a50f3c209d9bcc30b53cb346f3651f35.ppt
- Количество слайдов: 38
 Mutagenesis of Actinomycetes Workshop July 11 – 15 2005 University of Wales Swansea Actino. GEN SIXTH FRAMEWORK PROGRAMME PRIORITY 1 LIFE SCIENCES, GENOMICS AND BIOTECHNOLOGY FOR HEALTH
Mutagenesis of Actinomycetes Workshop July 11 – 15 2005 University of Wales Swansea Actino. GEN SIXTH FRAMEWORK PROGRAMME PRIORITY 1 LIFE SCIENCES, GENOMICS AND BIOTECHNOLOGY FOR HEALTH
 Actinomycetes are an important resource for new antibiotics • techniques to manipulate actinomycete genes are vital to exploiting this resource • precursor biosynthesis • regulatory networks • antibiotic biosynthetic genes • exemplified in S. coelicolor
Actinomycetes are an important resource for new antibiotics • techniques to manipulate actinomycete genes are vital to exploiting this resource • precursor biosynthesis • regulatory networks • antibiotic biosynthetic genes • exemplified in S. coelicolor

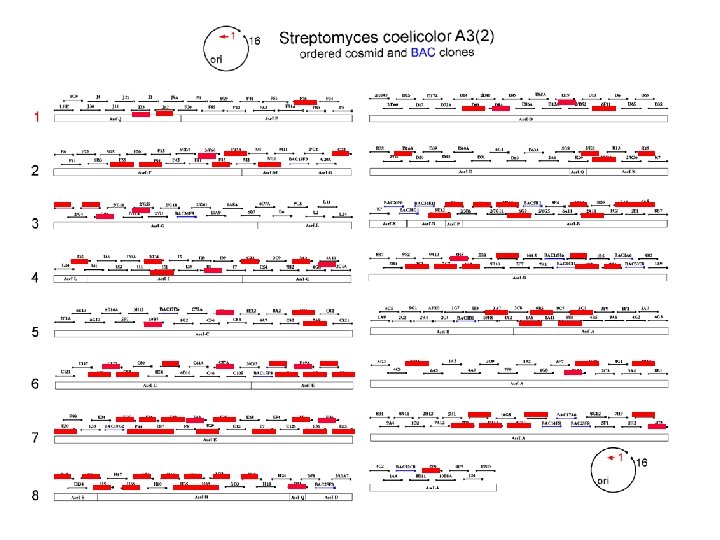
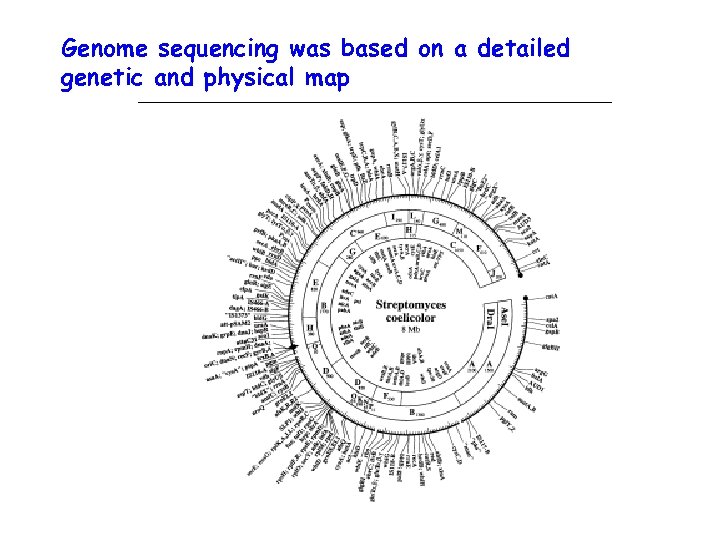 Genome sequencing was based on a detailed genetic and physical map
Genome sequencing was based on a detailed genetic and physical map
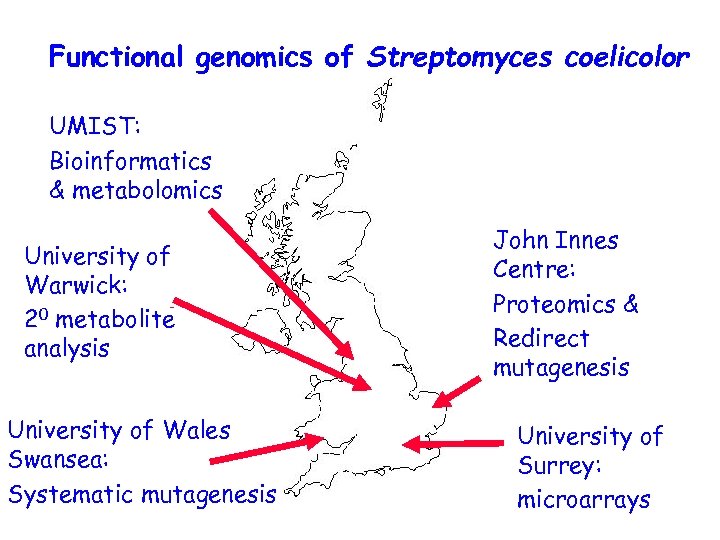 Functional genomics of Streptomyces coelicolor UMIST: Bioinformatics & metabolomics University of Warwick: 20 metabolite analysis University of Wales Swansea: Systematic mutagenesis John Innes Centre: Proteomics & Redirect mutagenesis University of Surrey: microarrays
Functional genomics of Streptomyces coelicolor UMIST: Bioinformatics & metabolomics University of Warwick: 20 metabolite analysis University of Wales Swansea: Systematic mutagenesis John Innes Centre: Proteomics & Redirect mutagenesis University of Surrey: microarrays
 Mutagenesis Three techniques that exploit the genome sequence: (1) In vitro transposon mutagenesis – systematic (2) In vivo transposon mutagenesis – identify genes of related function (3) PCR targetting (Redirect) – functional analysis of a set of genes
Mutagenesis Three techniques that exploit the genome sequence: (1) In vitro transposon mutagenesis – systematic (2) In vivo transposon mutagenesis – identify genes of related function (3) PCR targetting (Redirect) – functional analysis of a set of genes
