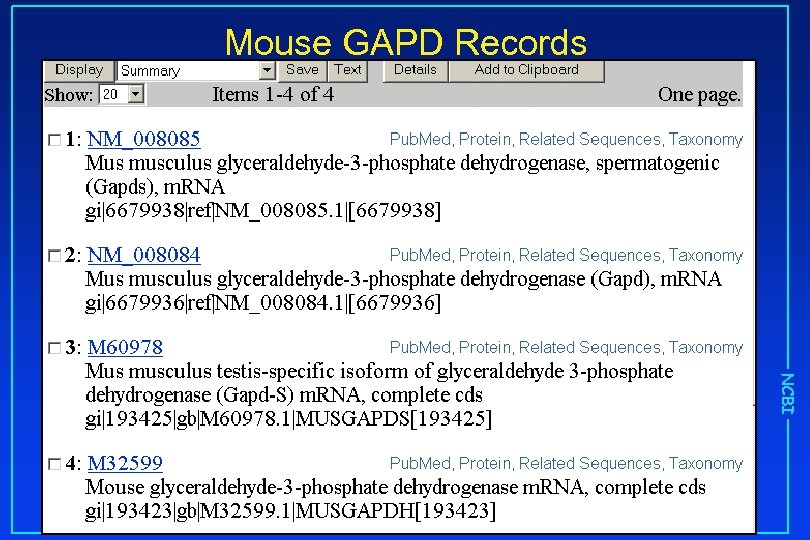
Mouse GAPD Records NCBI
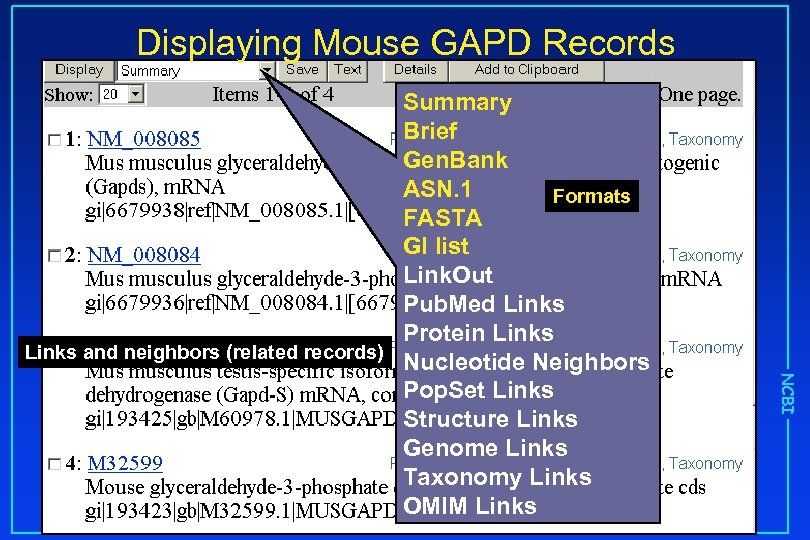
Displaying Mouse GAPD Records NCBI Summary Brief Gen. Bank ASN. 1 Formats FASTA GI list Link. Out Pub. Med Links Protein Links and neighbors (related records) Nucleotide Neighbors Pop. Set Links Structure Links Genome Links Taxonomy Links OMIM Links
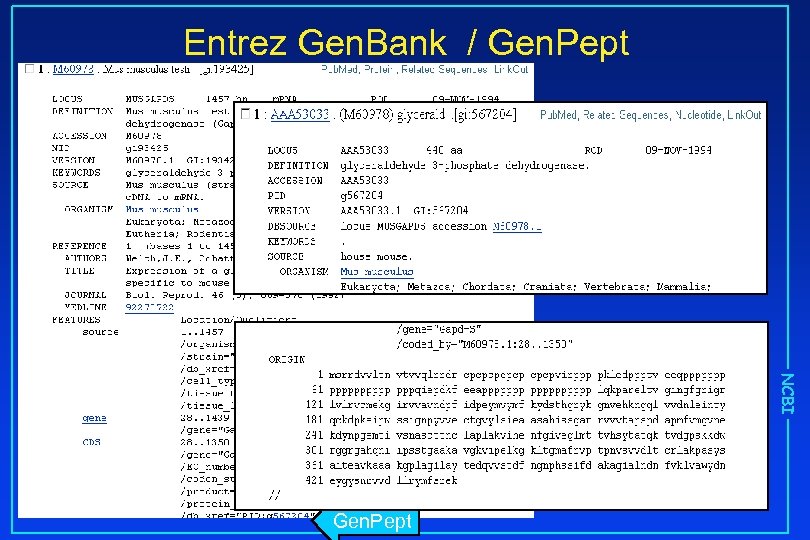
Entrez Gen. Bank / Gen. Pept NCBI Gen. Pept

FASTA Format >gi|193425|gb|M 60978. 1|MUSGAPDS Mus musculus testis-specific isoform of glycerald GGCAGCCAGGCCATGAGATCTTAGGCCATGTCGAGACGTGGTCCTTACCAATGTTACTGTTGTCC AGCTACGGCGGGACCGATGCCCATGCCCATGTCCATGCCCTGTGATCAGACC ACCTCCACCCAAGCTTGAGGATCCACCACGGTTGAAGAACAGCCACCGCCGCCGCCACCTCCACCACCACCTCCTCCTCCACCCCAGATAGAGCCAGACAAGTTTGAAGAGGCTCCCCCTCCTCCTCCCCCTCCTCCACCACTCCAAAAGCCAGCTAGAGAGCT GACAGTGGGTATCAATGGATTTGGACGCATTGGTCGTCTGGTGCTGCGAGTCTGCATGGAGAAGGGCATT > AGGGTGGTAGCAGTGAATGACCCATTGATCCAGAATACATGGTTTACATGTTCAAATATGACTCCA CACATGGTAGATACAAAGGAAACGTGGAACATAAGAATGGACAACTAGTTGTGGACAACCTTGAGATCAA CACGTACCAGTGCAAAGACCCTAAAGAAATCCCCTGGAGCTCTATAGGGAATCCCTACGTGGTGGAGTGT gi number