e49bda1731c8d4fca1dd792ecb495836.ppt
- Количество слайдов: 94

NCBI Field. Guide NCBI Molecular Biology Resources A Field Guide Part 1 January 12, 2007
• The NCBI Entrez System • NCBI Sequence Databases – Primary data: Gen. Bank – Derivative data: Ref. Seq, Gene • Protein Structure and Function • Sequence polymorphisms and phenotypes ** Intermission ** • NCBI Genomic Resources • BLAST NCBI Field. Guide NCBI Resources

NCBI Field. Guide The National Center for Biotechnology Information Bethesda, MD Created in 1988 as a part of the National Library of Medicine at NIH – national resource for molecular biology information (biological information direct from organisms) – gather data both nationally and internationally – develop new information technologies to aid in the understanding of fundamental molecular and genetic processes that control health and disease
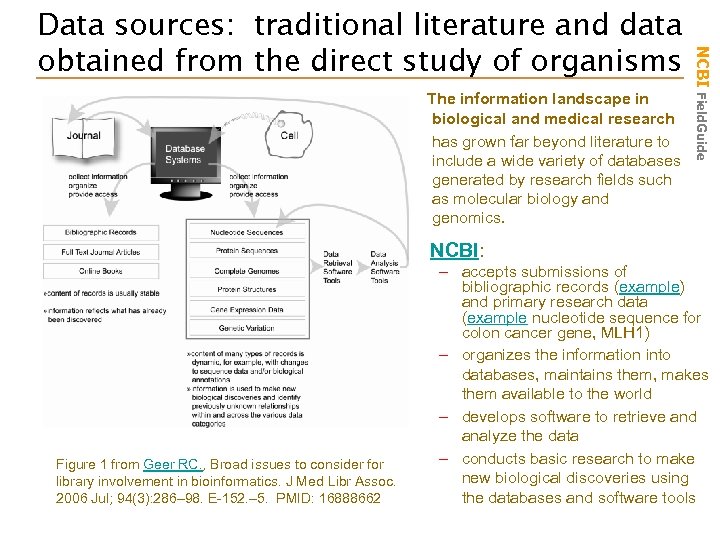
The information landscape in biological and medical research has grown far beyond literature to include a wide variety of databases generated by research fields such as molecular biology and genomics. NCBI Field. Guide Data sources: traditional literature and data obtained from the direct study of organisms NCBI: Figure 1 from Geer RC. , Broad issues to consider for library involvement in bioinformatics. J Med Libr Assoc. 2006 Jul; 94(3): 286– 98. E-152. – 5. PMID: 16888662 – accepts submissions of bibliographic records (example) and primary research data (example nucleotide sequence for colon cancer gene, MLH 1) – organizes the information into databases, maintains them, makes them available to the world – develops software to retrieve and analyze the data – conducts basic research to make new biological discoveries using the databases and software tools

• NCBI accepts submissions of primary data • NCBI develops tools to analyze these data • NCBI uses these tools to create derivative databases based on the primary data • NCBI provides free search, link, and retrieval of these data, primarily through the Entrez system NCBI Field. Guide What does NCBI do?
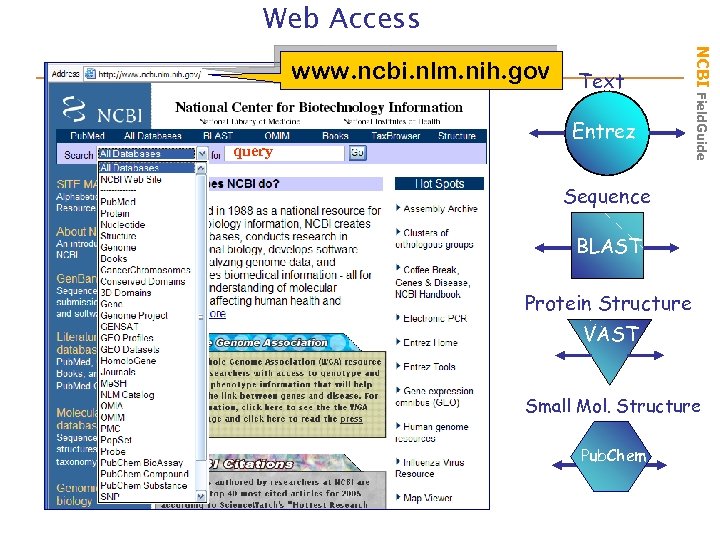
Web Access query Text Entrez NCBI Field. Guide www. ncbi. nlm. nih. gov Sequence BLAST Protein Structure VAST Small Mol. Structure Pub. Chem

30, 000 files per day 620 Gigabytes per day NCBI Field. Guide The NCBI ftp site
Help for Programmers NCBI-like functionality into their programs. Three main parts: Data Model, Data Encoding and Programming Libraries. NCBI Field. Guide NCBI Toolbox: In-house source code useful for incorporating • Examples: BLAST, Cn 3 D, Sequin, Data format conversion scripts http: //www. ncbi. nlm. nih. gov/IEB/Tool. Box/index. cgi E-Utilities: Guidelines for Entrez “URL calls” used to access data. Designed for use in scripts. • Examples: ESearch, EPost, ESummary, EFetch and ELink http: //www. ncbi. nih. gov/entrez/query/static/eutils_help. html Caution: Overuse may result in blocked IPs!
![Global Entrez Search Page NCBI Field. Guide All[Filter] Global Entrez Search Page NCBI Field. Guide All[Filter]](https://present5.com/presentation/e49bda1731c8d4fca1dd792ecb495836/image-9.jpg)
Global Entrez Search Page NCBI Field. Guide All[Filter]

• • • A system of 31 linked databases A text search engine A tool for finding biologically linked data A retrieval engine A virtual workspace for manipulating large datasets NCBI Field. Guide What is Entrez?
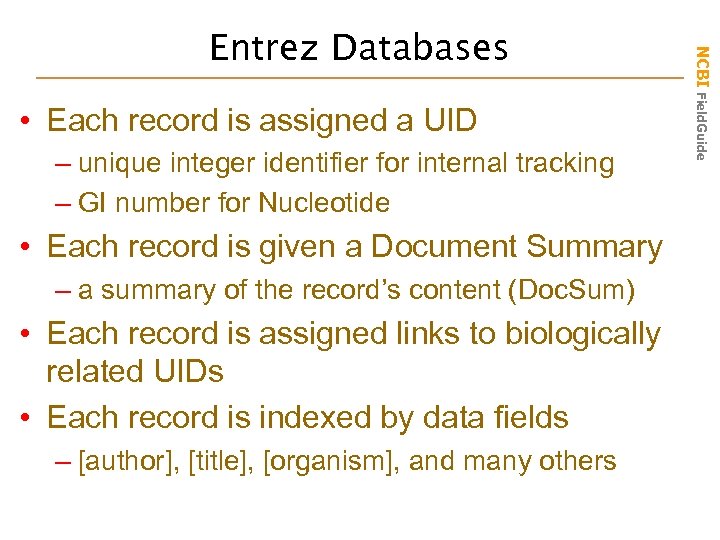
• Each record is assigned a UID – unique integer identifier for internal tracking – GI number for Nucleotide • Each record is given a Document Summary – a summary of the record’s content (Doc. Sum) • Each record is assigned links to biologically related UIDs • Each record is indexed by data fields – [author], [title], [organism], and many others NCBI Field. Guide Entrez Databases
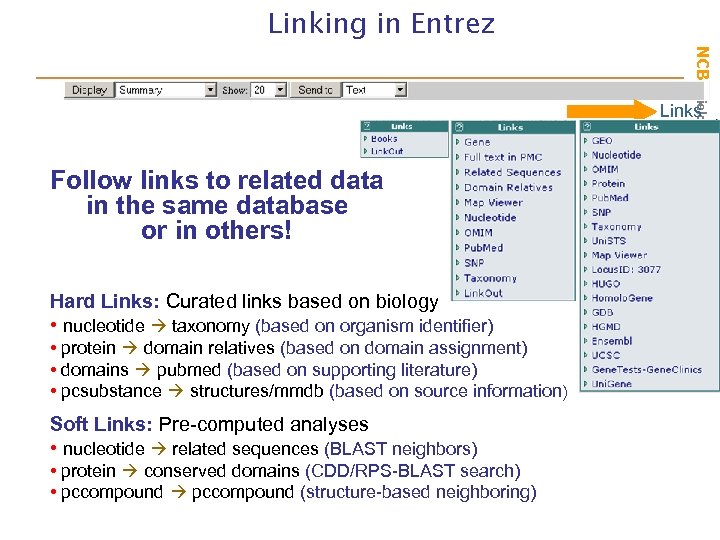
NCBI Field. Guide Linking in Entrez Links Follow links to related data in the same database or in others! Hard Links: Curated links based on biology • nucleotide taxonomy (based on organism identifier) • protein domain relatives (based on domain assignment) • domains pubmed (based on supporting literature) • pcsubstance structures/mmdb (based on source information) Soft Links: Pre-computed analyses • nucleotide related sequences (BLAST neighbors) • protein conserved domains (CDD/RPS-BLAST search) • pccompound (structure-based neighboring)
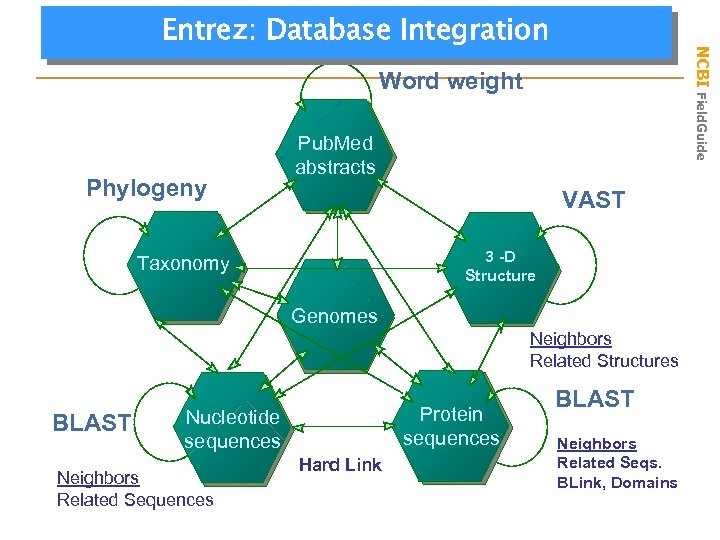
NCBI Field. Guide Entrez: Database Integration Word weight Phylogeny Pub. Med abstracts VAST 3 -D 3 -D Structure Taxonomy Genomes Neighbors Related Structures BLAST Protein sequences Nucleotide sequences Neighbors Related Sequences Hard Link BLAST Neighbors Related Seqs. BLink, Domains
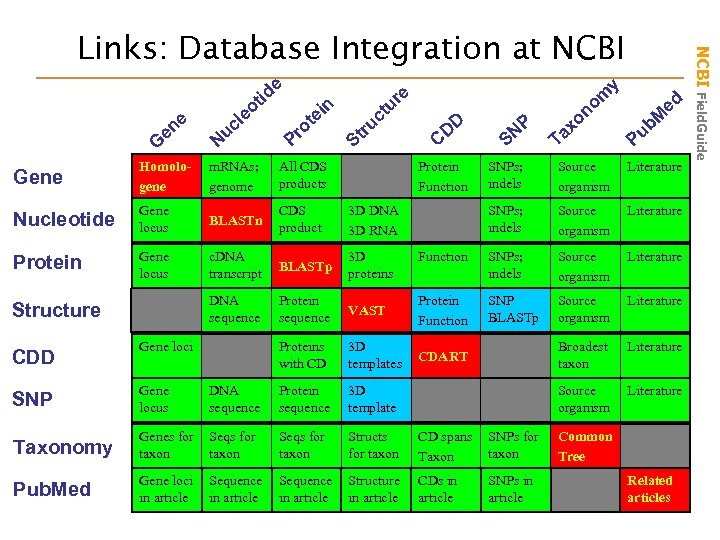
e ne e G tid eo l uc N n ei t ro P u ct u tr S Gene Homologene m. RNAs; genome All CDS products Nucleotide Gene locus BLASTn CDS product Protein Gene locus c. DNA transcript BLASTp 3 D proteins DNA sequence Protein sequence D D C 3 D DNA 3 D RNA Structure CDD y m o re SN ax T b. M u P SNPs; indels Source organism Literature Function SNPs; indels Source organism Literature VAST Protein Function SNP BLASTp Source organism Literature Proteins with CD Gene loci Protein Function P ed on 3 D templates CDART Broadest taxon Literature Source organism Literature SNP Gene locus DNA sequence Protein sequence 3 D template Taxonomy Genes for taxon Seqs for taxon Structs for taxon CD spans Taxon SNPs for taxon Pub. Med Gene loci in article Sequence in article Structure in article CDs in article SNPs in article Common Tree Related articles NCBI Field. Guide Links: Database Integration at NCBI
• Primary Databases – Original submissions by experimentalists – Content controlled by the submitter • Examples: Gen. Bank, db. SNP, GEO, Pub. Chem Substance and Pub. Chem Bioassays • Derivative Databases – Built from primary data – Content controlled by third party (NCBI) • Examples: Refseq, Ref. SNP, GEO Datasets, Pub. Chem Compound NCBI Field. Guide Types of Databases
• Gen. Bank: Primary Data (98. 2%) – original submissions by experimentalists – submitters retain editorial control of records – archival in nature • Ref. Seq: Derivative Data (1. 8%) – curated by NCBI staff – NCBI retains editorial control of records – record content is updated continually NCBI Field. Guide An Entrez Database - Nucleotide

NCBI Field. Guide Literature Databases

Books NCBI Field. Guide NM_000249: Pub. Med
NCBI Field. Guide Books Link

NCBI Field. Guide

Part 2. Data Flow and Processing Part 3. Querying and Linking the Data Part 4. User Support A part of the NCBI Bookshelf NCBI Field. Guide Part 1. The Databases

NCBI Field. Guide
NCBI Field. Guide

NCBI Field. Guide
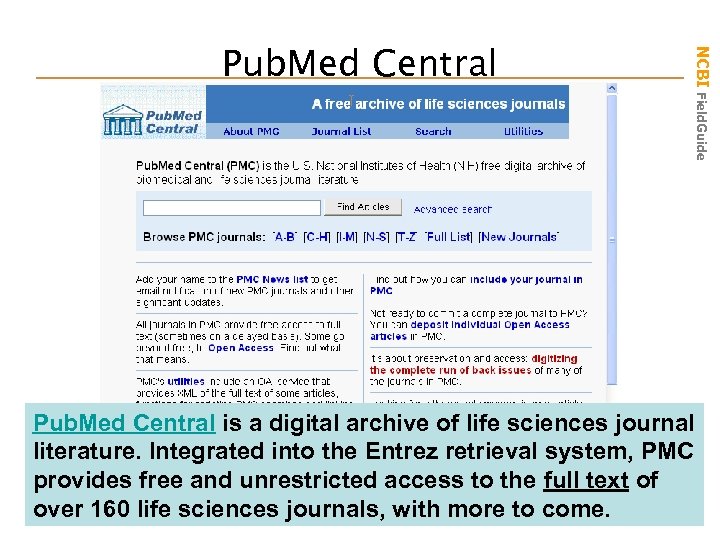
NCBI Field. Guide Pub. Med Central is a digital archive of life sciences journal literature. Integrated into the Entrez retrieval system, PMC provides free and unrestricted access to the full text of over 160 life sciences journals, with more to come.