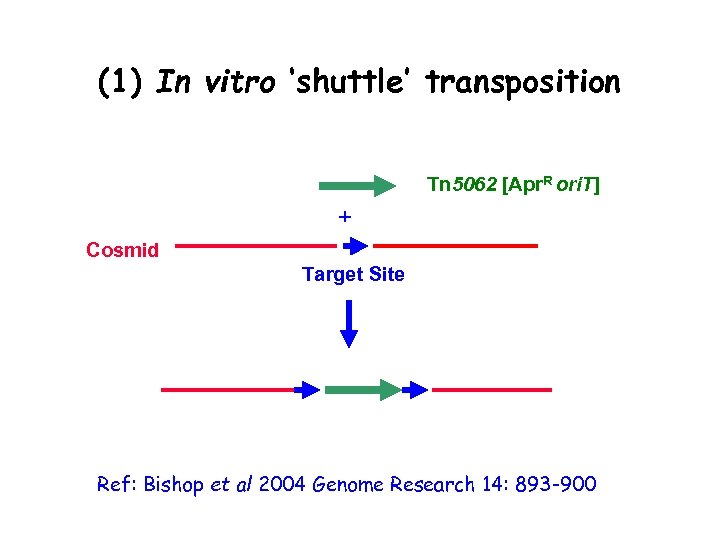 (1) In vitro ‘shuttle’ transposition • Transposition is (fairly) random • Target site is duplicated and Insertion Sequence integrated Tn 5062 [Apr. R ori. T] + Cosmid Target Site Ref: Bishop et al 2004 Genome Research 14: 893 -900
(1) In vitro ‘shuttle’ transposition • Transposition is (fairly) random • Target site is duplicated and Insertion Sequence integrated Tn 5062 [Apr. R ori. T] + Cosmid Target Site Ref: Bishop et al 2004 Genome Research 14: 893 -900
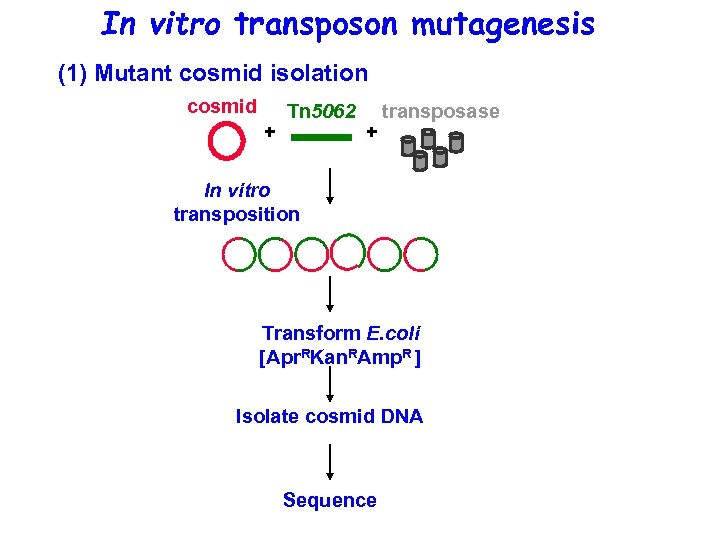 In vitro transposon mutagenesis (1) Mutant cosmid isolation cosmid + Tn 5062 + transposase In vitro transposition Transform E. coli [Apr. RKan. RAmp. R ] Isolate cosmid DNA Sequence
In vitro transposon mutagenesis (1) Mutant cosmid isolation cosmid + Tn 5062 + transposase In vitro transposition Transform E. coli [Apr. RKan. RAmp. R ] Isolate cosmid DNA Sequence
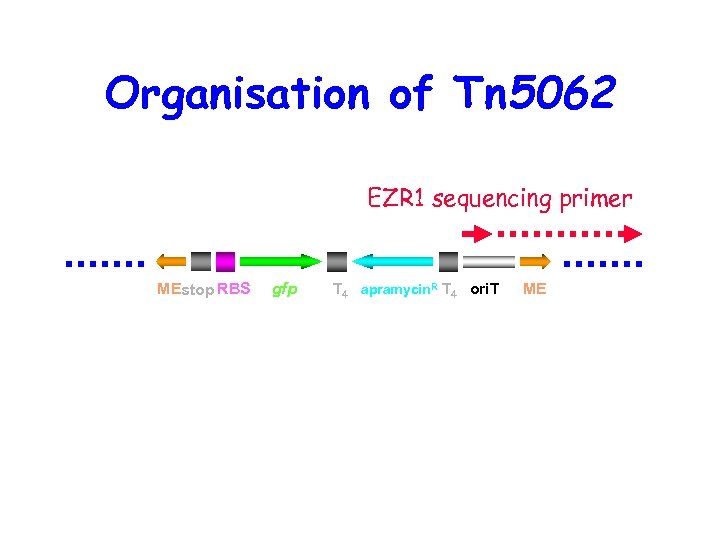 Organisation of Tn 5062 EZR 1 sequencing primer MEstop RBS gfp T 4 apramycin. R T 4 ori. T ME
Organisation of Tn 5062 EZR 1 sequencing primer MEstop RBS gfp T 4 apramycin. R T 4 ori. T ME
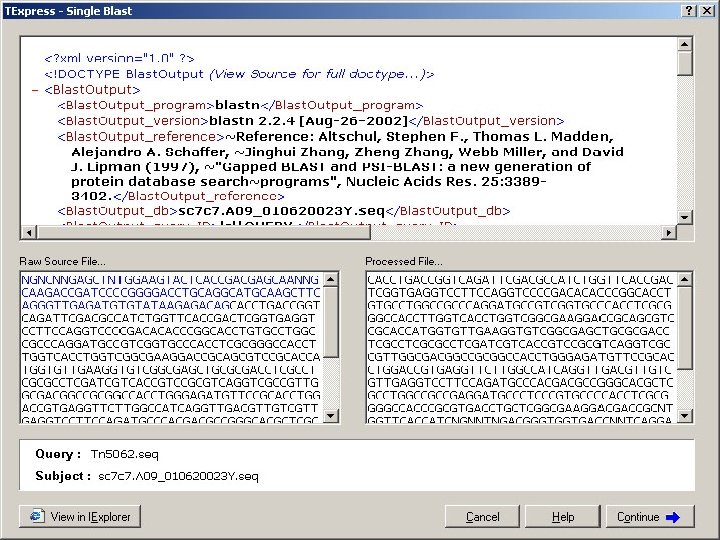
 Analysis of Tn 5062 insertions • sequence files are directly processed using Transposon Express software • finds boundary of Tn 5062 sequence • compares succeeding sequence with cosmid or genome sequence • reports coordinates of insertion and identity of disrupted gene Ref: Herron et al 2004 Nucleic Acids Res 32: e 113
Analysis of Tn 5062 insertions • sequence files are directly processed using Transposon Express software • finds boundary of Tn 5062 sequence • compares succeeding sequence with cosmid or genome sequence • reports coordinates of insertion and identity of disrupted gene Ref: Herron et al 2004 Nucleic Acids Res 32: e 113
 Transposon Express
Transposon Express
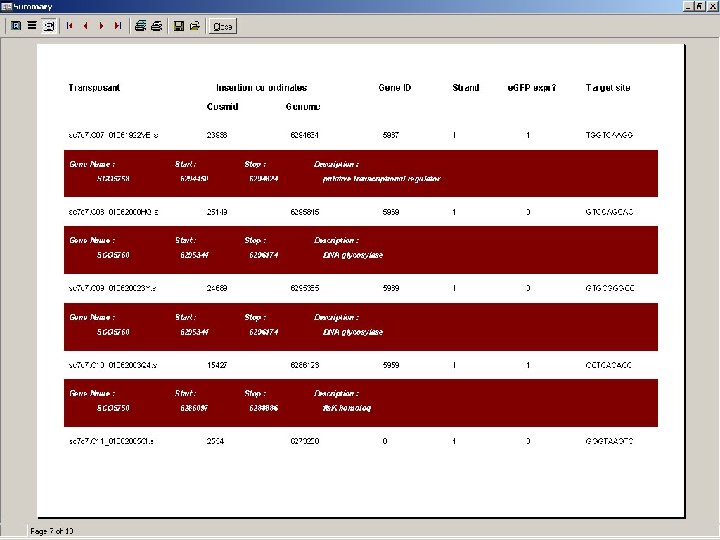
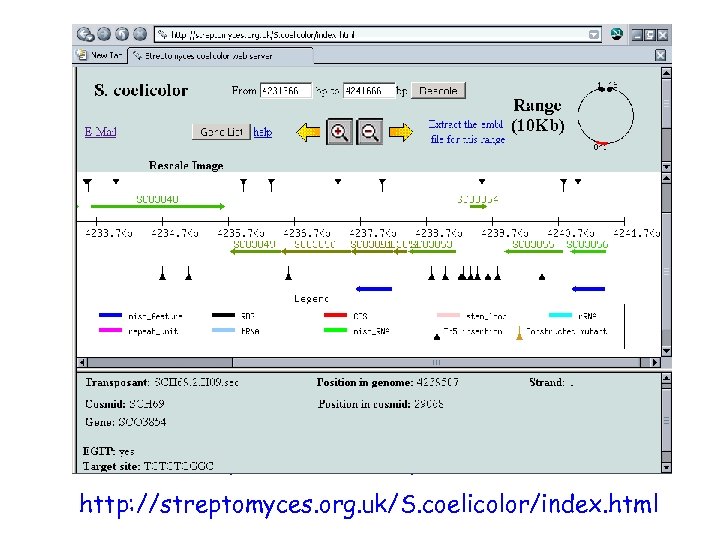 • location and description of each insertion provided at: http: //streptomyces. org. uk/S. coelicolor/index. html
• location and description of each insertion provided at: http: //streptomyces. org. uk/S. coelicolor/index. html
