e8433335758c693e8315a495ac617caa.ppt
- Количество слайдов: 13
 National Cancer Institute HPC and Life Sciences Jack Collins Advanced Biomedical Computing Center Advanced Technology Program SAIC-Frederick, Inc. National Cancer Institute at Frederick April 15, 2008
National Cancer Institute HPC and Life Sciences Jack Collins Advanced Biomedical Computing Center Advanced Technology Program SAIC-Frederick, Inc. National Cancer Institute at Frederick April 15, 2008
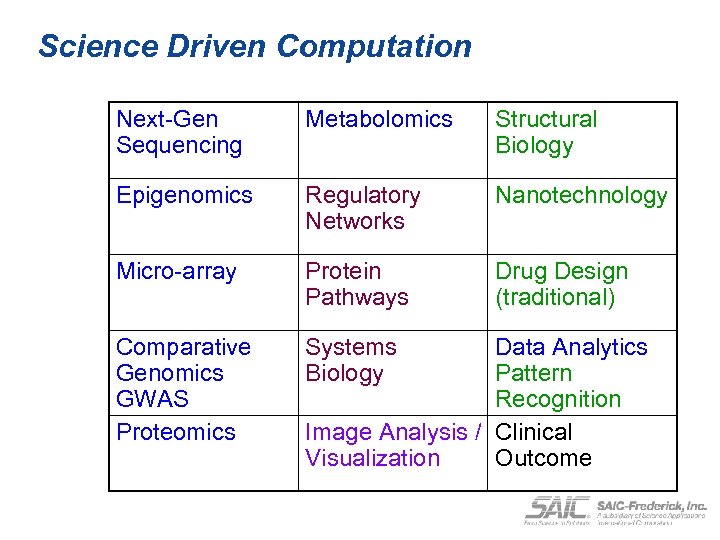 Science Driven Computation Next-Gen Sequencing Metabolomics Structural Biology Epigenomics Regulatory Networks Nanotechnology Micro-array Protein Pathways Drug Design (traditional) Comparative Genomics GWAS Proteomics Systems Biology Data Analytics Pattern Recognition Image Analysis / Clinical Visualization Outcome
Science Driven Computation Next-Gen Sequencing Metabolomics Structural Biology Epigenomics Regulatory Networks Nanotechnology Micro-array Protein Pathways Drug Design (traditional) Comparative Genomics GWAS Proteomics Systems Biology Data Analytics Pattern Recognition Image Analysis / Clinical Visualization Outcome
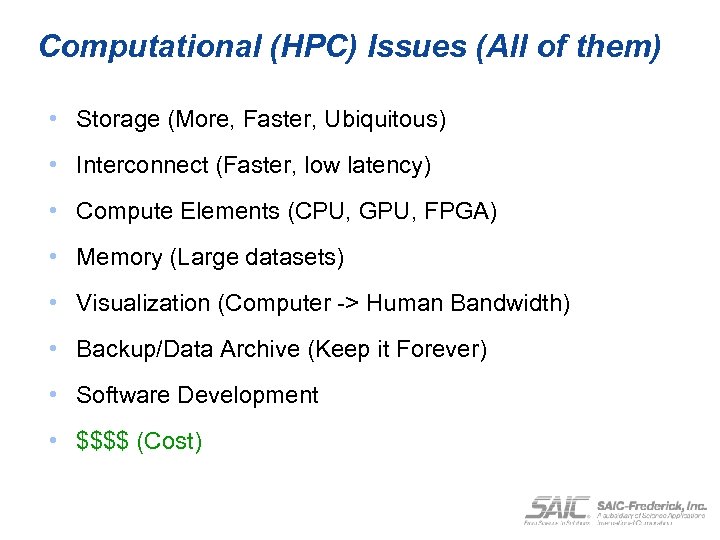 Computational (HPC) Issues (All of them) • Storage (More, Faster, Ubiquitous) • Interconnect (Faster, low latency) • Compute Elements (CPU, GPU, FPGA) • Memory (Large datasets) • Visualization (Computer -> Human Bandwidth) • Backup/Data Archive (Keep it Forever) • Software Development • $$$$ (Cost)
Computational (HPC) Issues (All of them) • Storage (More, Faster, Ubiquitous) • Interconnect (Faster, low latency) • Compute Elements (CPU, GPU, FPGA) • Memory (Large datasets) • Visualization (Computer -> Human Bandwidth) • Backup/Data Archive (Keep it Forever) • Software Development • $$$$ (Cost)
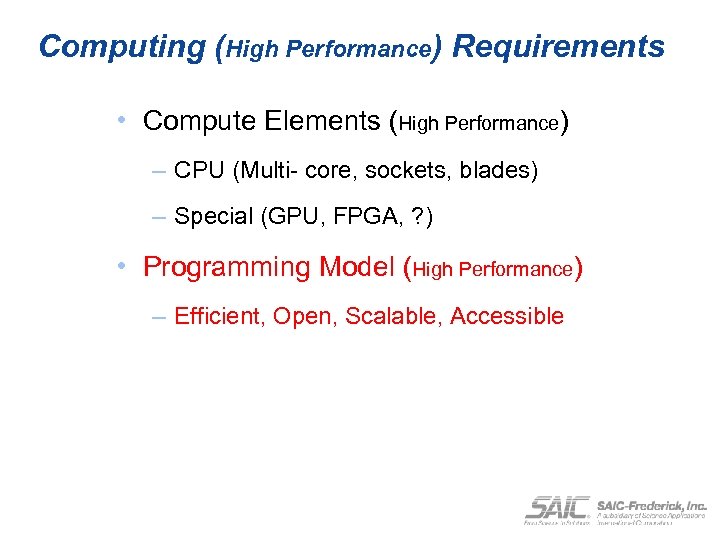 Computing (High Performance) Requirements • Compute Elements (High Performance) – CPU (Multi- core, sockets, blades) – Special (GPU, FPGA, ? ) • Programming Model (High Performance) – Efficient, Open, Scalable, Accessible
Computing (High Performance) Requirements • Compute Elements (High Performance) – CPU (Multi- core, sockets, blades) – Special (GPU, FPGA, ? ) • Programming Model (High Performance) – Efficient, Open, Scalable, Accessible
 Power to the People! • People solve problems – Scientists, Engineers, Doctors, etc. • People write Software – Ready access to personal computers drove Linux Development and Open Source Software (Paradigm Shift) • Ready Access to HPC will drive HPC Development • People will use HPC when they are exposed to HPC and have access early in their career/life