Detailed journal information NCBI Field. Guide NCBI Journal Database
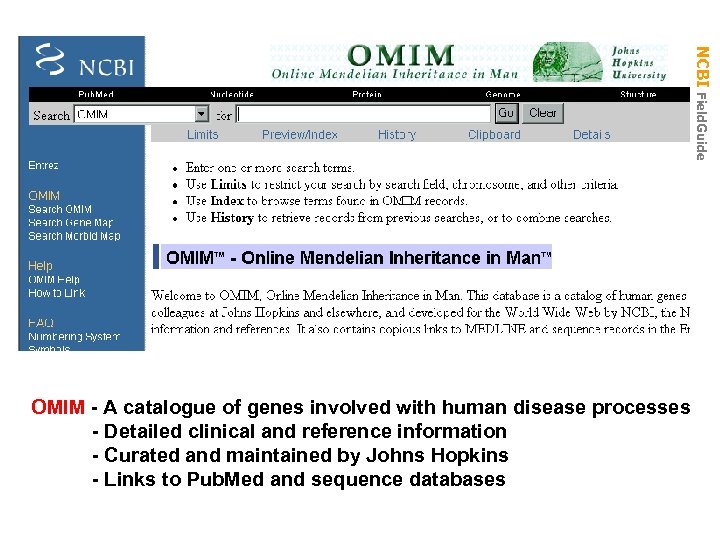
NCBI Field. Guide OMIM - A catalogue of genes involved with human disease processes - Detailed clinical and reference information - Curated and maintained by Johns Hopkins - Links to Pub. Med and sequence databases
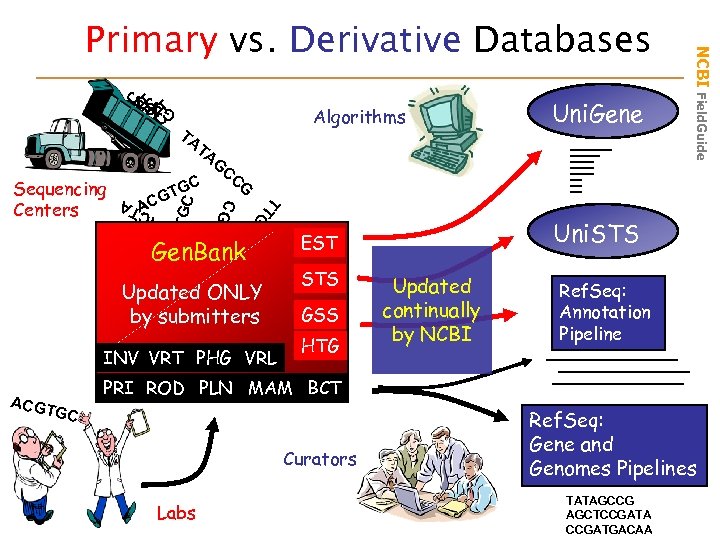
C TC T ATC TC A Algorithms TA Uni. Gene GAGAG G A A TA TGC AT T A AC T G ACG T TG A CA C GTG A Sequencing Centers G CC G GC GT AC Gen. Bank Updated ONLY by submitters INV VRT PHG VRL ACGT GC Uni. STS EST STS GSS HTG NCBI Field. Guide Primary vs. Derivative Databases Updated continually by NCBI Ref. Seq: Annotation Pipeline PRI ROD PLN MAM BCT Curators Labs Ref. Seq: Gene and Genomes Pipelines TATAGCCG AGCTCCGATA CCGATGACAA
NCBI’s Primary Sequence Database • • Nucleotide only sequence database Archival in nature Each record is assigned a stable accession number Gen. Bank Data – Direct submissions (traditional records ) – Batch submissions (EST, GSS, STS) – ftp accounts (genome data) • Three collaborating databases – Gen. Bank – DNA Database of Japan (DDBJ) – European Molecular Biology Laboratory (EMBL) Database NCBI Field. Guide What is Gen. Bank?
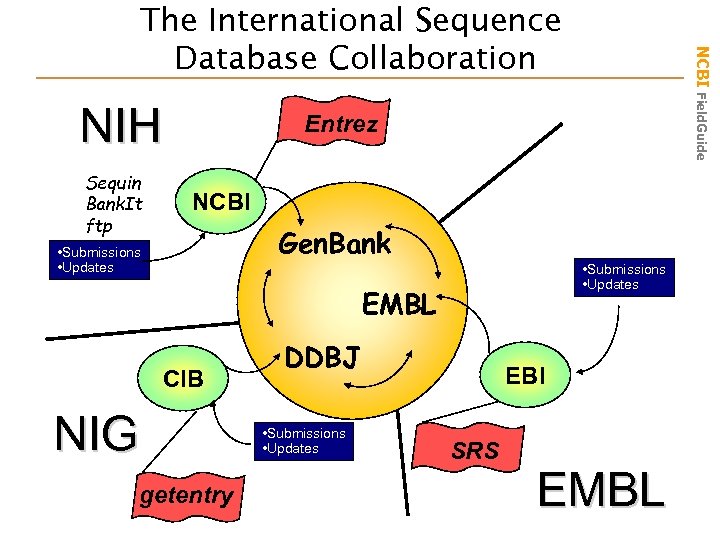
NIH Sequin Bank. It ftp NCBI Field. Guide The International Sequence Database Collaboration Entrez NCBI Gen. Bank • Submissions • Updates EMBL CIB NIG getentry DDBJ • Submissions • Updates EBI SRS EMBL
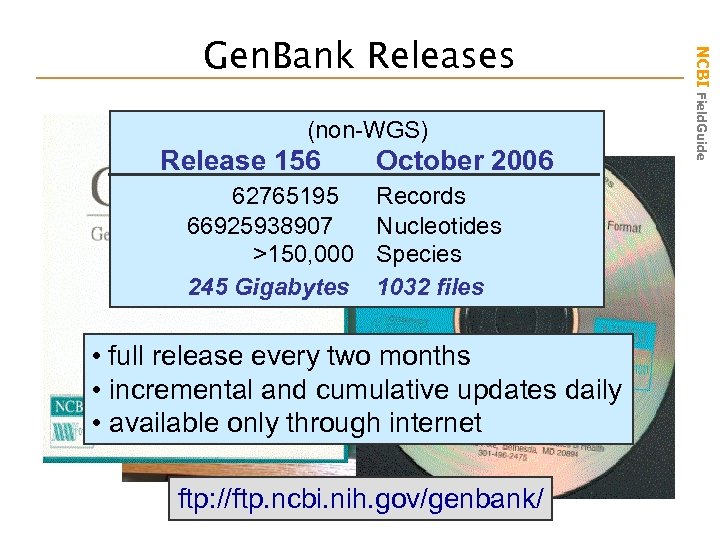
(non-WGS) Release 156 October 2006 62765195 Records 66925938907 Nucleotides >150, 000 Species 245 Gigabytes 1032 files • full release every two months • incremental and cumulative updates daily • available only through internet ftp: //ftp. ncbi. nih. gov/genbank/ NCBI Field. Guide Gen. Bank Releases
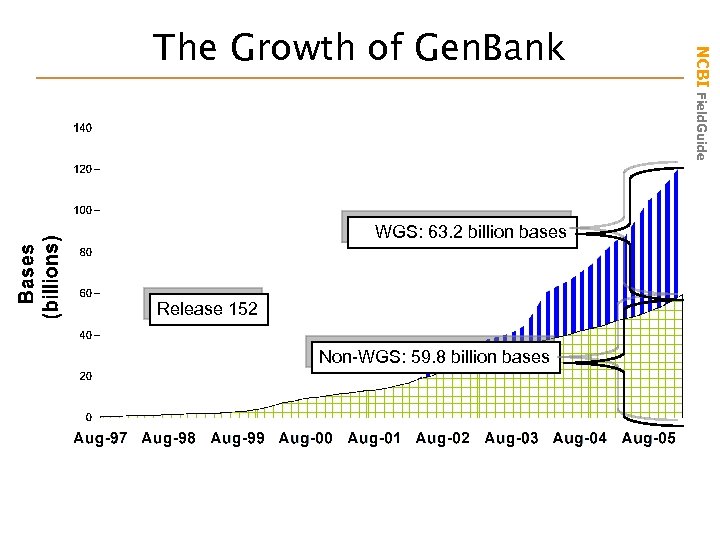
WGS: 63. 2 billion bases Release 152 Non-WGS: 59. 8 billion bases NCBI Field. Guide The Growth of Gen. Bank
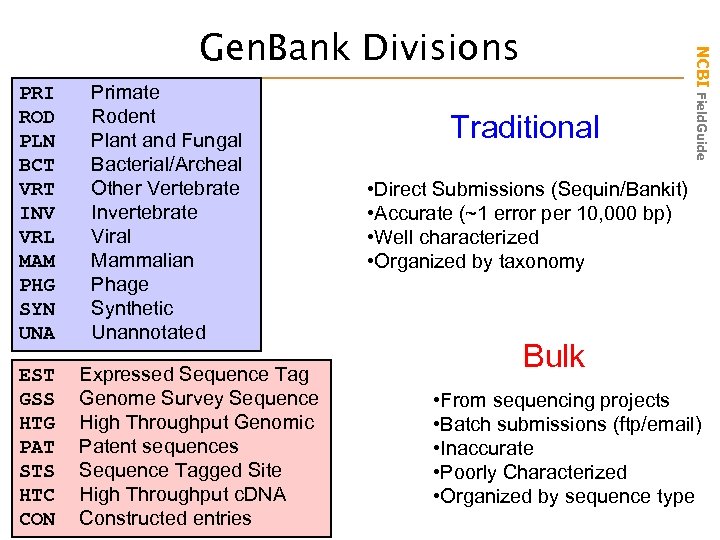
PRI Primate ROD Rodent PLN Plant and Fungal BCT Bacterial/Archeal VRT Other Vertebrate INV Invertebrate VRL Viral MAM Mammalian PHG Phage SYN Synthetic UNA Unannotated EST Expressed Sequence Tag GSS Genome Survey Sequence HTG High Throughput Genomic PAT Patent sequences STS Sequence Tagged Site HTC High Throughput c. DNA CON Constructed entries Traditional NCBI Field. Guide Gen. Bank Divisions • Direct Submissions (Sequin/Bankit) • Accurate (~1 error per 10, 000 bp) • Well characterized • Organized by taxonomy Bulk • From sequencing projects • Batch submissions (ftp/email) • Inaccurate • Poorly Characterized • Organized by sequence type
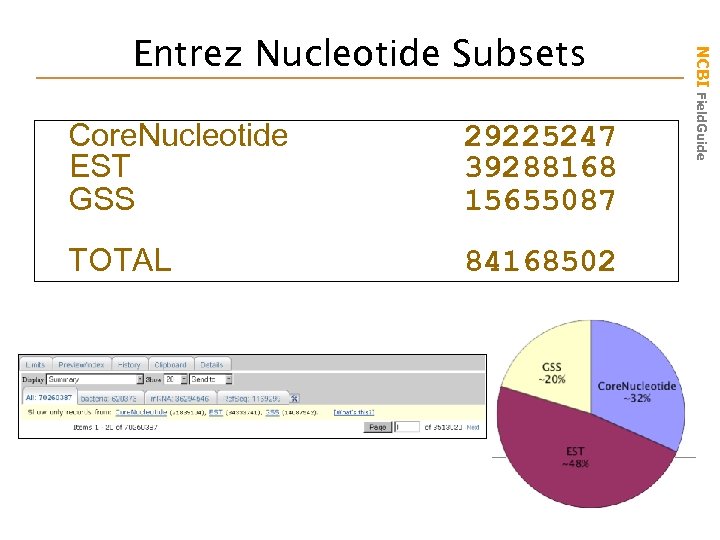
Core. Nucleotide EST GSS 29225247 39288168 15655087 TOTAL 84168502 NCBI Field. Guide Entrez Nucleotide Subsets
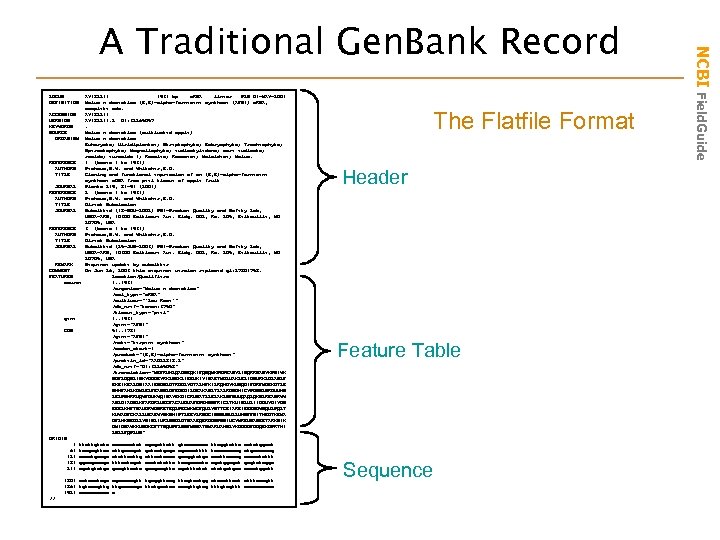
LOCUS AY 182241 1931 bp m. RNA linear PLN 04 -MAY-2004 DEFINITION Malus x domestica (E, E)-alpha-farnesene synthase (AFS 1) m. RNA, complete cds. ACCESSION AY 182241 VERSION AY 182241. 2 GI: 32265057 KEYWORDS . SOURCE Malus x domestica (cultivated apple) ORGANISM Malus x domestica Eukaryota; Viridiplantae; Streptophyta; Embryophyta; Tracheophyta; Spermatophyta; Magnoliophyta; eudicotyledons; core eudicots; rosids; eurosids I; Rosales; Rosaceae; Maloideae; Malus. REFERENCE 1 (bases 1 to 1931) AUTHORS Pechous, S. W. and Whitaker, B. D. TITLE Cloning and functional expression of an (E, E)-alpha-farnesene synthase c. DNA from peel tissue of apple fruit JOURNAL Planta 219, 84 -94 (2004) REFERENCE 2 (bases 1 to 1931) AUTHORS Pechous, S. W. and Whitaker, B. D. TITLE Direct Submission JOURNAL Submitted (18 -NOV-2002) PSI-Produce Quality and Safety Lab, USDA-ARS, 10300 Baltimore Ave. Bldg. 002, Rm. 205, Beltsville, MD 20705, USA REFERENCE 3 (bases 1 to 1931) AUTHORS Pechous, S. W. and Whitaker, B. D. TITLE Direct Submission JOURNAL Submitted (25 -JUN-2003) PSI-Produce Quality and Safety Lab, USDA-ARS, 10300 Baltimore Ave. Bldg. 002, Rm. 205, Beltsville, MD 20705, USA REMARK Sequence update by submitter COMMENT On Jun 26, 2003 this sequence version replaced gi: 27804758. FEATURES Location/Qualifiers source 1. . 