Locus Name ACAGGCGTCTATCTGTCCATCGAGGCAGCTTCGGCACATATTTCATCTGGTGCCAGGCGTGTGGTGGTCA CTGCACCCTCCCCCGATGCACCCATGTTTGTCATGGGAGTGAACGAGAAGGACTATAACCCTGGCTCTAT Database Identifiers GACCATTGTCAGCAATGCATCCTGTACCACCAACTGCCTGGCTCCTCTCGCCAAGGTTATTCATGAAAAC Accession number gb Gen. Bank TTCGGGATCGTGGAAGGGCTAATGACCACAGTCCATTCCTACACAGCCACTCAGAAGACAGTGGATGGGC CATCAAAGAAGGACTGGCGAGGTGGCCGCGGCGCTCACCAAAACATCATCCCATCGTCCACTGGGGCTGC emb EMBL CAAGGCTGTAGGCAAAGTCATCCCAGAGCTCAAAGGGAAGCTAACAGGAATGGCATTCCGGGTGCCAACC dbj DDBJ CCAAACGTGTCAGTTGTGGACCTGCCGCCTGGCCAAGCCTGCTTCTTACTCGGCTATCACGGAGG sp SWISS-PROT CTGTGAAAGCTGCAGCCAAGGGACCTTTGGCATCCTTGCTTACACAGAGGACCAGGTGGTCTCCAC GGACTTTAACGGCAATCCCCATTCTTCCATCTTTGATGCTAAGGCTGGAATTGCCCTCAATGACAACTTC pdb Protein Databank GTGAAGCTTGTTGCCTGGTACGACAACGAATATGGCTACAGTAACCGAGTGGTCGACCTCCTCCGCTACA pir PIR TGTTTAGCCGAGAGAAGTAACACAAAAGGCCCCTCCTTGCTCCCCTGCGCACCTCGCGTTCCTGACTTCG prf PRF GCTTCCACTCAAAGGCGCCGCCACCGGGTCAACAATGAAATAAAAACGAGAATGCGC FASTA Definition Line >gi|193425|gb|M 60978. 1|MUSGAPDS Ref. Seq NCBI ref
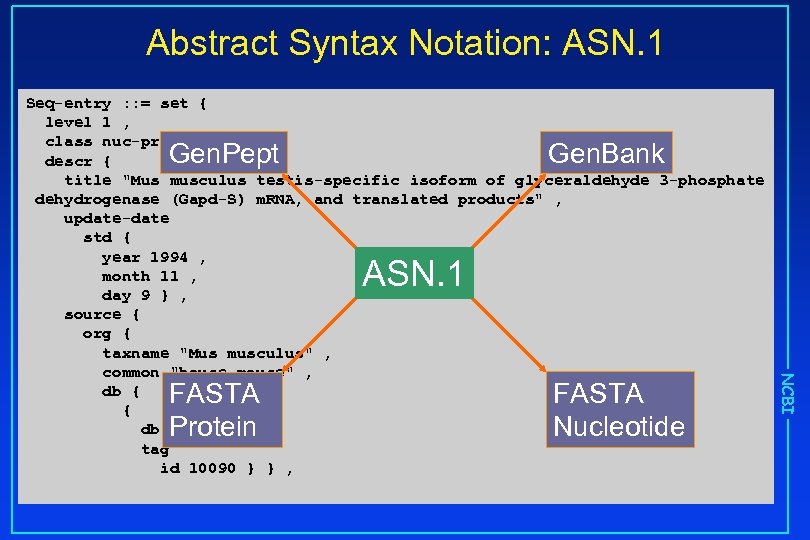