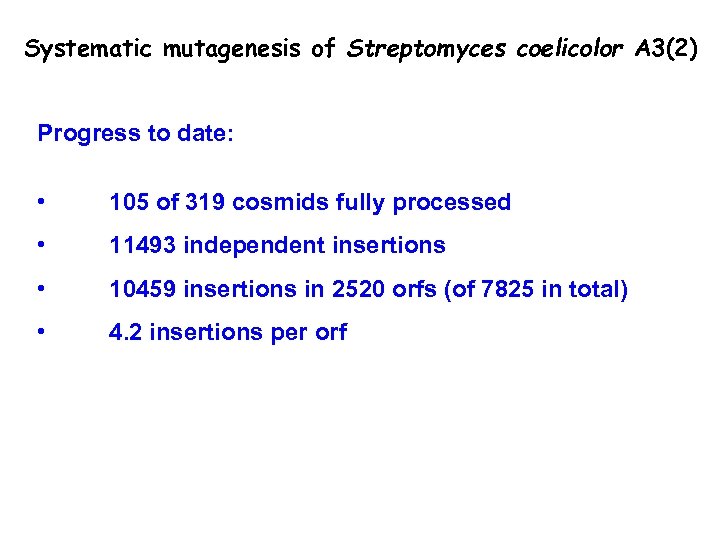 Systematic mutagenesis of Streptomyces coelicolor A 3(2) Progress to date: • 105 of 319 cosmids fully processed • 11493 independent insertions • 10459 insertions in 2520 orfs (of 7825 in total) • 4. 2 insertions per orf
Systematic mutagenesis of Streptomyces coelicolor A 3(2) Progress to date: • 105 of 319 cosmids fully processed • 11493 independent insertions • 10459 insertions in 2520 orfs (of 7825 in total) • 4. 2 insertions per orf
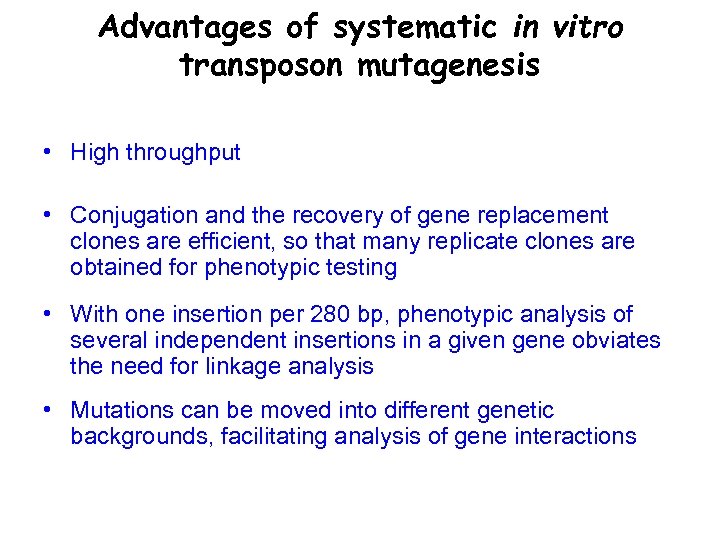 Advantages of systematic in vitro transposon mutagenesis • High throughput • Conjugation and the recovery of gene replacement clones are efficient, so that many replicate clones are obtained for phenotypic testing • With one insertion per 280 bp, phenotypic analysis of several independent insertions in a given gene obviates the need for linkage analysis • Mutations can be moved into different genetic backgrounds, facilitating analysis of gene interactions
Advantages of systematic in vitro transposon mutagenesis • High throughput • Conjugation and the recovery of gene replacement clones are efficient, so that many replicate clones are obtained for phenotypic testing • With one insertion per 280 bp, phenotypic analysis of several independent insertions in a given gene obviates the need for linkage analysis • Mutations can be moved into different genetic backgrounds, facilitating analysis of gene interactions
 Advantages of systematic in vitro transposon mutagenesis • Mutations can be stored and shipped as: Ø cosmid DNA Ø E coli containing cosmids Ø Streptomyces mutants • A Tn 5062 insertion can be manipulated to: Ø change resistance marker (eg switch Apr. R to Hyg. R ) Ø leave an in-frame deletion Ø induce transcription of downstream genes
Advantages of systematic in vitro transposon mutagenesis • Mutations can be stored and shipped as: Ø cosmid DNA Ø E coli containing cosmids Ø Streptomyces mutants • A Tn 5062 insertion can be manipulated to: Ø change resistance marker (eg switch Apr. R to Hyg. R ) Ø leave an in-frame deletion Ø induce transcription of downstream genes