1931 /organism="Malus x domestica" /mol_type="m. RNA" /cultivar="'Law Rome'" /db_xref="taxon: 3750" /tissue_type="peel" gene 1. . 1931 /gene="AFS 1" CDS 54. . 1784 /gene="AFS 1" /note="terpene synthase" /codon_start=1 /product="(E, E)-alpha-farnesene synthase" /protein_id="AAO 22848. 2" /db_xref="GI: 32265058" /translation="MEFRVHLQADNEQKIFQNQMKPEPEASYLINQRRSANYKPNIWK NDFLDQSLISKYDGDEYRKLSEKLIEEVKIYISAETMDLVAKLELIDSVRKLGLANLF EKEIKEALDSIAAIESDNLGTRDDLYGTALHFKILRQHGYKVSQDIFGRFMDEKGTLE NHHFAHLKGMLELFEASNLGFEGEDILDEAKASLTLALRDSGHICYPDSNLSRDVVHS LELPSHRRVQWFDVKWQINAYEKDICRVNATLLELAKLNFNVVQAQLQKNLREASRWW ANLGIADNLKFARDRLVECFACAVGVAFEPEHSSFRICLTKVINLVLIIDDVYDIYGS EEELKHFTNAVDRWDSRETEQLPECMKMCFQVLYNTTCEIAREIEEENGWNQVLPQLT KVWADFCKALLVEAEWYNKSHIPTLEEYLRNGCISSSVSVLLVHSFFSITHEGTKEMA DFLHKNEDLLYNISLIVRLNNDLGTSAAEQERGDSPSSIVCYMREVNASEETARKNIK GMIDNAWKKVNGKCFTTNQVPFLSSFMNNATNMARVAHSLYKDGDGFGDQEKGPRTHI LSLLFQPLVN" ORIGIN 1 ttcttgtatc ccaaacatct cgagcttctt gtacaccaaa ttaggtattc actatggaat 61 tcagagttca cttgcaagct gataatgagc agaaaatttt tcaaaaccag atgaaacccg 121 aacctgaagc ctcttacttg attaatcaaa gacggtctgc aaattacaag ccaaatattt 181 ggaagaacga tttcctagat caatctctta tcagcaaata cgatggagat gagtatcgga 241 agctgtctga gaagttaata gaagaagtta agatttatat atctgctgaa acaatggatt 1801 aatagc agcaaaagtt tgcggttcag ttcgtcatgg ataaattaat ctttacagtt 1861 tgtaacgttg ttgccaaaga ttatgaataa aaagttgtag tttgtcgttt aaaaa 1921 aaaaa a // The Flatfile Format Header Feature Table Sequence NCBI Field. Guide A Traditional Gen. Bank Record
![Indexing for Nucleotide UID 4680720 Field Indexed Terms [primary accession] [title] [organism] [sequence length] Indexing for Nucleotide UID 4680720 Field Indexed Terms [primary accession] [title] [organism] [sequence length]](https://present5.com/presentation/e49bda1731c8d4fca1dd792ecb495836/image-36.jpg)
Indexing for Nucleotide UID 4680720 Field Indexed Terms [primary accession] [title] [organism] [sequence length] [modification date] [properties] M 17755 Homo sapiens thyroid peroxidase (TPO) m. RNA… Homo sapiens 3060 1999/04/26 biomol mrna gbdiv pri srcdb genbank NCBI Field. Guide An Example Record – M 17755
![NCBI Field. Guide M 17755: Feature Table TPO [gene name] CDS position in bp NCBI Field. Guide M 17755: Feature Table TPO [gene name] CDS position in bp](https://present5.com/presentation/e49bda1731c8d4fca1dd792ecb495836/image-37.jpg)
NCBI Field. Guide M 17755: Feature Table TPO [gene name] CDS position in bp thyroiditis [text word] thyroid peroxidase [protein name] protein accession
The sequence itself is not indexed… Use BLAST for that! NCBI Field. Guide Sequence: 99. 99% Accurate
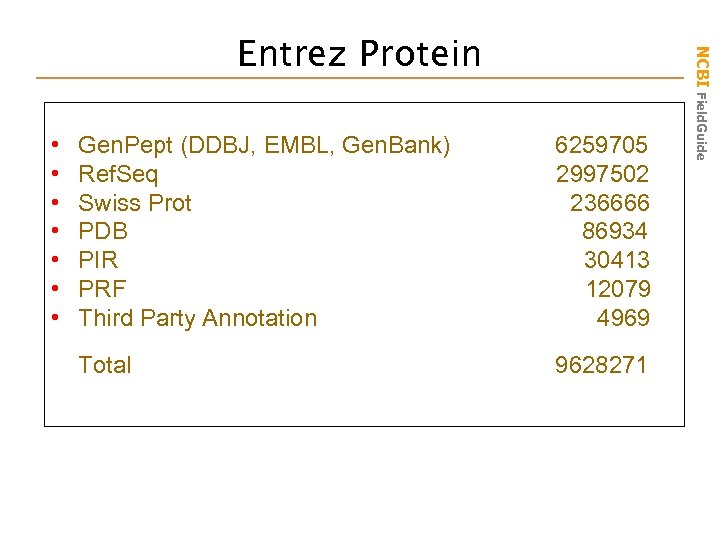
• Gen. Pept (DDBJ, EMBL, Gen. Bank) 6259705 • Ref. Seq 2997502 • Swiss Prot 236666 • PDB 86934 • PIR 30413 • PRF 12079 • Third Party Annotation 4969 Total 9628271 NCBI Field. Guide Entrez Protein
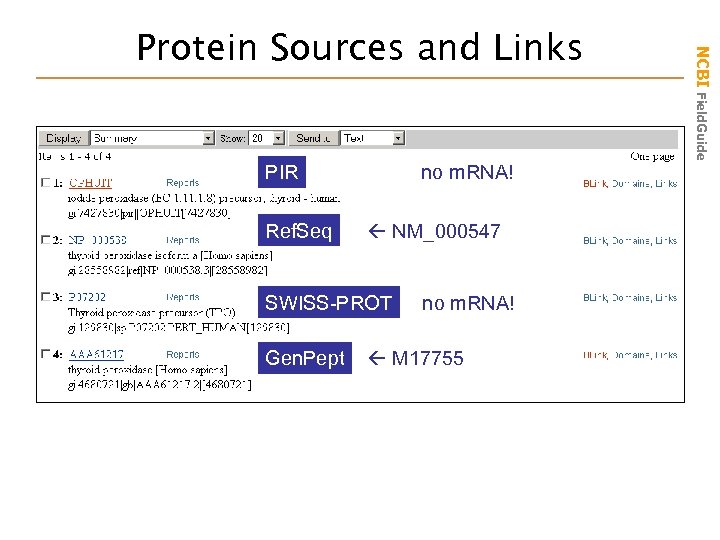
PIR Ref. Seq no m. RNA! NM_000547 SWISS-PROT Gen. Pept no m. RNA! M 17755 NCBI Field. Guide Protein Sources and Links
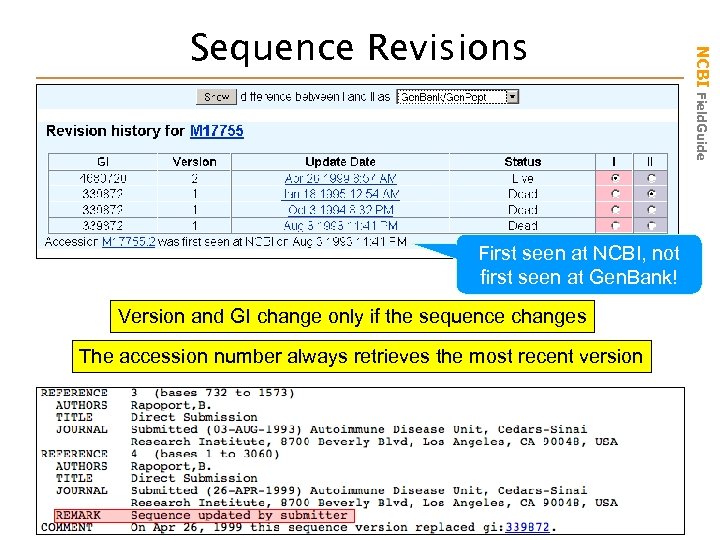
First seen at NCBI, not first seen at Gen. Bank! Version and GI change only if the sequence changes The accession number always retrieves the most recent version NCBI Field. Guide Sequence Revisions
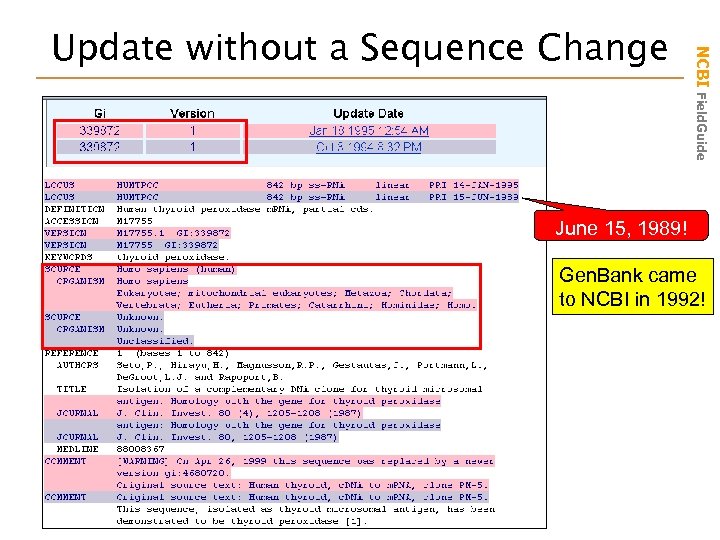
NCBI Field. Guide Update without a Sequence Change June 15, 1989! Gen. Bank came to NCBI in 1992!