Abstract Syntax Notation: ASN. 1 Seq-entry : : = set { level 1 , class nuc-prot , descr { title "Mus musculus testis-specific isoform of glyceraldehyde 3 -phosphate dehydrogenase (Gapd-S) m. RNA, and translated products" , update-date std { year 1994 , month 11 , day 9 } , source { org { taxname "Mus musculus" , common "house mouse" , db { { db "taxon" , tag id 10090 } } , Gen. Pept Gen. Bank ASN. 1 FASTA Nucleotide NCBI FASTA Protein
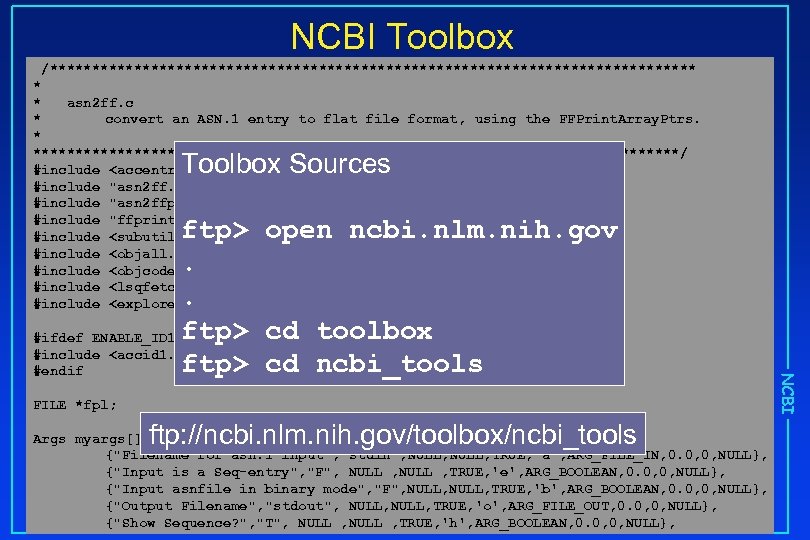
NCBI Toolbox /*************************************** * * asn 2 ff. c * convert an ASN. 1 entry to flat file format, using the FFPrint. Array. Ptrs. * ***************************************/ #include <accentr. h> #include "asn 2 ff. h" #include "asn 2 ffp. h" #include "ffprint. h" #include <subutil. h> #include <objall. h> #include <objcode. h> #include <lsqfetch. h> #include <explore. h> Toolbox Sources FILE *fpl; ftp: //ncbi. nlm. nih. gov/toolbox/ncbi_tools Args myargs[] = { {"Filename for asn. 1 input", "stdin", NULL, TRUE, 'a', ARG_FILE_IN, 0. 0, 0, NULL}, {"Input is a Seq-entry", "F", NULL , TRUE, 'e', ARG_BOOLEAN, 0. 0, 0, NULL}, {"Input asnfile in binary mode", "F", NULL, TRUE, 'b', ARG_BOOLEAN, 0. 