Power to the People! • People solve problems – Scientists, Engineers, Doctors, etc. • People write Software – Ready access to personal computers drove Linux Development and Open Source Software (Paradigm Shift) • Ready Access to HPC will drive HPC Development • People will use HPC when they are exposed to HPC and have access early in their career/life
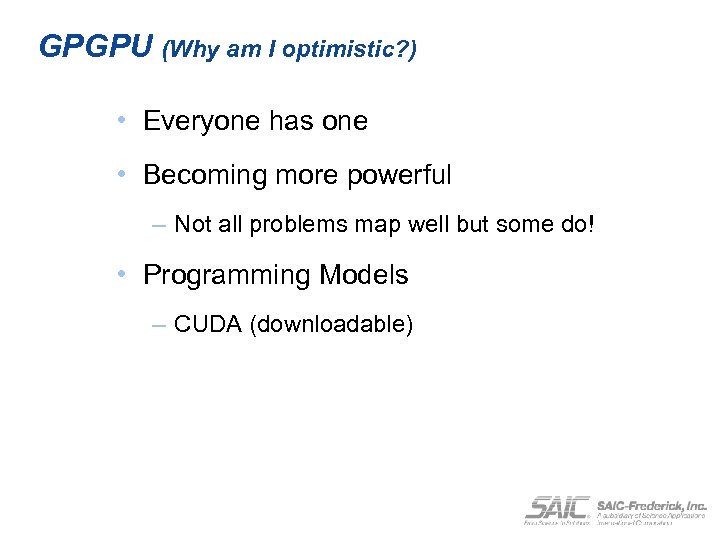 GPGPU (Why am I optimistic? ) • Everyone has one • Becoming more powerful – Not all problems map well but some do! • Programming Models – CUDA (downloadable)
GPGPU (Why am I optimistic? ) • Everyone has one • Becoming more powerful – Not all problems map well but some do! • Programming Models – CUDA (downloadable)
 CUDA (at the price of a download) • The CUDAェ Toolkit is a C language development environment for CUDA-enabled GPUs • The CUDA development environment includes: – nvcc C compiler – CUDA FFT and BLAS libraries for the GPU – Profiler – gdb debugger for the GPU (alpha available in March, 2008) – CUDA runtime driver (now also available in the standard NVIDIA GPU driver) – CUDA programming manual
CUDA (at the price of a download) • The CUDAェ Toolkit is a C language development environment for CUDA-enabled GPUs • The CUDA development environment includes: – nvcc C compiler – CUDA FFT and BLAS libraries for the GPU – Profiler – gdb debugger for the GPU (alpha available in March, 2008) – CUDA runtime driver (now also available in the standard NVIDIA GPU driver) – CUDA programming manual
 CUDA Examples • Smith-Waterman S. Manavski, G. Valle, CRIBI Genomics March 2008 A Neural Network on GPU Billconan, Kavinguy March 2008 MDGPU: Molecular Dynamics simulation J. A. van Meel, A. Arnold October 2007 Interactive Visualization of Volumetric White Matter Connectivity in DT-MRI Won-Ki Jeong, P. Thomas Fletcher, Ran Tao, and Ross T. Whitaker October 2007 Astrophysical simulations based on smoothed particle hydrodynamics: Fourier Volume Rendering Andrew Corrigan and John Wallin, Computational and Data Sciences, George Mason University July 2007 Computational Astrophysics Lab, RIKEN: Astrophysical N-body simulation: The Chamomile Scheme Tsuyoshi Hamada and Toshiaki Iitaka July 2007 Computational biology string matching: CMATCH Michael C. Schatz and Cole Trapnell, Center for Bioinformatics & Computational