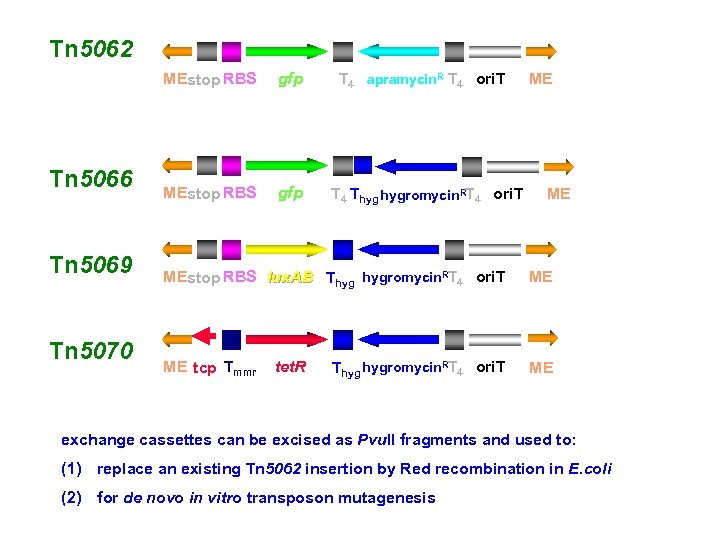 Tn 5062 MEstop RBS Tn 5066 Tn 5069 Tn 5070 gfp MEstop RBS gfp T 4 apramycin. R T 4 ori. T T 4 Thyg hygromycin. RT 4 ori. T ME ME MEstop RBS lux. AB Thyg hygromycin. RT 4 ori. T ME ME tcp Tmmr ME tet. R Thyg hygromycin. RT 4 ori. T exchange cassettes can be excised as Pvu. II fragments and used to: (1) replace an existing Tn 5062 insertion by Red recombination in E. coli (2) for de novo in vitro transposon mutagenesis
Tn 5062 MEstop RBS Tn 5066 Tn 5069 Tn 5070 gfp MEstop RBS gfp T 4 apramycin. R T 4 ori. T T 4 Thyg hygromycin. RT 4 ori. T ME ME MEstop RBS lux. AB Thyg hygromycin. RT 4 ori. T ME ME tcp Tmmr ME tet. R Thyg hygromycin. RT 4 ori. T exchange cassettes can be excised as Pvu. II fragments and used to: (1) replace an existing Tn 5062 insertion by Red recombination in E. coli (2) for de novo in vitro transposon mutagenesis
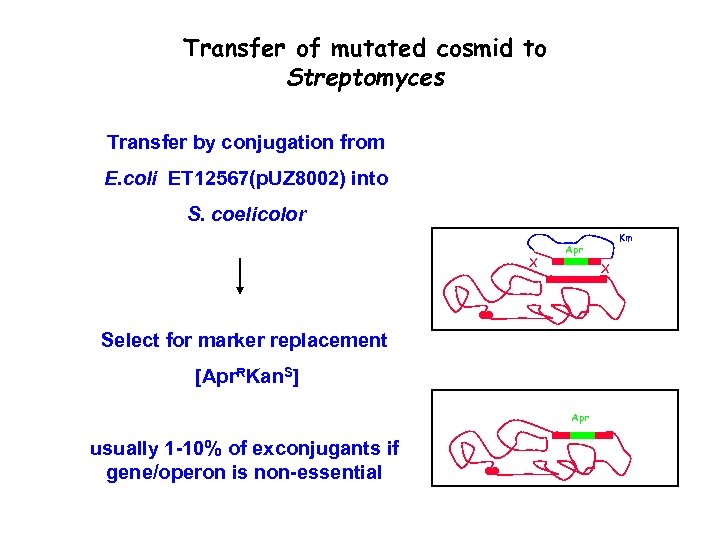 Transfer of mutated cosmid to Streptomyces Transfer by conjugation from E. coli ET 12567(p. UZ 8002) into S. coelicolor X X Select for marker replacement [Apr. RKan. S] Apr usually 1 -10% of exconjugants if gene/operon is non-essential Km Apr
Transfer of mutated cosmid to Streptomyces Transfer by conjugation from E. coli ET 12567(p. UZ 8002) into S. coelicolor X X Select for marker replacement [Apr. RKan. S] Apr usually 1 -10% of exconjugants if gene/operon is non-essential Km Apr
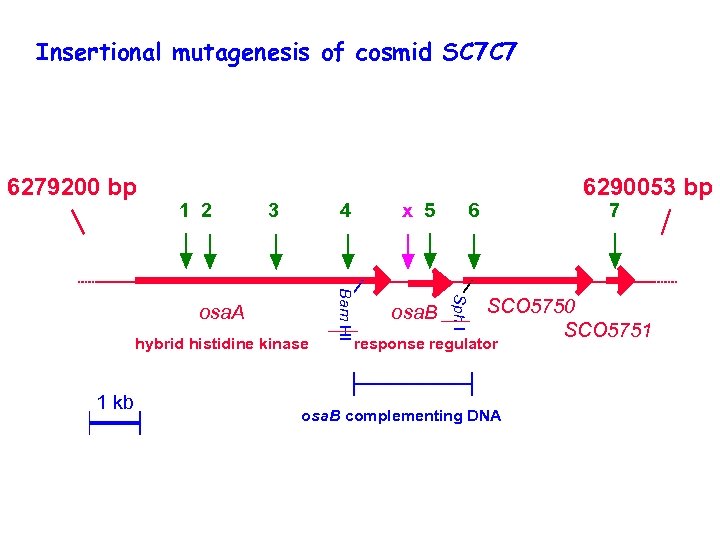 Insertional mutagenesis of cosmid SC 7 C 7 1 2 3 osa. A hybrid histidine kinase 1 kb x 5 osa. B 6290053 bp 6 Sph I 4 Bam HI 6279200 bp 7 SCO 5750 SCO 5751 response regulator osa. B complementing DNA
Insertional mutagenesis of cosmid SC 7 C 7 1 2 3 osa. A hybrid histidine kinase 1 kb x 5 osa. B 6290053 bp 6 Sph I 4 Bam HI 6279200 bp 7 SCO 5750 SCO 5751 response regulator osa. B complementing DNA
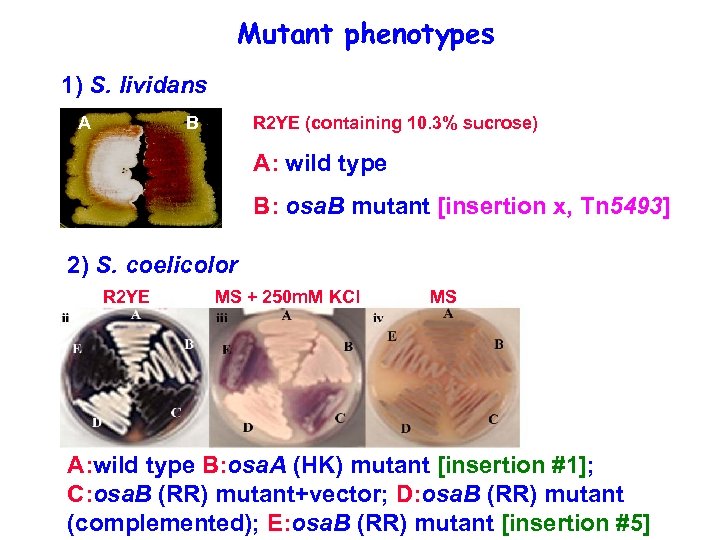 Mutant phenotypes 1) S. lividans A B R 2 YE (containing 10. 3% sucrose) A: wild type B: osa. B mutant [insertion x, Tn 5493] 2) S. coelicolor R 2 YE MS + 250 m. M KCl MS A: wild type B: osa. A (HK) mutant [insertion #1]; C: osa. B (RR) mutant+vector; D: osa. B (RR) mutant (complemented); E: osa. B (RR) mutant [insertion #5]
Mutant phenotypes 1) S. lividans A B R 2 YE (containing 10. 3% sucrose) A: wild type B: osa. B mutant [insertion x, Tn 5493] 2) S. coelicolor R 2 YE MS + 250 m. M KCl MS A: wild type B: osa. A (HK) mutant [insertion #1]; C: osa. B (RR) mutant+vector; D: osa. B (RR) mutant (complemented); E: osa. B (RR) mutant [insertion #5]