0, 0, NULL}, {"Output Filename", "stdout", NULL, TRUE, 'o', ARG_FILE_OUT, 0. 0, 0, NULL}, {"Show Sequence? ", "T", NULL , TRUE, 'h', ARG_BOOLEAN, 0. 0, 0, NULL}, NCBI ftp> open ncbi. nlm. nih. gov. . #ifdef ENABLE_ID 1 ftp> cd toolbox #include <accid 1. h> ftp> cd ncbi_tools #endif

Protein Neighbors-Structure Links Related Proteins Cn 3 D GAPD Structure Links NCBI
Advanced Neighbors: BLink NCBI
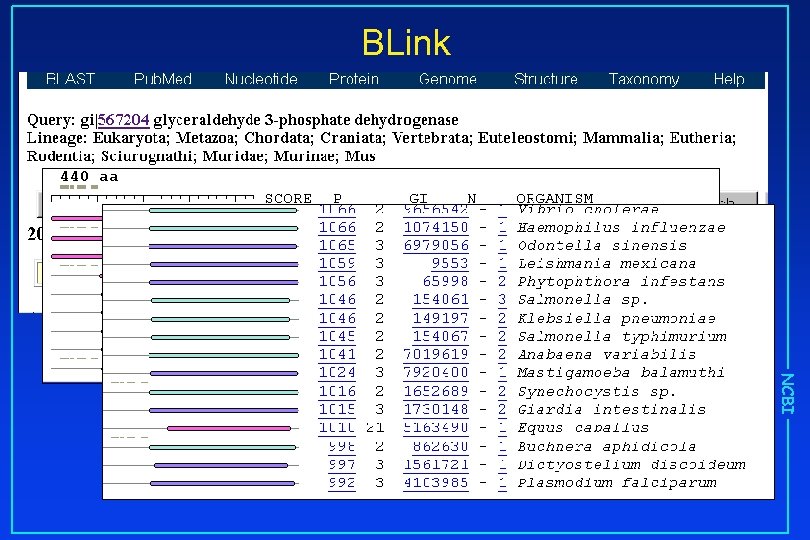
BLink NCBI

Pub. Med Link NCBI
Online Books NCBI
Entrez Structures Molecular Modeling Database (MMDB) and Cn 3 D NCBI

MMDB: Molecular Modeling Data Base n Derived from experimentally determined PDB records n Value added to PDB records including: • • Structure neighbors determined by Vector Alignment Search Tool (VAST) NCBI n Addition of explicit chemical graph information Validation Inclusion of Taxonomy, Citation, and other information Conversion to parseable ASN. 1 data description language
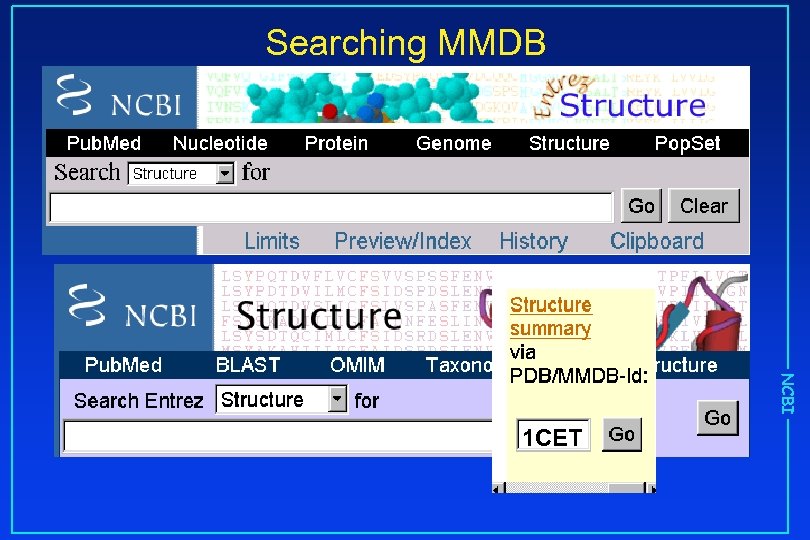
Searching MMDB NCBI 1 CET
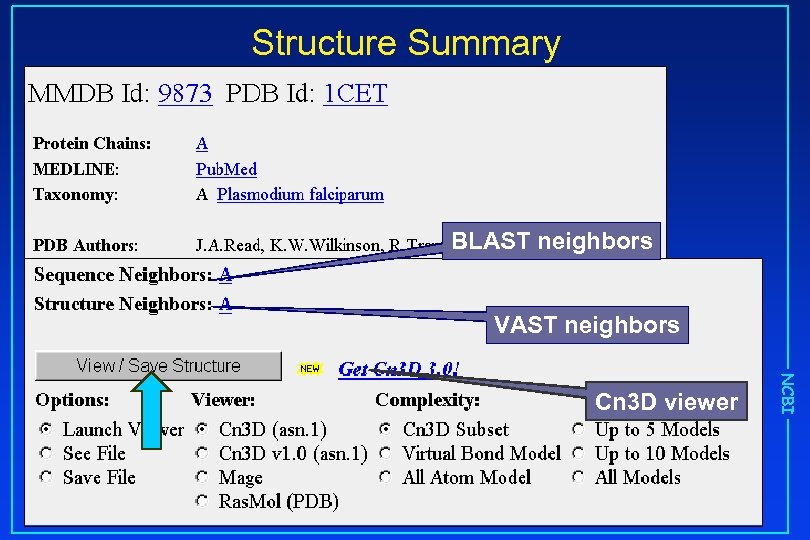