Biology, University of Maryland May 2007 Simulation Open Framework Architecture (SOFA) for real-time simulation with an emphasis on medical simulation. INRIA and CIMIT February 2007 Visual Molecular Dynamics: VMD Beckman Institute, NIH, NSF, University of Illinois at Urbana. Champaign 2007 Scalable Molecular Dynamics: NAMD Beckman Institute, NIH, NSF, University of Illinois at Urbana-Champaign 2007 NVIDIA Texture Tools 2 Alpha. Source Code NVIDIA 2007 Py. Stream: Python interface to CUDA, CUBLAS and CUFFT Tech-X Corporation 2007 Highly Optimized Objectoriented Molecular Dynamics: HOOMD Joshua A. Anderson, Chris D. Lorenz, and Alex Travesset: Iowa State University 2007 The Schroedinger project: portable libraries for the high quality Dirac video codec created by BBC R&D. Wladimir J. van der Laan, BBC R&D, Fluendo 2007
CUDA Examples • Smith-Waterman S. Manavski, G. Valle, CRIBI Genomics March 2008 A Neural Network on GPU Billconan, Kavinguy March 2008 MDGPU: Molecular Dynamics simulation J. A. van Meel, A. Arnold October 2007 Interactive Visualization of Volumetric White Matter Connectivity in DT-MRI Won-Ki Jeong, P. Thomas Fletcher, Ran Tao, and Ross T. Whitaker October 2007 Astrophysical simulations based on smoothed particle hydrodynamics: Fourier Volume Rendering Andrew Corrigan and John Wallin, Computational and Data Sciences, George Mason University July 2007 Computational Astrophysics Lab, RIKEN: Astrophysical N-body simulation: The Chamomile Scheme Tsuyoshi Hamada and Toshiaki Iitaka July 2007 Computational biology string matching: CMATCH Michael C. Schatz and Cole Trapnell, Center for Bioinformatics & Computational Biology, University of Maryland May 2007 Simulation Open Framework Architecture (SOFA) for real-time simulation with an emphasis on medical simulation. INRIA and CIMIT February 2007 Visual Molecular Dynamics: VMD Beckman Institute, NIH, NSF, University of Illinois at Urbana. Champaign 2007 Scalable Molecular Dynamics: NAMD Beckman Institute, NIH, NSF, University of Illinois at Urbana-Champaign 2007 NVIDIA Texture Tools 2 Alpha. Source Code NVIDIA 2007 Py. Stream: Python interface to CUDA, CUBLAS and CUFFT Tech-X Corporation 2007 Highly Optimized Objectoriented Molecular Dynamics: HOOMD Joshua A. Anderson, Chris D. Lorenz, and Alex Travesset: Iowa State University 2007 The Schroedinger project: portable libraries for the high quality Dirac video codec created by BBC R&D. Wladimir J. van der Laan, BBC R&D, Fluendo 2007