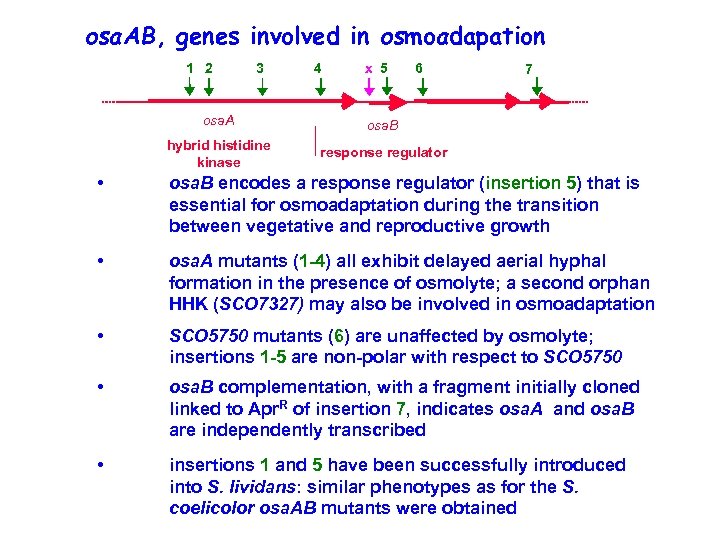 osa. AB, genes involved in osmoadapation 1 2 3 4 x 5 6 osa. A osa. B hybrid histidine kinase 7 response regulator • osa. B encodes a response regulator (insertion 5) that is essential for osmoadaptation during the transition between vegetative and reproductive growth • osa. A mutants (1 -4) all exhibit delayed aerial hyphal formation in the presence of osmolyte; a second orphan HHK (SCO 7327) may also be involved in osmoadaptation • SCO 5750 mutants (6) are unaffected by osmolyte; insertions 1 -5 are non-polar with respect to SCO 5750 • osa. B complementation, with a fragment initially cloned linked to Apr. R of insertion 7, indicates osa. A and osa. B are independently transcribed • insertions 1 and 5 have been successfully introduced into S. lividans: similar phenotypes as for the S. coelicolor osa. AB mutants were obtained
osa. AB, genes involved in osmoadapation 1 2 3 4 x 5 6 osa. A osa. B hybrid histidine kinase 7 response regulator • osa. B encodes a response regulator (insertion 5) that is essential for osmoadaptation during the transition between vegetative and reproductive growth • osa. A mutants (1 -4) all exhibit delayed aerial hyphal formation in the presence of osmolyte; a second orphan HHK (SCO 7327) may also be involved in osmoadaptation • SCO 5750 mutants (6) are unaffected by osmolyte; insertions 1 -5 are non-polar with respect to SCO 5750 • osa. B complementation, with a fragment initially cloned linked to Apr. R of insertion 7, indicates osa. A and osa. B are independently transcribed • insertions 1 and 5 have been successfully introduced into S. lividans: similar phenotypes as for the S. coelicolor osa. AB mutants were obtained
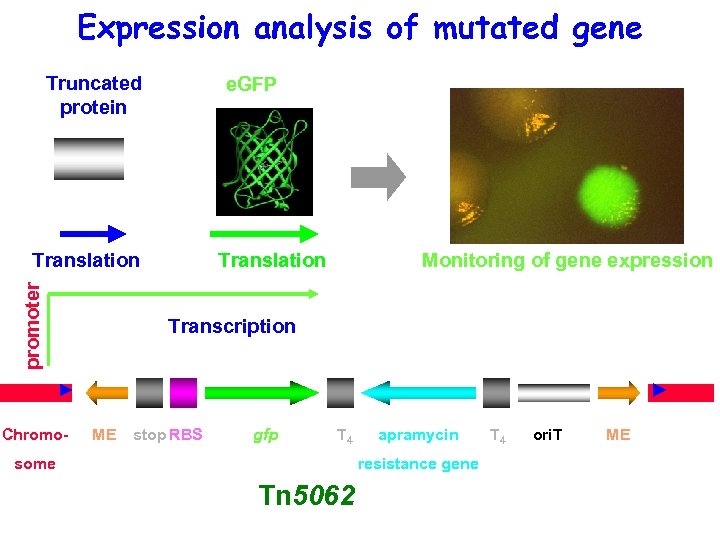 Expression analysis of mutated gene Truncated protein e. GFP promoter Translation Chromo- Translation Monitoring of gene expression Transcription ME stop RBS gfp T 4 some apramycin resistance gene Tn 5062 T 4 ori. T ME
Expression analysis of mutated gene Truncated protein e. GFP promoter Translation Chromo- Translation Monitoring of gene expression Transcription ME stop RBS gfp T 4 some apramycin resistance gene Tn 5062 T 4 ori. T ME