Structure Summary BLAST neighbors VAST neighbors NCBI Cn 3 D viewer
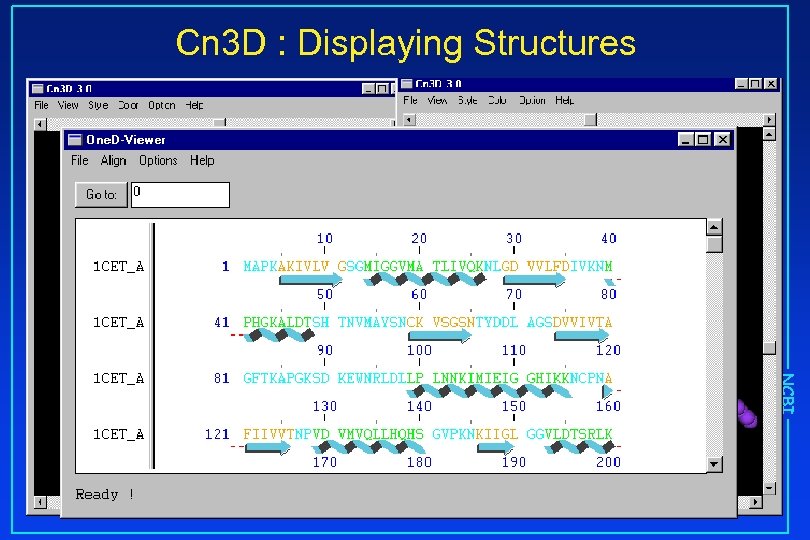
Cn 3 D : Displaying Structures NCBI Chloroquine
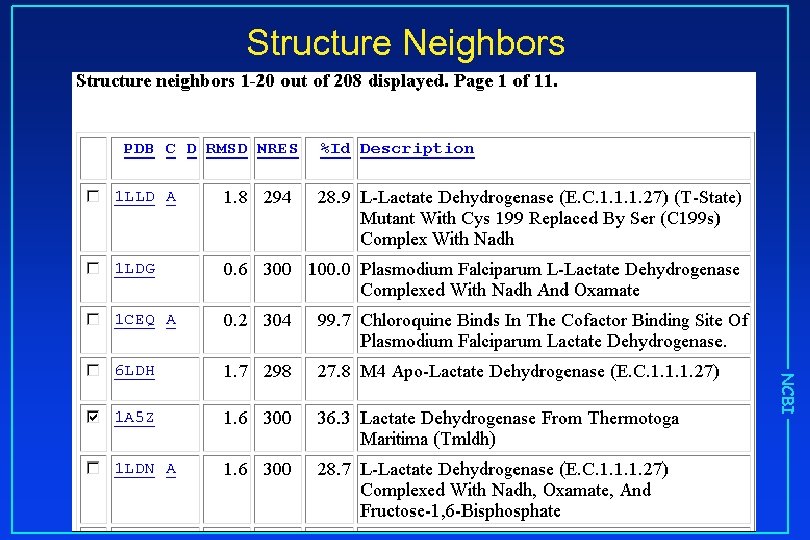
Structure Neighbors NCBI
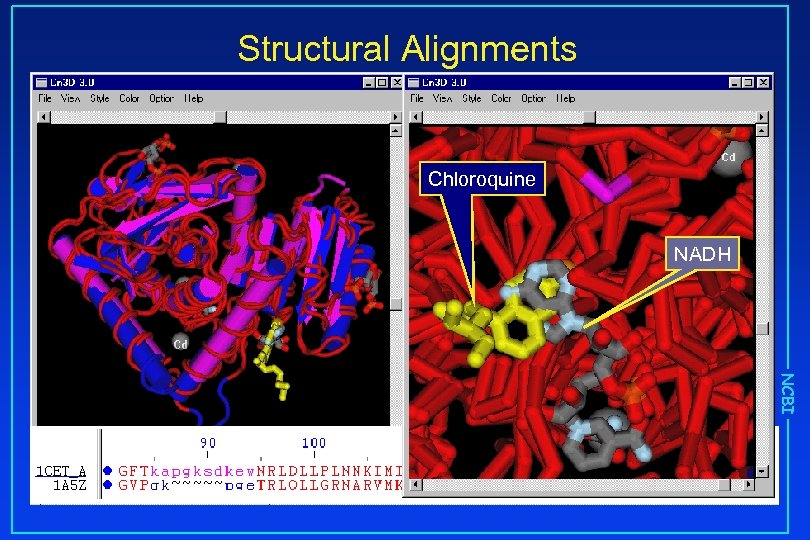
Structural Alignments Chloroquine NADH NCBI

Why do we need similarity searching? èIdentification and annotation • Incomplete or no annotations (Gen. Bank) • Incorrectly annotated sequences but it ain’t necessarily so! NCBI è Evolutionary relationships homologous molecules may have similar functions

Basic Local Alignment Search Tool n Widely used similarity search tool n Heuristic approach based on Smith Waterman algorithm n Finds best local alignments n Provides statistical significance n All combinations (DNA/Protein) query and database. n DNA vs DNA translation vs Protein vs DNA translation www, email server, standalone, and network clients NCBI • • •
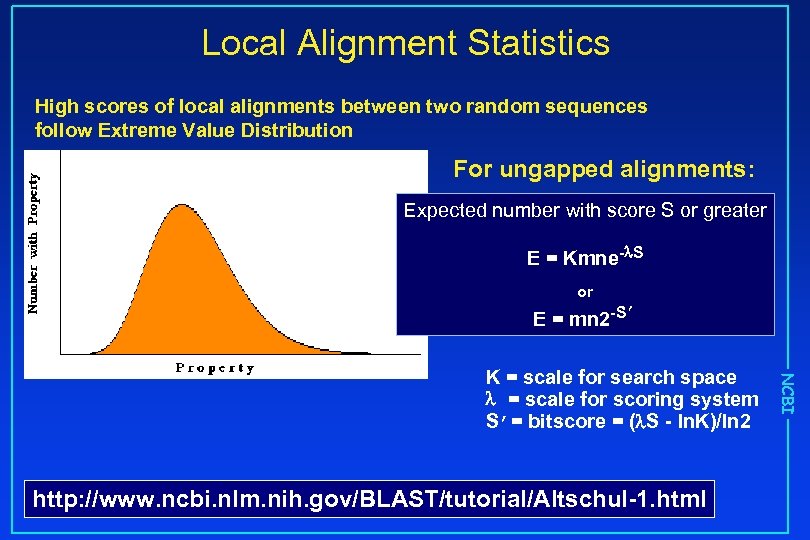
Local Alignment Statistics High scores of local alignments between two random sequences follow Extreme Value Distribution For ungapped alignments: Expected number with score S or