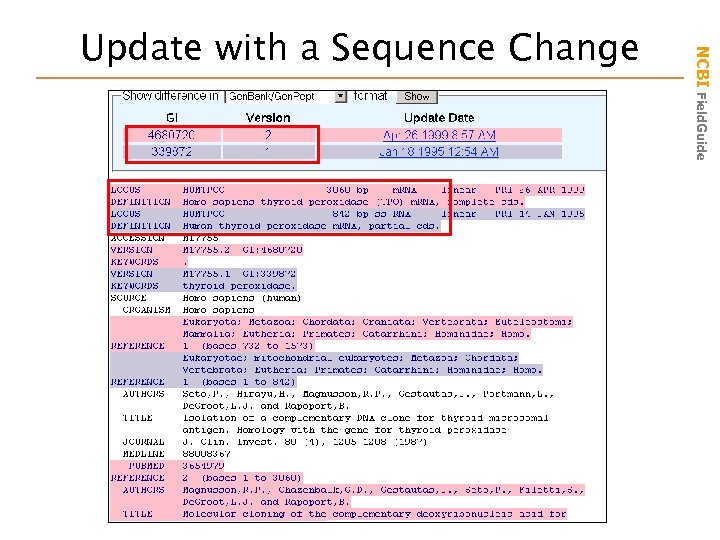
NCBI Field. Guide Update with a Sequence Change
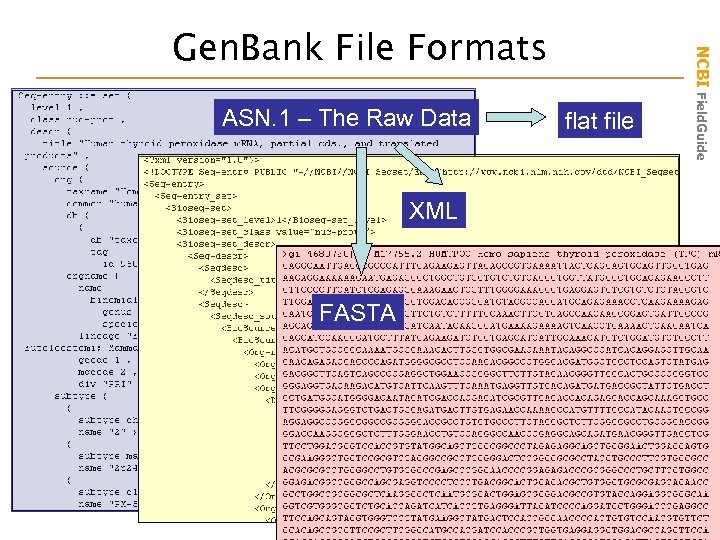
ASN. 1 – The Raw Data XML FASTA flat file NCBI Field. Guide Gen. Bank File Formats
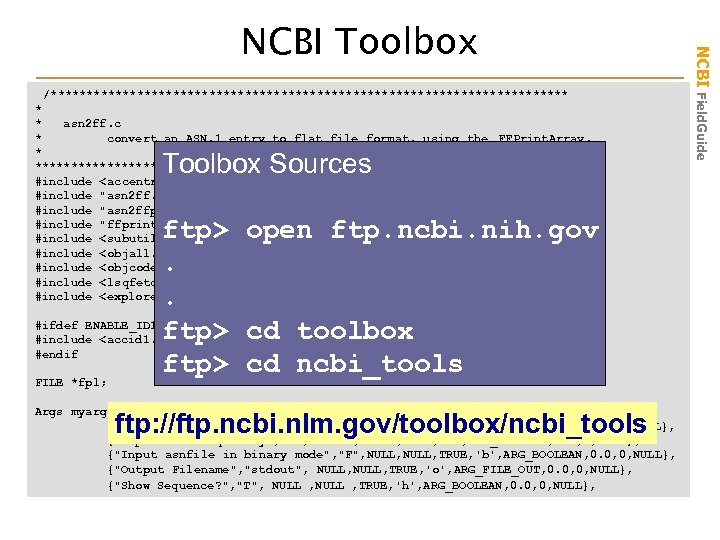
/************************************ * * asn 2 ff. c * convert an ASN. 1 entry to flat file format, using the FFPrint. Array. * *************************************/ #include <accentr. h> #include "asn 2 ff. h" #include "asn 2 ffp. h" #include "ffprint. h" #include <subutil. h> #include <objall. h> #include <objcode. h> #include <lsqfetch. h> #include <explore. h> Toolbox Sources ftp> open ftp. ncbi. nih. gov. . #ifdef ENABLE_ID 1 ftp> cd toolbox #include <accid 1. h> #endif ftp> cd ncbi_tools FILE *fpl; Args myargs[] = { {"Filename for asn. 1 input", "stdin", NULL, TRUE, 'a', ARG_FILE_IN, 0. 0, 0, NULL}, {"Input is a Seq-entry", "F", NULL , TRUE, 'e', ARG_BOOLEAN, 0. 0, 0, NULL}, {"Input asnfile in binary mode", "F", NULL, TRUE, 'b', ARG_BOOLEAN, 0. 0, 0, NULL}, {"Output Filename", "stdout", NULL, TRUE, 'o', ARG_FILE_OUT, 0. 0, 0, NULL}, {"Show Sequence? ", "T", NULL , TRUE, 'h', ARG_BOOLEAN, 0. 0, 0, NULL}, ftp: //ftp. ncbi. nlm. gov/toolbox/ncbi_tools NCBI Field. Guide NCBI Toolbox
![Text Queries in Entrez term 1[limit] OP term 2[limit] OP … where limit = Text Queries in Entrez term 1[limit] OP term 2[limit] OP … where limit =](https://present5.com/presentation/e49bda1731c8d4fca1dd792ecb495836/image-46.jpg)
Text Queries in Entrez term 1[limit] OP term 2[limit] OP … where limit = Entrez indexing field (organism, author, …) OP = Boolean operator = AND, OR, NOT Wildcards: Ranges: 1: 200[MW] cancer[title] vs. cancer*[title] Complex queries: ((A[limit 1] OR B[limit 2]) AND C[limit 3]) NOT D[limit 4] NCBI Field. Guide term 1 term 2
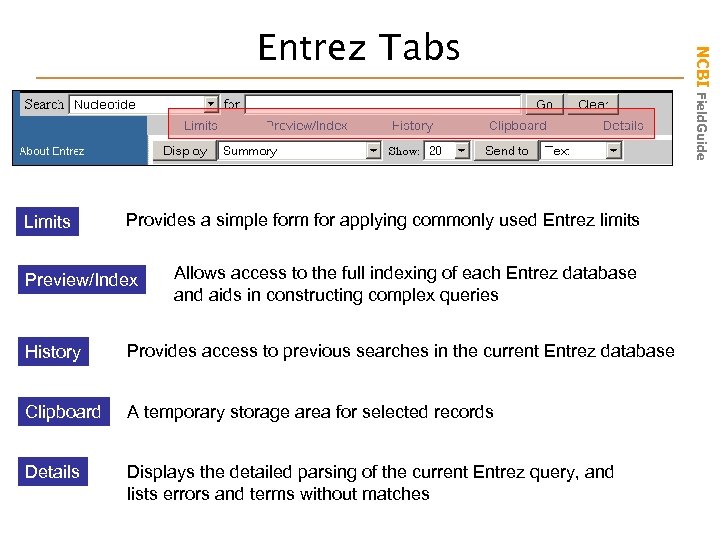
Limits Provides a simple form for applying commonly used Entrez limits Preview/Index Allows access to the full indexing of each Entrez database and aids in constructing complex queries History Provides access to previous searches in the current Entrez database Clipboard A temporary storage area for selected records Details Displays the detailed parsing of the current Entrez query, and lists errors and terms without matches NCBI Field. Guide Entrez Tabs
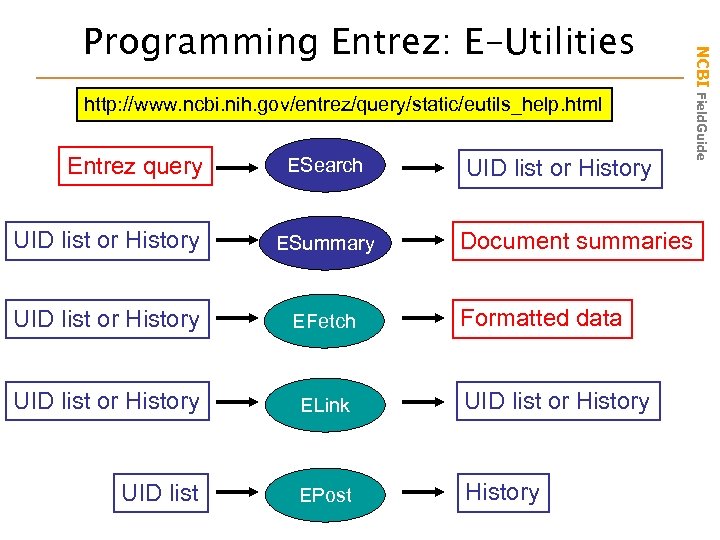
http: //www. ncbi. nih. gov/entrez/query/static/eutils_help. html Entrez query ESearch UID list or History ESummary UID list or History EFetch UID list or History ELink UID list or History UID list EPost History Document summaries Formatted data NCBI Field. Guide Programming Entrez: E-Utilities
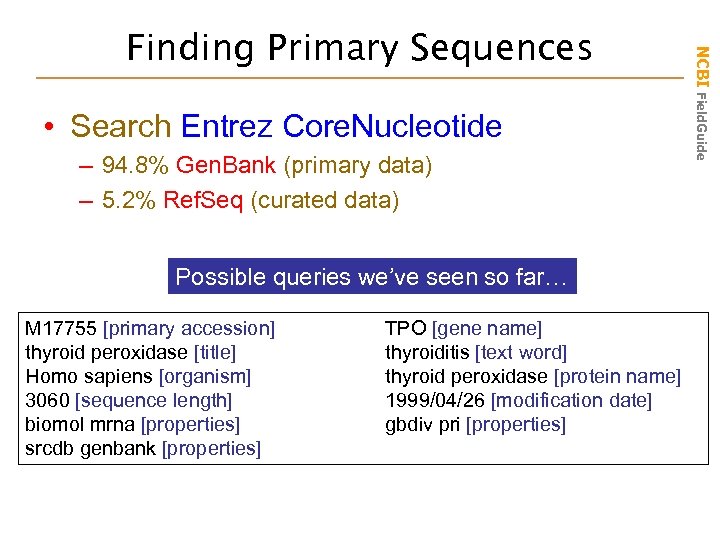
• Search Entrez Core. Nucleotide – 94. 8% Gen. Bank (primary data) – 5. 2% Ref. Seq (curated data) Possible queries we’ve seen so far… M 17755 [primary accession] thyroid peroxidase [title] Homo sapiens [organism] 3060 [sequence length] biomol mrna [properties] srcdb genbank [properties] TPO [gene name] thyroiditis [text word] thyroid peroxidase [protein name] 1999/04/26 [modification date] gbdiv pri [properties] NCBI Field. Guide Finding Primary Sequences