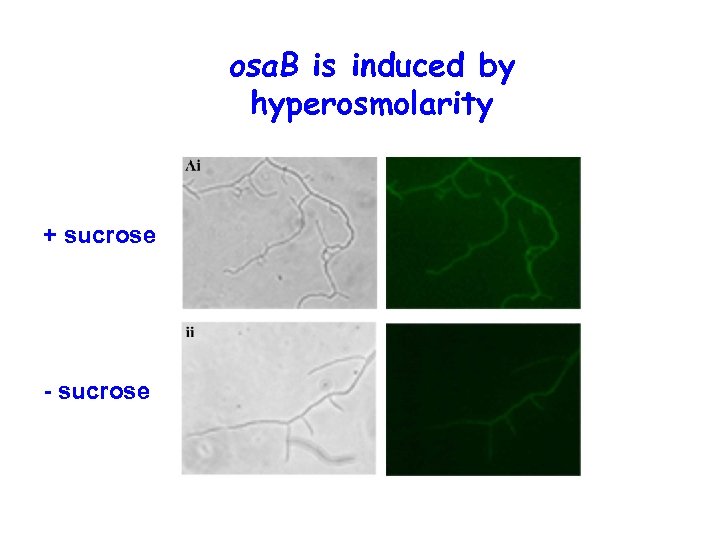 osa. B is induced by hyperosmolarity + sucrose - sucrose
osa. B is induced by hyperosmolarity + sucrose - sucrose
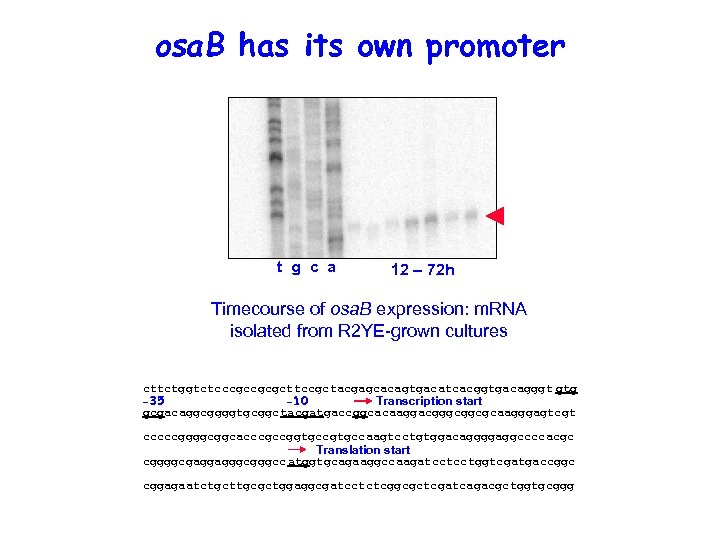 osa. B has its own promoter t g c a 12 – 72 h Timecourse of osa. B expression: m. RNA isolated from R 2 YE-grown cultures 6285056 chromosome position…… cttctggtctcccgccgcgcttccgctacgagcacagtgacatcacggtgacagggt gtg Transcription start -35 -10 gcgacaggcggggtgcggctacgatgaccggcacaaggacgggcggcgcaagggagtcgt cccccggggcggcacccgccggtgccaagtcctgtggacaggggaggccccacgc Translation start cggggcgaggagggccatggtgcagaaggccaagatcctcctggtcgatgaccggc cggagaatctgcttgcgctggaggcgatcctctcggcgctcgatcagacgctggtgcggg
osa. B has its own promoter t g c a 12 – 72 h Timecourse of osa. B expression: m. RNA isolated from R 2 YE-grown cultures 6285056 chromosome position…… cttctggtctcccgccgcgcttccgctacgagcacagtgacatcacggtgacagggt gtg Transcription start -35 -10 gcgacaggcggggtgcggctacgatgaccggcacaaggacgggcggcgcaagggagtcgt cccccggggcggcacccgccggtgccaagtcctgtggacaggggaggccccacgc Translation start cggggcgaggagggccatggtgcagaaggccaagatcctcctggtcgatgaccggc cggagaatctgcttgcgctggaggcgatcctctcggcgctcgatcagacgctggtgcggg
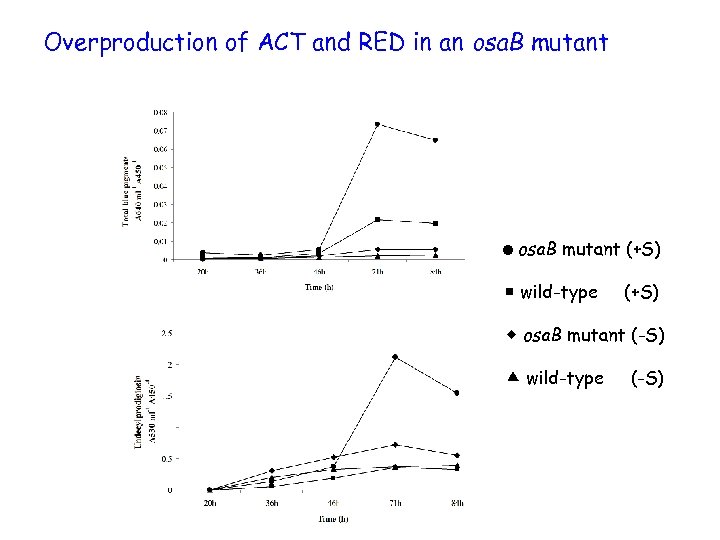 Overproduction of ACT and RED in an osa. B mutant =osa. B mutant (+S) wild-type (+S) osa. B mutant (-S) wild-type (-S)
Overproduction of ACT and RED in an osa. B mutant =osa. B mutant (+S) wild-type (+S) osa. B mutant (-S) wild-type (-S)
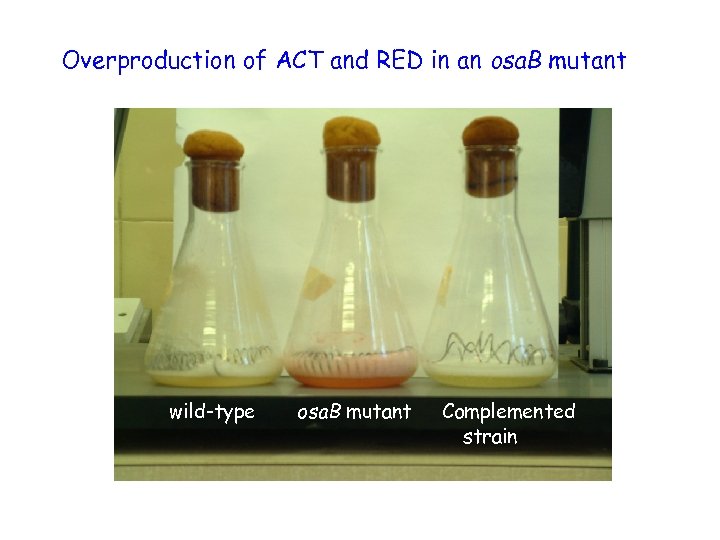 Overproduction of ACT and RED in an osa. B mutant wild-type osa. B mutant Complemented strain
Overproduction of ACT and RED in an osa. B mutant wild-type osa. B mutant Complemented strain
 Osmoadaptation – conclusions • the response regulator encoded by osa. B is essential for developmental osmoadaptation • osa. B impacts on antibiotic production in conditions of hyperosmolarity • unlike most sensory kinase-response regulator gene pairs, osa. B is independently transcribed • the sensory kinase encoded by osa. A is required for osmoadaptation, but not essential – another kinase may also interact with Osa. B