greater E = Kmne- S or E = mn 2 -S’ http: //www. ncbi. nlm. nih. gov/BLAST/tutorial/Altschul-1. html NCBI K = scale for search space = scale for scoring system S’= bitscore = ( S - ln. K)/ln 2

Scoring Systems • Nucleic acids identity matrix • Proteins • Position Independent Matrices • PAM Matrices (Percent Accepted Mutation) • Implicit model of evolution • Higher PAM number all calculated from PAM 1 • PAM 250 widely used • BLOSUM Matrices (BLOck SUbstition Matrices) • Position Specific Score Matrices (PSSM) • PSI and RPS BLAST NCBI • Empirically determined from alignment of conserved blocks • Each includes information up to a certain level of identity • BLOSUM 62 widely used
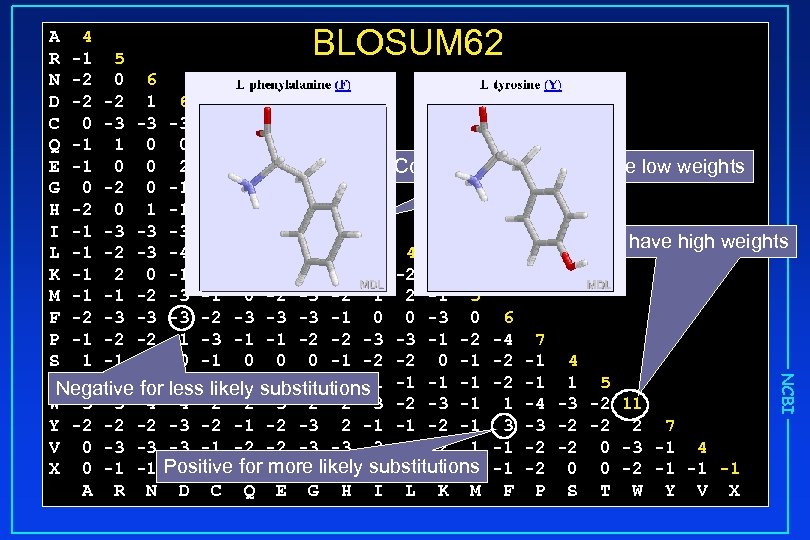
BLOSUM 62 NCBI A 4 R -1 5 N -2 0 6 D -2 -2 1 6 C 0 -3 -3 -3 9 Q -1 1 0 0 -3 5 E -1 0 0 2 -4 2 5 Common amino acids have low weights G 0 -2 0 -1 -3 -2 -2 6 H -2 0 1 -1 -3 0 0 -2 8 I -1 -3 -3 -3 -1 -3 -3 -4 -3 4 Rare amino acids have high weights L -1 -2 -3 -4 -3 2 4 K -1 2 0 -1 -3 1 1 -2 -1 -3 -2 5 M -1 -1 -2 -3 -1 0 -2 -3 -2 1 2 -1 5 F -2 -3 -3 -3 -1 0 0 -3 0 6 P -1 -2 -2 -1 -3 -1 -1 -2 -2 -3 -3 -1 -2 -4 7 S 1 -1 1 0 -1 0 0 0 -1 -2 -2 0 -1 -2 -1 4 T 0 -1 for -1 -1 -1 substitutions -1 -1 -1 -2 -1 1 5 Negative 0 less likely -1 -2 -2 -1 W -3 -3 -4 -4 -2 -2 -3 -1 1 -4 -3 -2 11 Y -2 -2 -2 -3 -2 -1 -2 -3 2 -1 -1 -2 -1 3 -3 -2 -2 2 7 V 0 -3 -3 -3 -1 -2 -2 -3 -3 3 1 -2 1 -1 -2 -2 0 -3 -1 4 X 0 -1 -1 Positive-1 -1 -1 likely-1 -1 -1 -2 0 0 -2 -1 -1 -2 for more -1 substitutions A R N D C Q E G H I L K M F P S T W Y V X
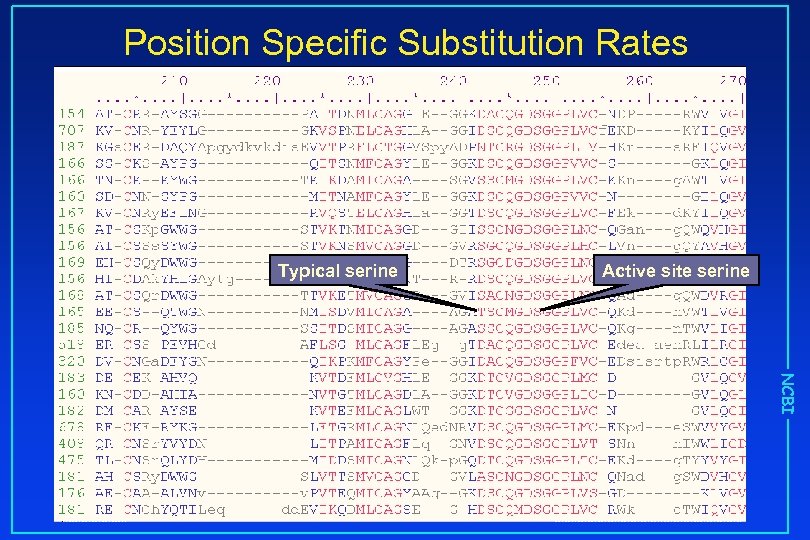
Position Specific Substitution Rates Typical serine Active site serine NCBI

Position Specific Score Matrix (PSSM) D G V I S S C N G D S G G P L N C Q A R N D C Q E G H I L K M -2 0 2 -4 2 4 -4 -3 -5 -4 0 -2 -1 0 -2 -4 -3 -3 6 -4 -5 -5 0 -2 1 -3 -3 -5 -1 -2 6 -1 -4 -5 1 -5 3 -3 -4 -6 0 -1 -4 -1 2 -4 6 -2 -5 0 8 -5 -3 -2 -1 -4 -7 -6 -4 -4 -1 -4 -2 -3 -3 -5 -4 -4 -7 -6 -7 12 -7 -7 -5 -6 -5 -5 -7 -5 Serine scored differently 0 2 -1 -6 7 0 -2 0 -6 -4 2 0 in -5 7 -4 -7 -7 -5 -3 -3 -4 -4 -4 these two positions -4 -5 -2 9 -7 -4 -1 -5 -5 -7 -7 -4 -2 -4 -4 -3 -3 -3 -4 -6 -6 -3 -5 -6 -4 -5 -6 8 -6 -8 -7 -5 -6 -6 -4 -5 -6 8 -6 -7 -7 -5 -6 Active