![Find nucleotide records for human thyroid peroxidase 276 records (("Homo sapiens“[Organism] OR human[All Fields]) Find nucleotide records for human thyroid peroxidase 276 records (("Homo sapiens“[Organism] OR human[All Fields])](https://present5.com/presentation/e49bda1731c8d4fca1dd792ecb495836/image-50.jpg)
Find nucleotide records for human thyroid peroxidase 276 records (("Homo sapiens“[Organism] OR human[All Fields]) AND thyroid peroxidase[All Fields]) Field Limit! human[organism] AND thyroid peroxidase 262 records ("Homo sapiens“[Organism] AND thyroid peroxidase[All Fields]) 14 records aren’t human sequences!! NCBI Field. Guide A Starting Query
![Entrez Nucleotide Gen. Bank Ref. Seq srcdb ddbj/embl/genbank[properties] NCBI Field. Guide Limit by Title Entrez Nucleotide Gen. Bank Ref. Seq srcdb ddbj/embl/genbank[properties] NCBI Field. Guide Limit by Title](https://present5.com/presentation/e49bda1731c8d4fca1dd792ecb495836/image-51.jpg)
Entrez Nucleotide Gen. Bank Ref. Seq srcdb ddbj/embl/genbank[properties] NCBI Field. Guide Limit by Title and Database srcdb refseq[properties] #1: thyroid peroxidase AND human[orgn] #2: thyroid peroxidase[title] AND human[orgn] #3: #2 AND srcdb refseq[properties] #4: #2 AND srcdb ddbj/embl/genbank[properties] primary data 262 55 5 50
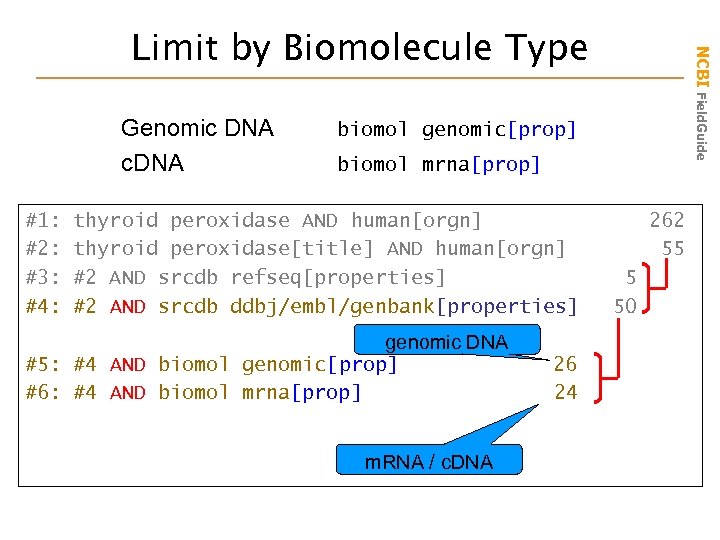
Genomic DNA c. DNA #1: #2: #3: #4: NCBI Field. Guide Limit by Biomolecule Type biomol genomic[prop] biomol mrna[prop] thyroid peroxidase AND human[orgn] thyroid peroxidase[title] AND human[orgn] #2 AND srcdb refseq[properties] #2 AND srcdb ddbj/embl/genbank[properties] genomic DNA #5: #4 AND biomol genomic[prop] #6: #4 AND biomol mrna[prop] m. RNA / c. DNA 26 24 262 55 5 50
![thyroid peroxidase[protein name] AND human[orgn] AND gbdiv pri[prop] AND biomol mrna[prop] 24 records [title] thyroid peroxidase[protein name] AND human[orgn] AND gbdiv pri[prop] AND biomol mrna[prop] 24 records [title]](https://present5.com/presentation/e49bda1731c8d4fca1dd792ecb495836/image-53.jpg)
thyroid peroxidase[protein name] AND human[orgn] AND gbdiv pri[prop] AND biomol mrna[prop] 24 records [title] 5 records [protein name] NCBI Field. Guide Limit by Protein Name
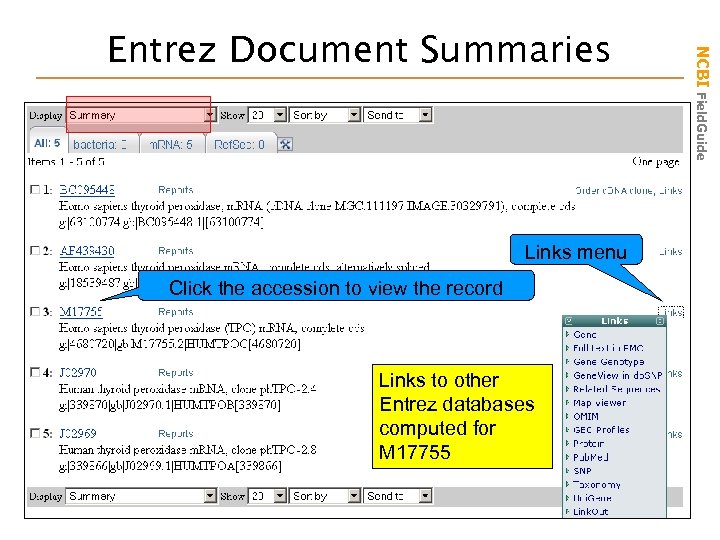
Links menu Click the accession to view the record Links to other Entrez databases computed for M 17755 NCBI Field. Guide Entrez Document Summaries
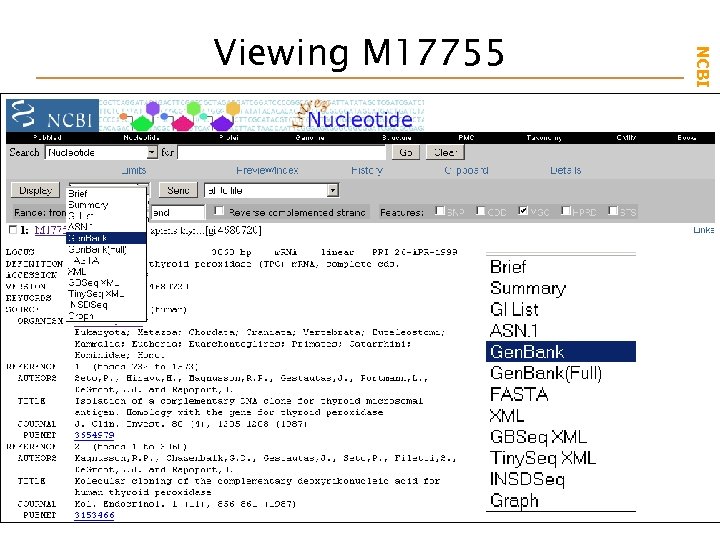
NCBI Field. Guide Viewing M 17755
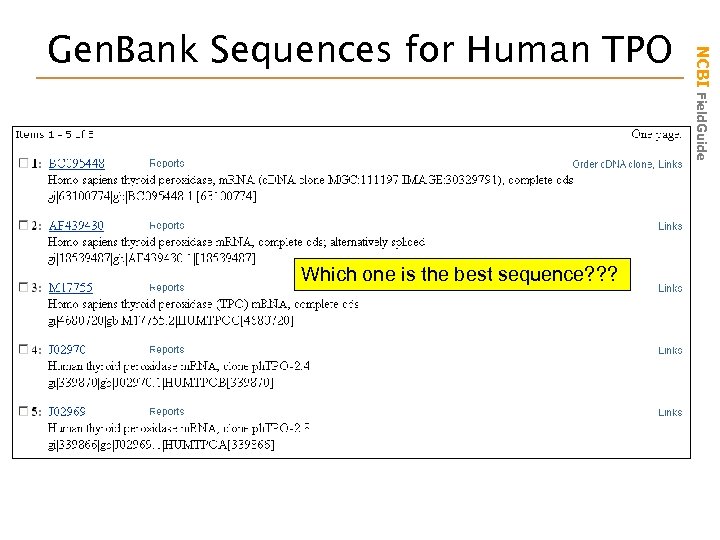
Which one is the best sequence? ? ? NCBI Field. Guide Gen. Bank Sequences for Human TPO
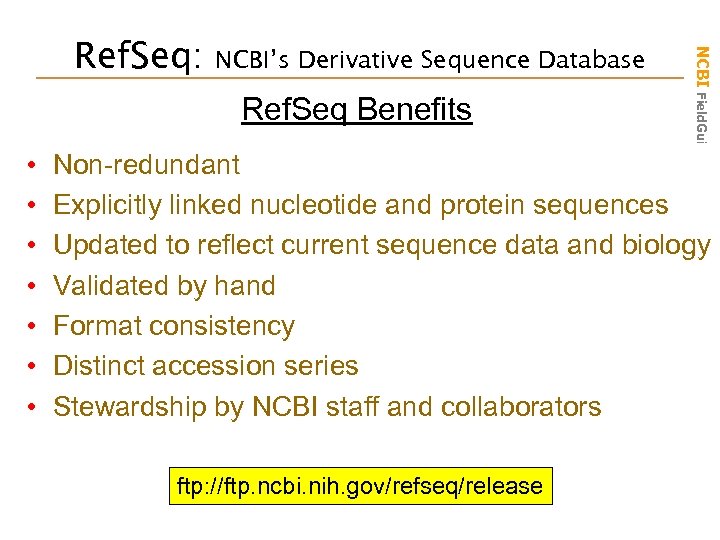
NCBI’s Derivative Sequence Database Ref. Seq Benefits • • NCBI Field. Guide Ref. Seq: Non-redundant Explicitly linked nucleotide and protein sequences Updated to reflect current sequence data and biology Validated by hand Format consistency Distinct accession series Stewardship by NCBI staff and collaborators ftp: //ftp. ncbi. nih. gov/refseq/release
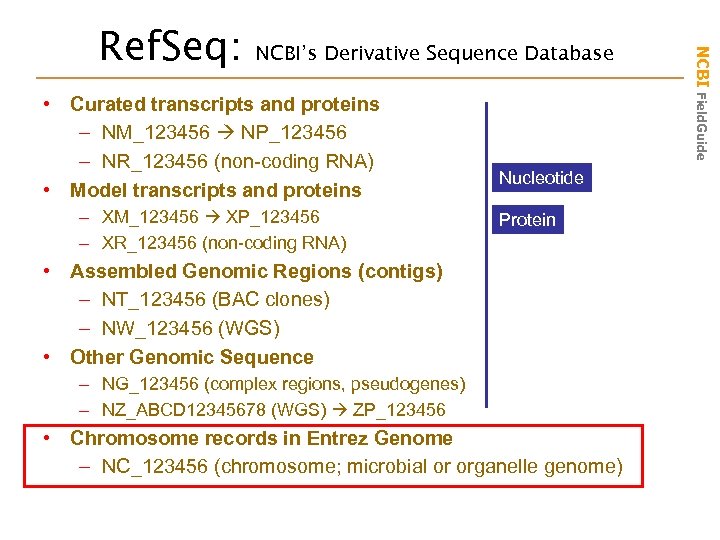