 Autodock (Drug Design) • Molecular Docking for Small Molecules – Open Source from Scripps Institute (Art Olsen, Garrett Morris) – Typical of many codes in biology – Not Designed for HPC • Single Threaded • Genetic Algorithm in iterative steps • Partnered with Silicon Informatics to enable Autodock on GPU and modern multi-core – Smart guys with experience
Autodock (Drug Design) • Molecular Docking for Small Molecules – Open Source from Scripps Institute (Art Olsen, Garrett Morris) – Typical of many codes in biology – Not Designed for HPC • Single Threaded • Genetic Algorithm in iterative steps • Partnered with Silicon Informatics to enable Autodock on GPU and modern multi-core – Smart guys with experience
 The “HPC” System (could buy for home)
The “HPC” System (could buy for home)
 Autodock Results (not NAMD 100 X+ but …) (Only using 1 GPU - Tesla)
Autodock Results (not NAMD 100 X+ but …) (Only using 1 GPU - Tesla)
 Motivation / Business Case • Little or no significant cost in the desktop workstation • Everyone has desktop with GPU • Can dramatically change workflow and thinking – A 10 X speedup can change an overnight (12 hour, 1 job/day) run (1 molecule) into ~4+ runs/workday thus increasing science productivity, greater interactivity – A small group of 10 staff (with Desktops) could now generate 100+ runs during off hours with no additional hardware cost – Success inspires bigger aspirations - so the HPC guys at the computing center could help us do 1, 000 or 10, 000 molecules a day with their big machines (so went the Walter Reed request).
Motivation / Business Case • Little or no significant cost in the desktop workstation • Everyone has desktop with GPU • Can dramatically change workflow and thinking – A 10 X speedup can change an overnight (12 hour, 1 job/day) run (1 molecule) into ~4+ runs/workday thus increasing science productivity, greater interactivity – A small group of 10 staff (with Desktops) could now generate 100+ runs during off hours with no additional hardware cost – Success inspires bigger aspirations - so the HPC guys at the computing center could help us do 1, 000 or 10, 000 molecules a day with their big machines (so went the Walter Reed request).
 Acknowledgements • Bob Keller, Silicon Informatics • Hemant Trivedi, Silicon Informatics • Sarangan “Ravi” Ravichandran, ABCC
Acknowledgements • Bob Keller, Silicon Informatics • Hemant Trivedi, Silicon Informatics • Sarangan “Ravi” Ravichandran, ABCC