site nucleophile -6 -6 -5 -5 -6 -6 -6 -7 -4 -6 -6 -7 -7 -5 -5 -6 -7 0 -1 6 -6 1 -6 0 -6 -4 -4 -6 -6 -1 3 0 -5 4 -4 -5 -5 10 -2 -5 -5 1 -1 -1 -5 0 1 4 2 -5 2 0 0 0 -4 -2 1 0 -1 1 3 -4 -1 1 4 -3 -1 -2 F -6 -3 -6 -5 -7 -5 0 -2 -4 -7 -6 -7 -7 -7 0 -3 -1 0 -2 P 1 -2 -4 -5 -5 -1 -7 -5 -6 -5 -4 -6 -6 9 -6 -6 -4 0 -3 S 0 -2 0 -3 1 4 -4 -1 -3 -4 7 -4 -2 -4 -6 -2 -1 -1 0 T -1 -1 -2 0 -3 3 -4 -3 -5 -4 -2 -5 -4 -4 -5 -1 0 -1 -2 W -6 0 -6 -1 -7 -6 -5 -3 -6 -8 -6 -6 -6 -7 -5 -6 -5 -3 -2 Y -4 -6 -4 -4 -5 -5 0 -4 -6 -7 -5 -7 -7 -7 -4 -1 0 -3 -2 V -1 -5 -2 0 -6 -3 -4 -3 -6 -7 -5 -7 -7 -6 0 -4 -3 NCBI 206 207 208 209 210 211 212 213 214 215 216 217 218 219 220 221 222 223 224 A 0 -2 -1 -3 -2 4 -4 -2 -2 -5 -2 -3 -3 -2 -4 -1 0 0 -1

Gapped Alignments NCBI • Gapping provides more biologically realistic alignments • Statistical behavior not completely understood for gapped alignments • Gapped BLAST parameters must be found by simulations for each matrix • Affine gap costs = -(a+bk) a = gap open penalty b = gap extend penalty A gap of length 1 receives the score -(a+b)

Intermission NCBI
8308095d65c54aee311c945fab2d86ee.ppt