NCBI’s Derivative Sequence Database • Curated transcripts and proteins – NM_123456 NP_123456 – NR_123456 (non-coding RNA) • Model transcripts and proteins – XM_123456 XP_123456 – XR_123456 (non-coding RNA) Nucleotide Protein • Assembled Genomic Regions (contigs) – NT_123456 (BAC clones) – NW_123456 (WGS) • Other Genomic Sequence – NG_123456 (complex regions, pseudogenes) – NZ_ABCD 12345678 (WGS) ZP_123456 • Chromosome records in Entrez Genome – NC_123456 (chromosome; microbial or organelle genome) NCBI Field. Guide Ref. Seq:
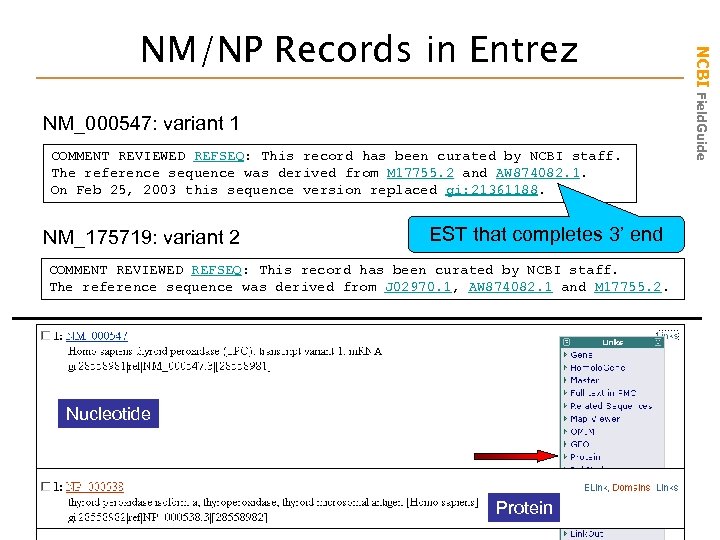
NM_000547: variant 1 COMMENT REVIEWED REFSEQ: This record has been curated by NCBI staff. The reference sequence was derived from M 17755. 2 and AW 874082. 1. On Feb 25, 2003 this sequence version replaced gi: 21361188. NM_175719: variant 2 EST that completes 3’ end COMMENT REVIEWED REFSEQ: This record has been curated by NCBI staff. The reference sequence was derived from J 02970. 1, AW 874082. 1 and M 17755. 2. Nucleotide Protein NCBI Field. Guide NM/NP Records in Entrez
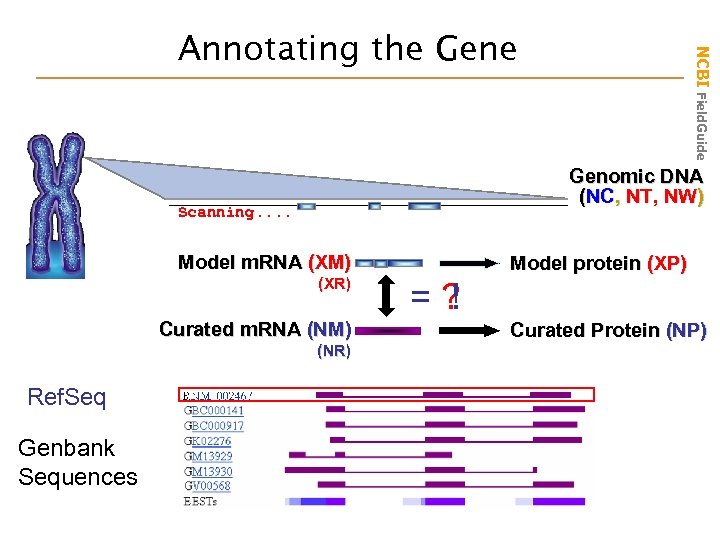
Genomic DNA (NC, NT, NW) Scanning. . Model m. RNA (XM) (XR) Curated m. RNA (NM) (NR) Ref. Seq Genbank Sequences NCBI Field. Guide Annotating the Gene Model protein (XP) = ! = ? Curated Protein (NP)
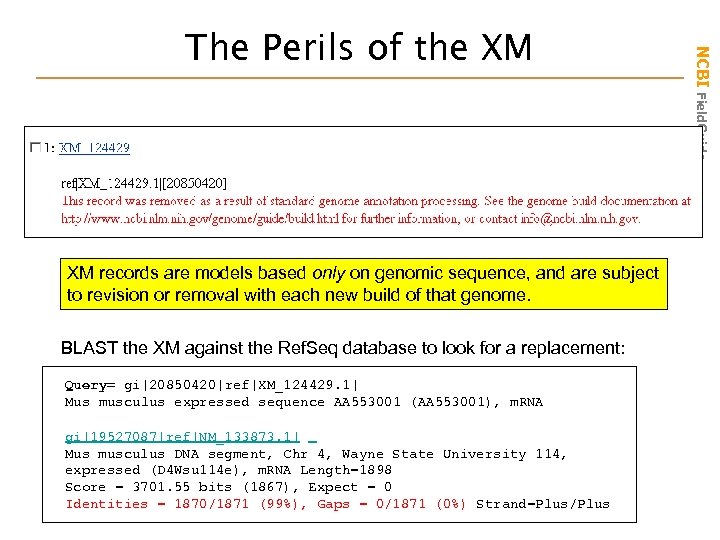
XM records are models based only on genomic sequence, and are subject to revision or removal with each new build of that genome. BLAST the XM against the Ref. Seq database to look for a replacement: Query= gi|20850420|ref|XM_124429. 1| Mus musculus expressed sequence AA 553001 (AA 553001), m. RNA gi|19527087|ref|NM_133873. 1| Mus musculus DNA segment, Chr 4, Wayne State University 114, expressed (D 4 Wsu 114 e), m. RNA Length=1898 Score = 3701. 55 bits (1867), Expect = 0 Identities = 1870/1871 (99%), Gaps = 0/1871 (0%) Strand=Plus/Plus NCBI Field. Guide The Perils of the XM
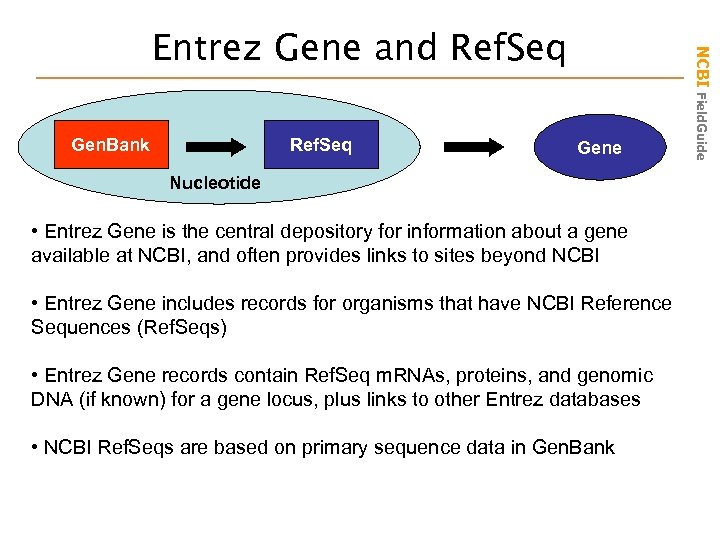
Gen. Bank Ref. Seq Gene Nucleotide • Entrez Gene is the central depository for information about a gene available at NCBI, and often provides links to sites beyond NCBI • Entrez Gene includes records for organisms that have NCBI Reference Sequences (Ref. Seqs) • Entrez Gene records contain Ref. Seq m. RNAs, proteins, and genomic DNA (if known) for a gene locus, plus links to other Entrez databases • NCBI Ref. Seqs are based on primary sequence data in Gen. Bank NCBI Field. Guide Entrez Gene and Ref. Seq
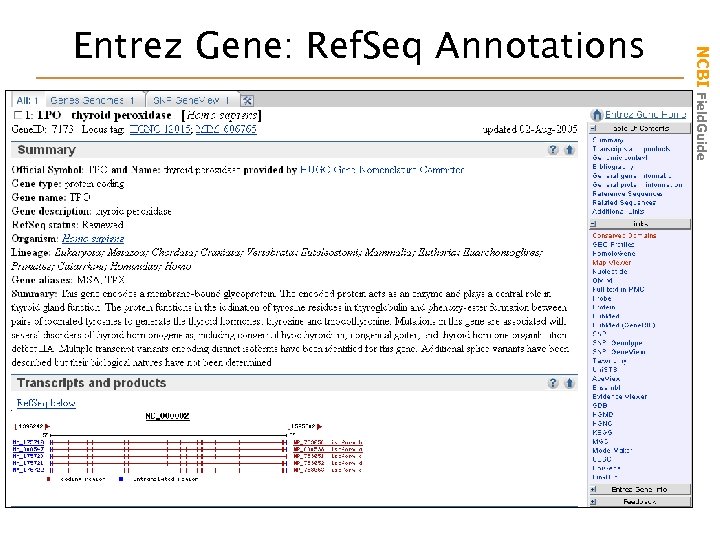
NCBI Field. Guide Entrez Gene: Ref. Seq Annotations
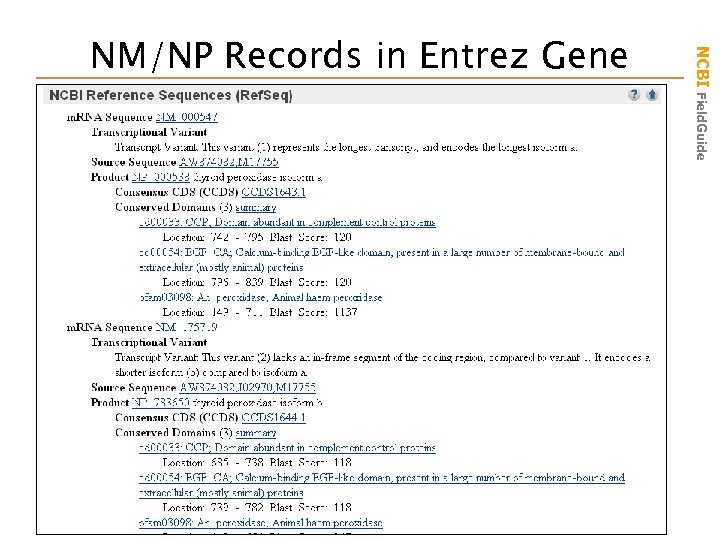
NCBI Field. Guide NM/NP Records in Entrez Gene
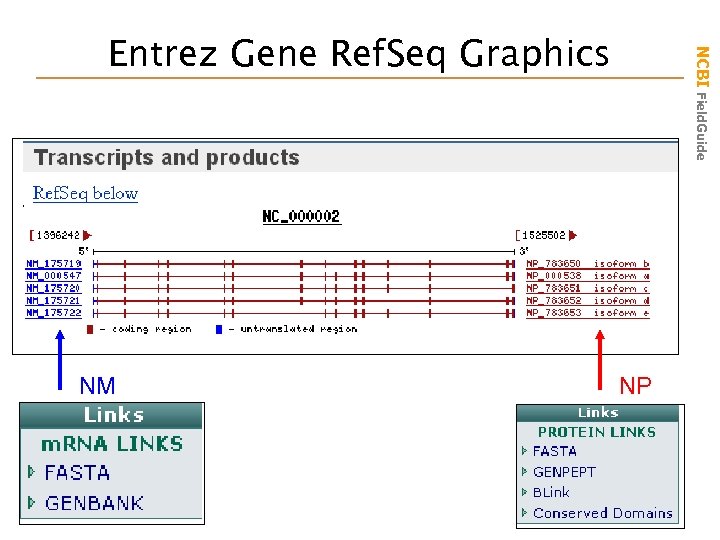
NM NCBI Field. Guide Entrez Gene Ref. Seq Graphics NP
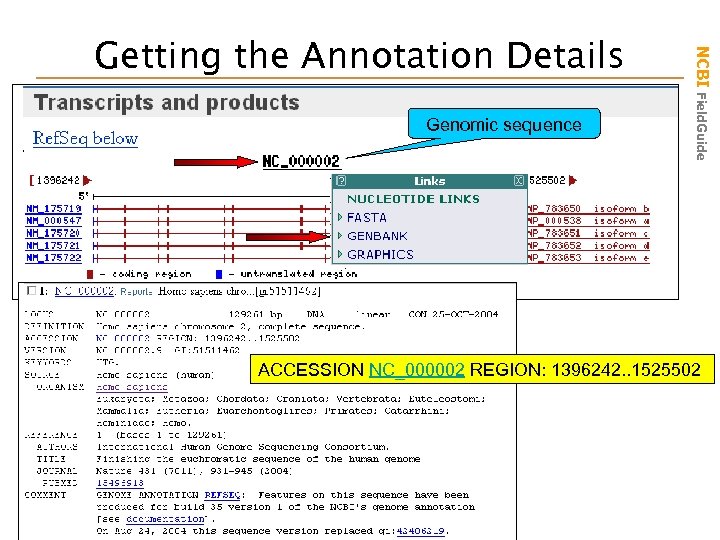
Genomic sequence NCBI Field. Guide Getting the Annotation Details ACCESSION NC_000002 REGION: 1396242. . 1525502
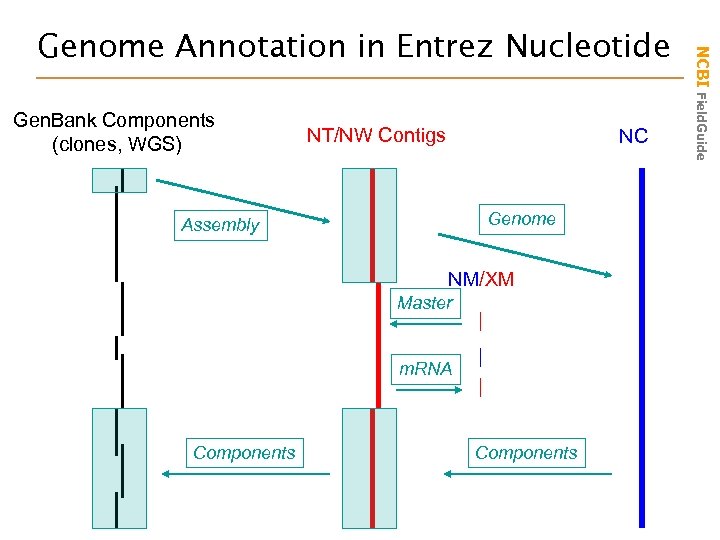
Gen. Bank Components (clones, WGS) NT/NW Contigs NC Genome Assembly NM/XM Master m. RNA Components NCBI Field. Guide Genome Annotation in Entrez Nucleotide
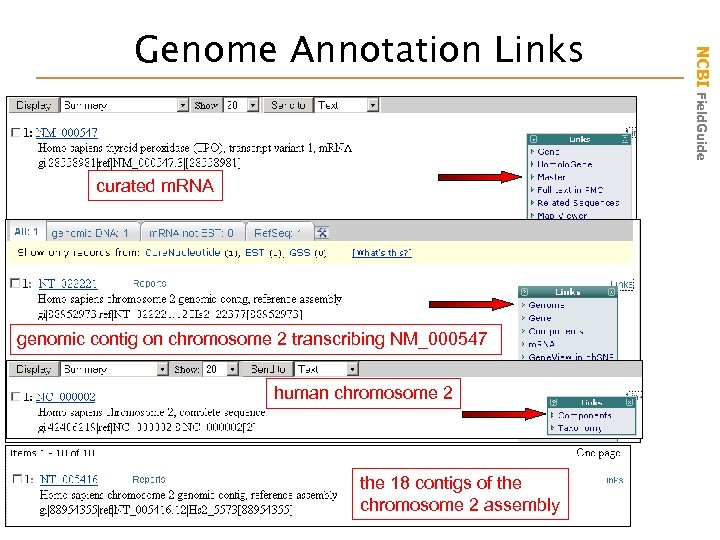
curated m. RNA genomic contig on chromosome 2 transcribing NM_000547 human chromosome 2 the 18 contigs of the chromosome 2 assembly NCBI Field. Guide Genome Annotation Links