Osmoadaptation – conclusions • the response regulator encoded by osa. B is essential for developmental osmoadaptation • osa. B impacts on antibiotic production in conditions of hyperosmolarity • unlike most sensory kinase-response regulator gene pairs, osa. B is independently transcribed • the sensory kinase encoded by osa. A is required for osmoadaptation, but not essential – another kinase may also interact with Osa. B
 (2) In vivo transposon mutagenesis Aim Generate a library of transposon induced, tagged mutants for gene function studies Kay Fowler
(2) In vivo transposon mutagenesis Aim Generate a library of transposon induced, tagged mutants for gene function studies Kay Fowler
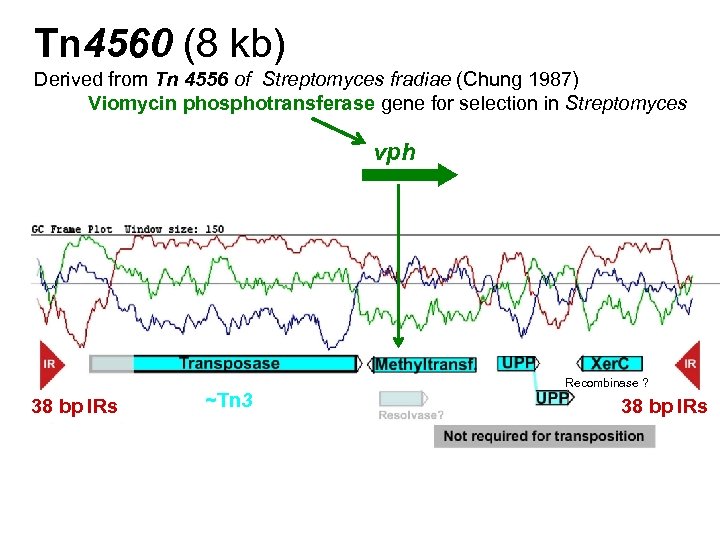 Tn 4560 (8 kb) Derived from Tn 4556 of Streptomyces fradiae (Chung 1987) Viomycin phosphotransferase gene for selection in Streptomyces vph 38 bp IRs ~Tn 3 Recombinase ? 38 bp IRs
Tn 4560 (8 kb) Derived from Tn 4556 of Streptomyces fradiae (Chung 1987) Viomycin phosphotransferase gene for selection in Streptomyces vph 38 bp IRs ~Tn 3 Recombinase ? 38 bp IRs
 Tn 4560 delivery plasmid p. KAY 1 • based on temperature-sensitive plasmid p. UC 1169 (derivative of p. IJ 101 containing Tn 4560) • p. OJ 260 (contains E. coli ori and ori. T) was cloned at the unique Bam. HI site • encodes a truncated Rep protein due to mutation at the unique Bst. BI site: -GCCCCGTTCGCGAACTCCTCGGATCGGGGACCTGA -Ala. Pro. Phe. Ala. Asn. Ser. Asp. Gly. Ser. Gly. Thr***
Tn 4560 delivery plasmid p. KAY 1 • based on temperature-sensitive plasmid p. UC 1169 (derivative of p. IJ 101 containing Tn 4560) • p. OJ 260 (contains E. coli ori and ori. T) was cloned at the unique Bam. HI site • encodes a truncated Rep protein due to mutation at the unique Bst. BI site: -GCCCCGTTCGCGAACTCCTCGGATCGGGGACCTGA -Ala. Pro. Phe. Ala. Asn. Ser. Asp. Gly. Ser. Gly. Thr***
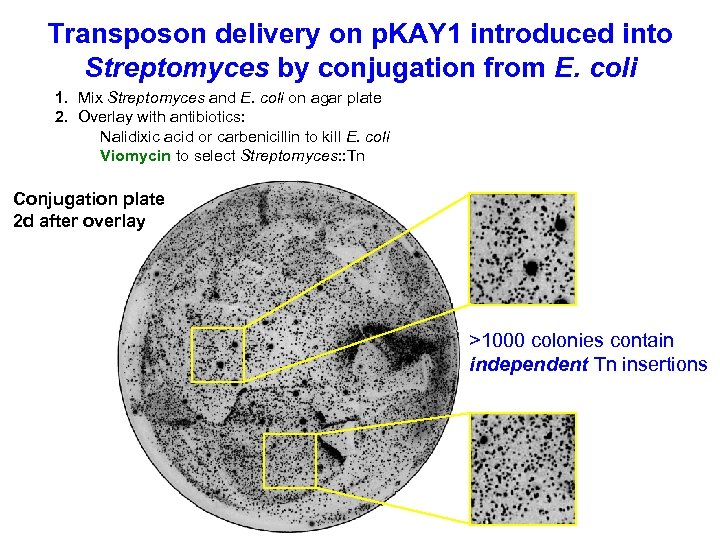 Transposon delivery on p. KAY 1 introduced into Streptomyces by conjugation from E. coli 1. Mix Streptomyces and E. coli on agar plate 2. Overlay with antibiotics: Nalidixic acid or carbenicillin to kill E. coli Viomycin to select Streptomyces: : Tn Conjugation plate 2 d after overlay >1000 colonies contain independent Tn insertions