![Gene symbol: human thyroid peroxidase (TPO) tpo [sym] AND human [organism] NCBI Field. Guide Gene symbol: human thyroid peroxidase (TPO) tpo [sym] AND human [organism] NCBI Field. Guide](https://present5.com/presentation/e49bda1731c8d4fca1dd792ecb495836/image-69.jpg)
Gene symbol: human thyroid peroxidase (TPO) tpo [sym] AND human [organism] NCBI Field. Guide Searching Entrez Gene Protein name: topoisomerase genes from Archaea topoisomerase[gene/protein name] AND archaea [organism] Chromosome and Links: genes on human chromosome 2 with OMIM links 2 [chromosome] AND gene omim [filter] AND human [organism] Ref. Seq status and variants: Reviewed Ref. Seqs with transcript variants srcdb refseq reviewed[prop] AND has transcript variants[prop] Disease and Gene Ontology: Membrane proteins linked to cancer integral to plasma membrane[gene ontology] AND cancer [dis]

NCBI now accepts the submission of new annotations of existing Gen. Bank sequences. NCBI Field. Guide Third Party Annotation (TPA) Database • Submissions must be published in a peer-reviewed journal. • Facilitates the annotation of sequences by experts. Examples of sequences appropriate for TPA are: Annotation of features on gene and/or m. RNA sequences Assembled “full length” genes and/or m. RNAs What should not be submitted to TPA? Synthetic constructs (such as cloning vectors) that use well-characterized, publicly available genes, promoters, or terminators Updates or changes to existing sequence data Sequence annotations without experimental evidence
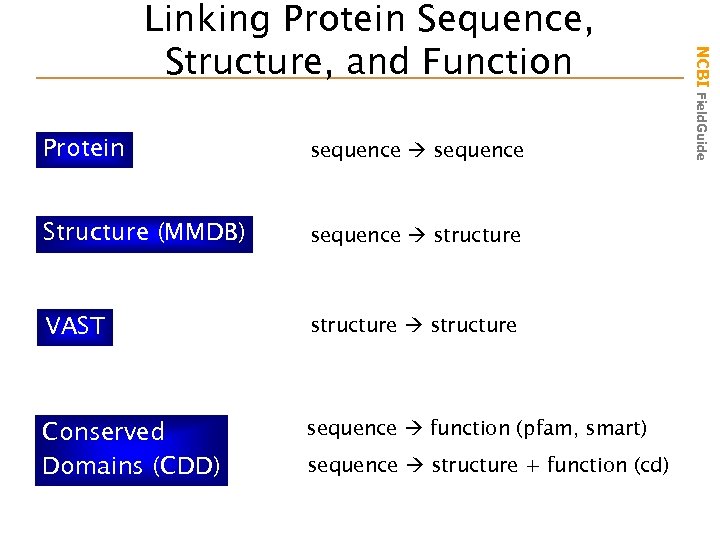
Protein sequence Structure (MMDB) sequence structure VAST structure Conserved Domains (CDD) sequence function (pfam, smart) sequence structure + function (cd) NCBI Field. Guide Linking Protein Sequence, Structure, and Function
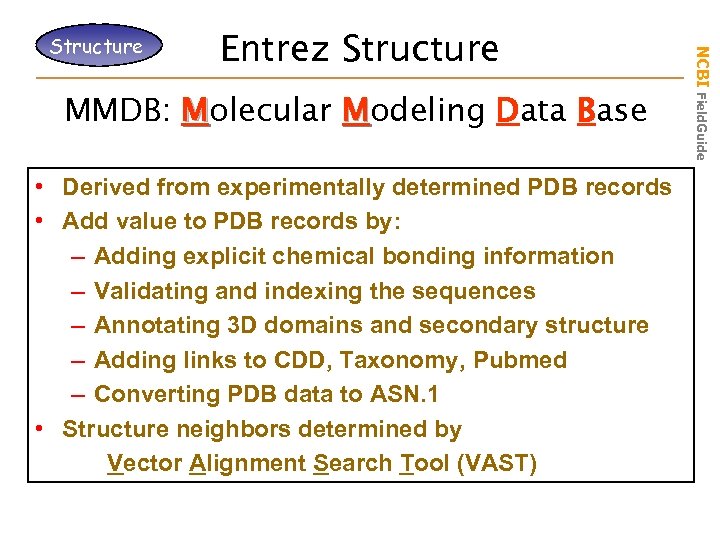
Entrez Structure MMDB: Molecular Modeling Data Base • Derived from experimentally determined PDB records • Add value to PDB records by: – Adding explicit chemical bonding information – Validating and indexing the sequences – Annotating 3 D domains and secondary structure – Adding links to CDD, Taxonomy, Pubmed – Converting PDB data to ASN. 1 • Structure neighbors determined by Vector Alignment Search Tool (VAST) NCBI Field. Guide Structure
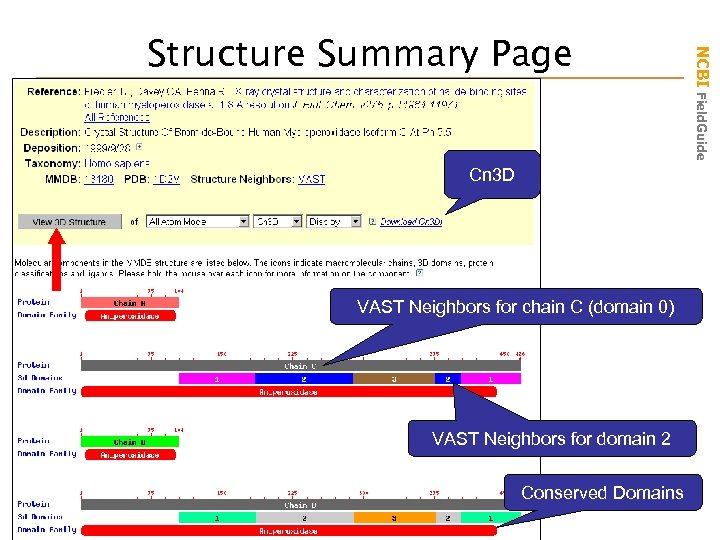
Cn 3 D VAST Neighbors for chain C (domain 0) VAST Neighbors for domain 2 Conserved Domains NCBI Field. Guide Structure Summary Page
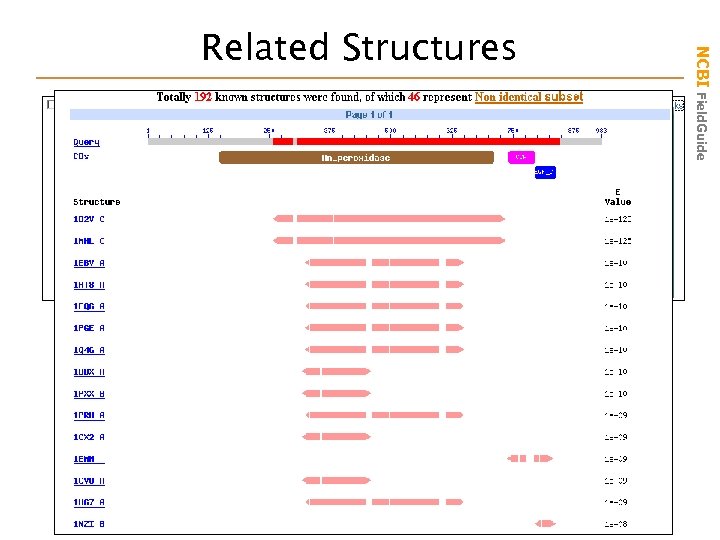
NCBI Field. Guide Related Structures
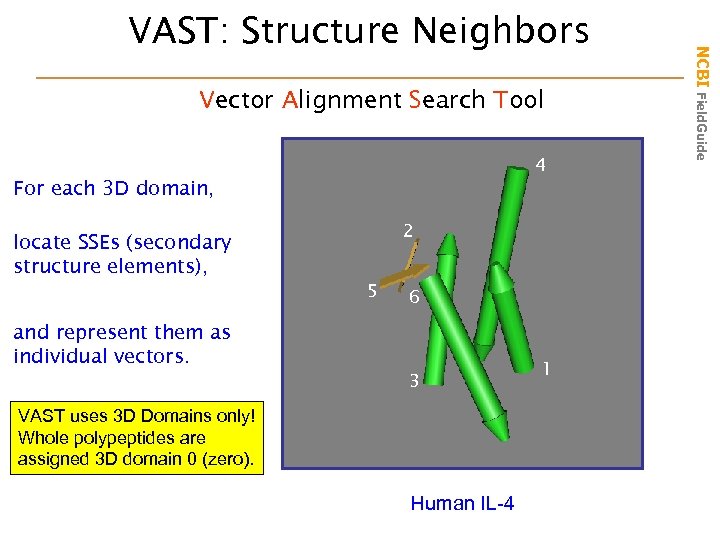
Vector Alignment Search Tool 4 For each 3 D domain, locate SSEs (secondary structure elements), and represent them as individual vectors. 2 5 6 3 VAST uses 3 D Domains only! Whole polypeptides are assigned 3 D domain 0 (zero). Human IL-4 1 NCBI Field. Guide VAST: Structure Neighbors
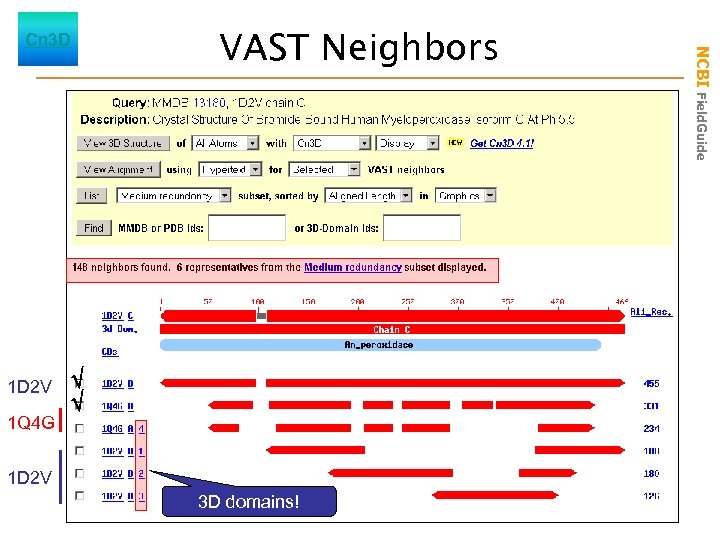
1 D 2 V 1 Q 4 G VAST Neighbors 1 D 2 V 3 D domains! NCBI Field. Guide Cn 3 D
NCBI Field. Guide Submitting a PDB File to VAST • Redesigned interface! • This is the best way to convert PDB into MMDB format! New!