Transposon delivery on p. KAY 1 introduced into Streptomyces by conjugation from E. coli 1. Mix Streptomyces and E. coli on agar plate 2. Overlay with antibiotics: Nalidixic acid or carbenicillin to kill E. coli Viomycin to select Streptomyces: : Tn Conjugation plate 2 d after overlay >1000 colonies contain independent Tn insertions
 In vivo transposon mutagenesis • wash off microcolonies • plate on SFM viomycin • harvest spores = Tn library • plate library using conditions to detect a specific phentype • isolate DNA from mutant • Ligation-mediated PCR
In vivo transposon mutagenesis • wash off microcolonies • plate on SFM viomycin • harvest spores = Tn library • plate library using conditions to detect a specific phentype • isolate DNA from mutant • Ligation-mediated PCR
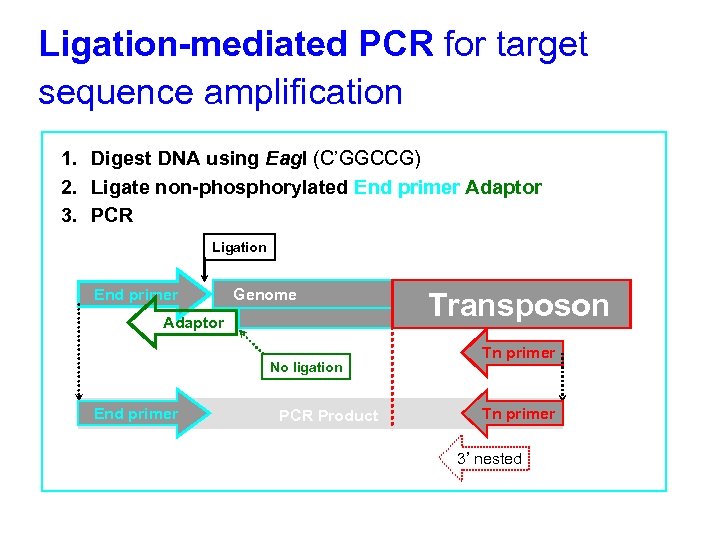 Ligation-mediated PCR for target sequence amplification 1. Digest DNA using Eag. I (C’GGCCG) 2. Ligate non-phosphorylated End primer/Adaptor 3. PCR Ligation End primer Genome Adaptor No ligation End primer PCR Product Transposon Tn primer 3’ nested
Ligation-mediated PCR for target sequence amplification 1. Digest DNA using Eag. I (C’GGCCG) 2. Ligate non-phosphorylated End primer/Adaptor 3. PCR Ligation End primer Genome Adaptor No ligation End primer PCR Product Transposon Tn primer 3’ nested
 Target sequence identification • Use TA cloning to clone PCR products • Sequence inserts • Blast sequence against genome to identify target gene
Target sequence identification • Use TA cloning to clone PCR products • Sequence inserts • Blast sequence against genome to identify target gene
 (3) PCR targetting (Redirect) Bertolt Gust Tübingen
(3) PCR targetting (Redirect) Bertolt Gust Tübingen
 Acknowledgements Swansea: Amy Bishop osa. AB Sue Fielding sequencing Paul Herron in vitro transposition Gareth Hughes Transposon Express Ricardo del Sol exchange cassettes Norwich: Govind Chandra Sco. DB Tobias Kieser in vivo transposon mutagenesis Kay Fowler in vivo transposon mutagenesis
Acknowledgements Swansea: Amy Bishop osa. AB Sue Fielding sequencing Paul Herron in vitro transposition Gareth Hughes Transposon Express Ricardo del Sol exchange cassettes Norwich: Govind Chandra Sco. DB Tobias Kieser in vivo transposon mutagenesis Kay Fowler in vivo transposon mutagenesis