VAST finds proteins that have similar 3 D folds CD-Search finds proteins that have similar sequences and similar functions Curated CDs = VAST + CD-Search Proteins that have similar 3 D folds, similar sequences and similar functions NCBI Field. Guide Structure + Function
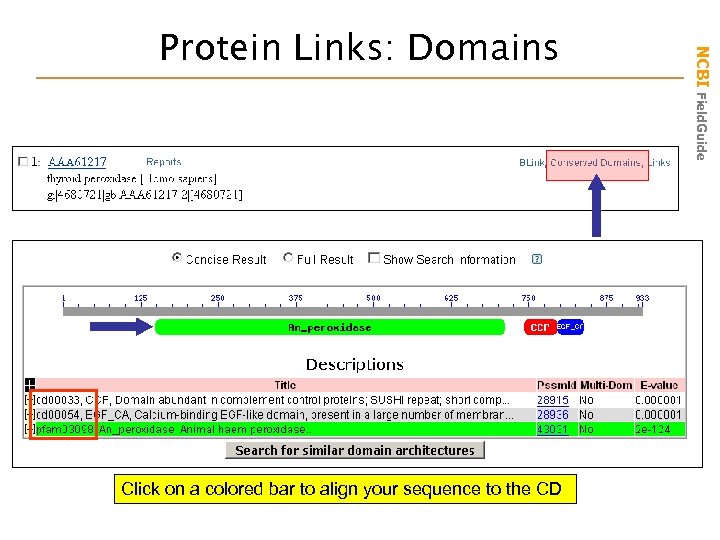
Click on a colored bar to align your sequence to the CD NCBI Field. Guide Protein Links: Domains
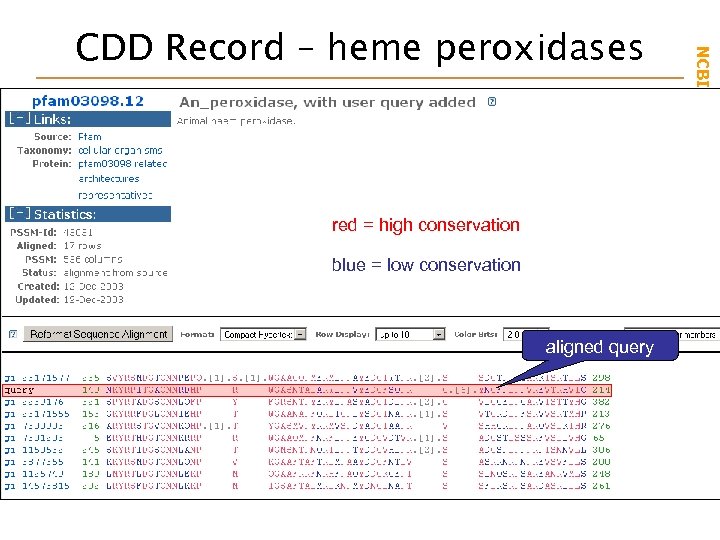
red = high conservation blue = low conservation aligned query NCBI Field. Guide CDD Record – heme peroxidases
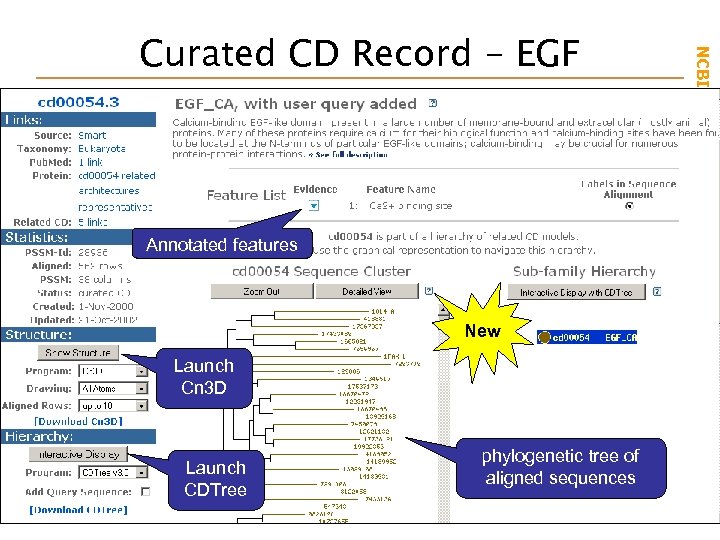
Annotated features New Launch Cn 3 D Launch CDTree phylogenetic tree of aligned sequences NCBI Field. Guide Curated CD Record - EGF
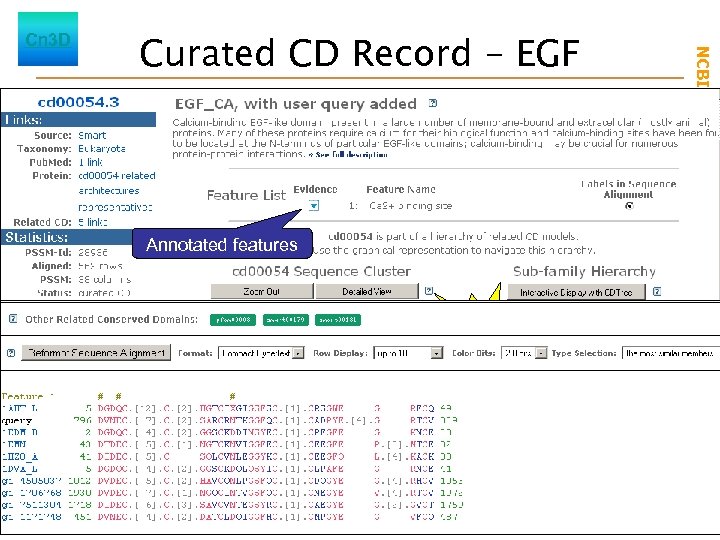
Curated CD Record - EGF Annotated features New Launch Cn 3 D Launch CDTree phylogenetic tree of aligned sequences NCBI Field. Guide Cn 3 D
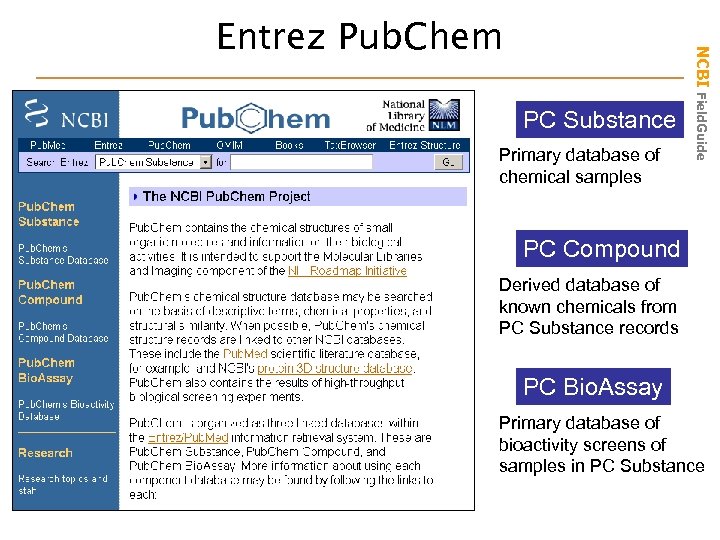
PC Substance Primary database of chemical samples NCBI Field. Guide Entrez Pub. Chem PC Compound Derived database of known chemicals from PC Substance records PC Bio. Assay Primary database of bioactivity screens of samples in PC Substance
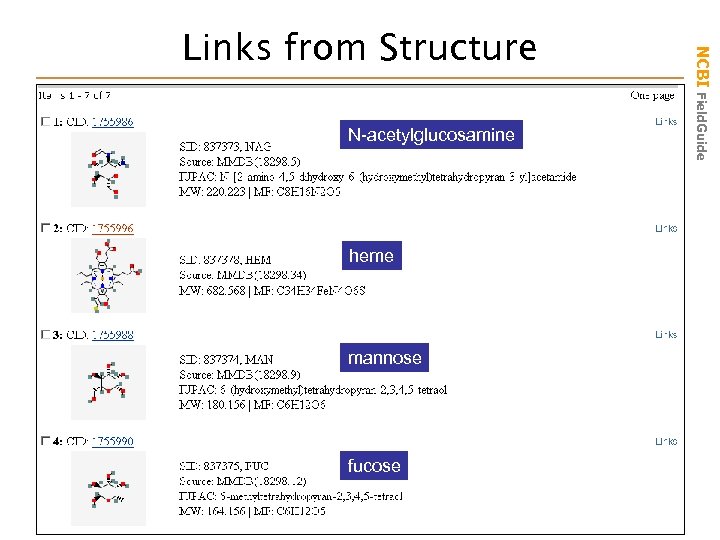
N-acetylglucosamine heme mannose fucose NCBI Field. Guide Links from Structure
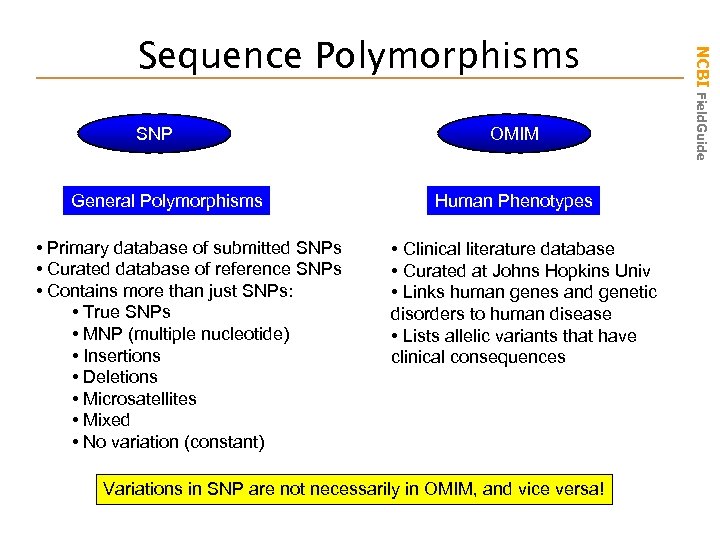
SNP General Polymorphisms • Primary database of submitted SNPs • Curated database of reference SNPs • Contains more than just SNPs: • True SNPs • MNP (multiple nucleotide) • Insertions • Deletions • Microsatellites • Mixed • No variation (constant) OMIM Human Phenotypes • Clinical literature database • Curated at Johns Hopkins Univ • Links human genes and genetic disorders to human disease • Lists allelic variants that have clinical consequences Variations in SNP are not necessarily in OMIM, and vice versa! NCBI Field. Guide Sequence Polymorphisms
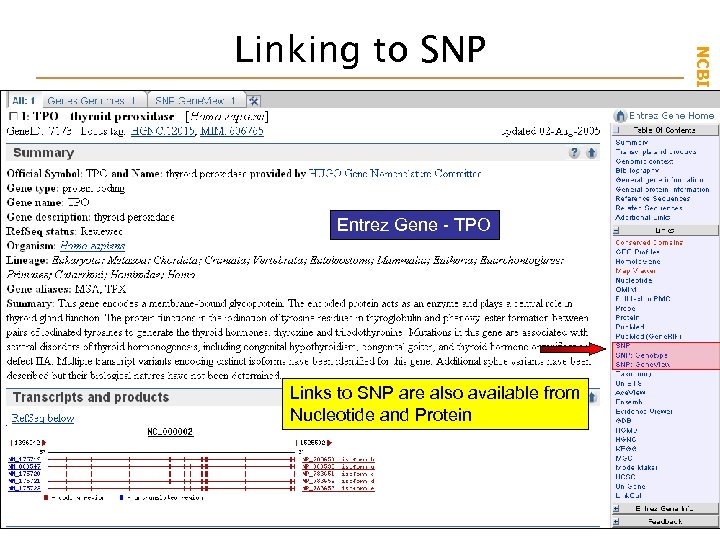
Entrez Gene - TPO Links to SNP are also available from Nucleotide and Protein NCBI Field. Guide Linking to SNP
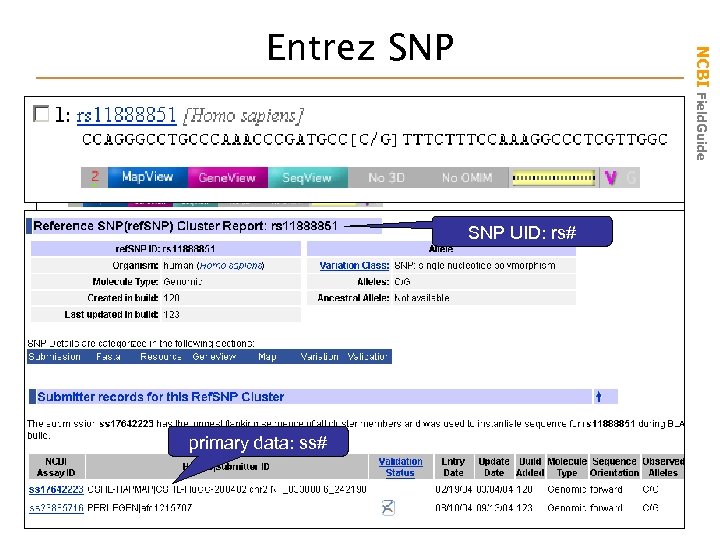
NCBI Field. Guide Entrez SNP UID: rs# primary data: ss#
![#7 AND coding nonsynon[Function Class] Function Class NCBI Field. Guide Find Non-synonymous SNPs #7 AND coding nonsynon[Function Class] Function Class NCBI Field. Guide Find Non-synonymous SNPs](https://present5.com/presentation/e49bda1731c8d4fca1dd792ecb495836/image-88.jpg)
#7 AND coding nonsynon[Function Class] Function Class NCBI Field. Guide Find Non-synonymous SNPs
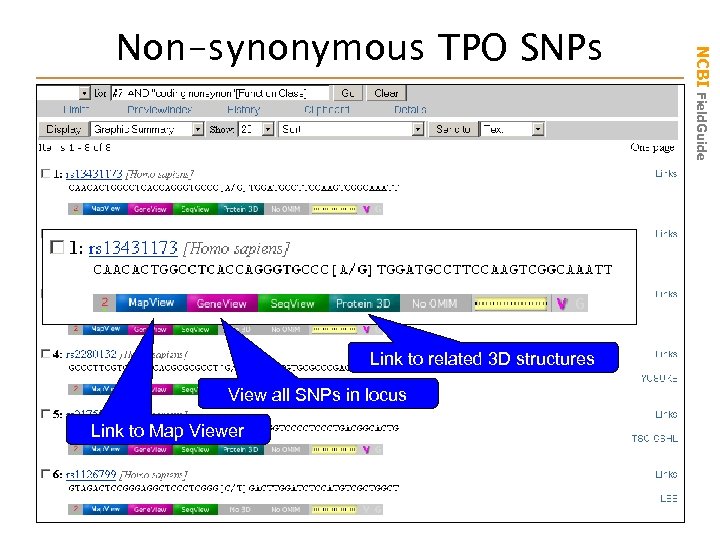
Link to related 3 D structures View all SNPs in locus Link to Map Viewer NCBI Field. Guide Non-synonymous TPO SNPs
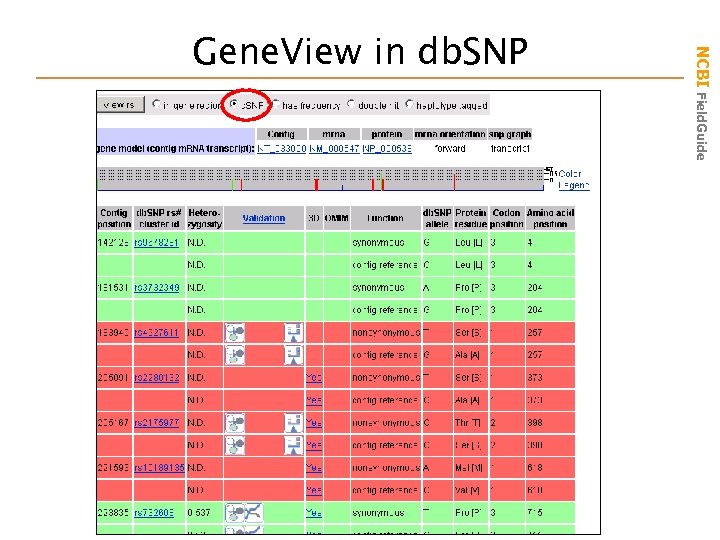
NCBI Field. Guide Gene. View in db. SNP
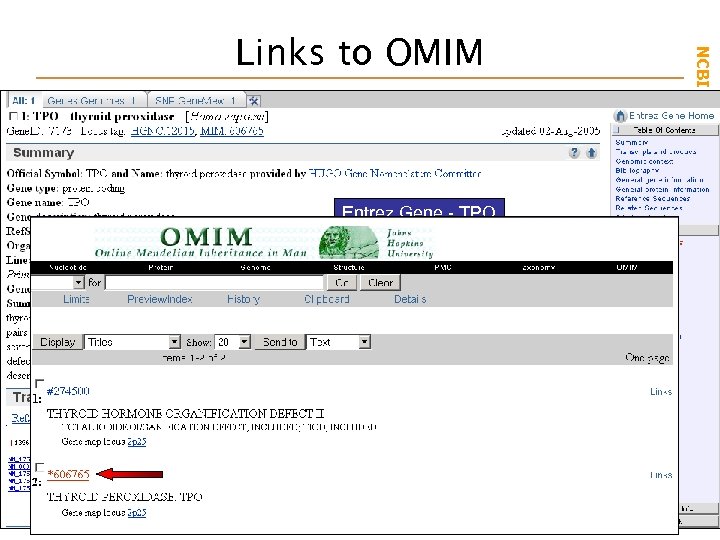
Entrez Gene - TPO NCBI Field. Guide Links to OMIM

NCBI Field. Guide OMIM Record
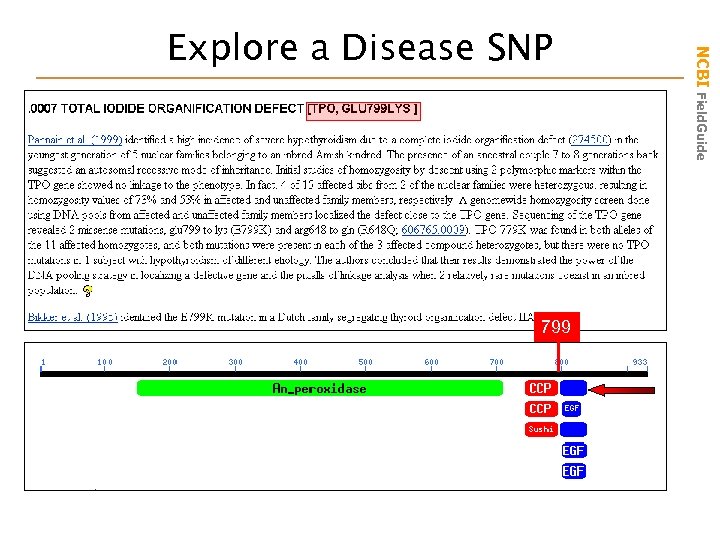
799 NCBI Field. Guide Explore a Disease SNP
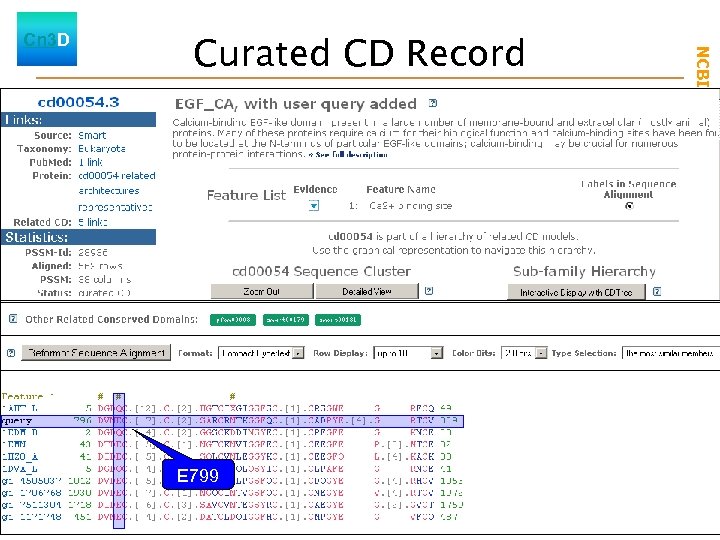
Curated CD Record Launch Cn 3 D Launch E 799 CDTree phylogenetic tree of aligned sequences NCBI Field. Guide Cn 3 D
e49bda1731c8d4fca1dd792ecb495836.ppt