ac7b8b2d416b93d351c0f9be0497a3e7.ppt
- Количество слайдов: 19
 Multiple Sequence Alignment Dynamic Programming
Multiple Sequence Alignment Dynamic Programming
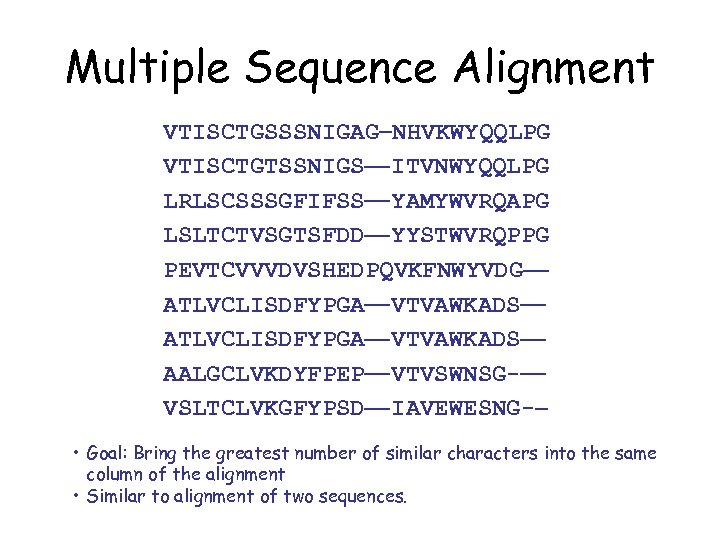 Multiple Sequence Alignment VTISCTGSSSNIGAG NHVKWYQQLPG VTISCTGTSSNIGS ITVNWYQQLPG LRLSCSSSGFIFSS YAMYWVRQAPG LSLTCTVSGTSFDD YYSTWVRQPPG PEVTCVVVDVSHEDPQVKFNWYVDG ATLVCLISDFYPGA VTVAWKADS AALGCLVKDYFPEP VTVSWNSG- VSLTCLVKGFYPSD IAVEWESNG- • Goal: Bring the greatest number of similar characters into the same column of the alignment • Similar to alignment of two sequences.
Multiple Sequence Alignment VTISCTGSSSNIGAG NHVKWYQQLPG VTISCTGTSSNIGS ITVNWYQQLPG LRLSCSSSGFIFSS YAMYWVRQAPG LSLTCTVSGTSFDD YYSTWVRQPPG PEVTCVVVDVSHEDPQVKFNWYVDG ATLVCLISDFYPGA VTVAWKADS AALGCLVKDYFPEP VTVSWNSG- VSLTCLVKGFYPSD IAVEWESNG- • Goal: Bring the greatest number of similar characters into the same column of the alignment • Similar to alignment of two sequences.
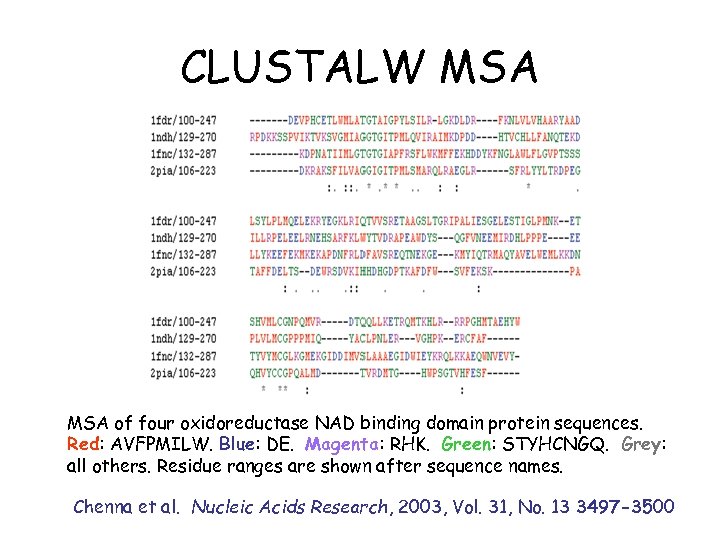 CLUSTALW MSA of four oxidoreductase NAD binding domain protein sequences. Red: AVFPMILW. Blue: DE. Magenta: RHK. Green: STYHCNGQ. Grey: all others. Residue ranges are shown after sequence names. Chenna et al. Nucleic Acids Research, 2003, Vol. 31, No. 13 3497 -3500
CLUSTALW MSA of four oxidoreductase NAD binding domain protein sequences. Red: AVFPMILW. Blue: DE. Magenta: RHK. Green: STYHCNGQ. Grey: all others. Residue ranges are shown after sequence names. Chenna et al. Nucleic Acids Research, 2003, Vol. 31, No. 13 3497 -3500
 Multiple Sequence Alignment: Motivation • Correspondence. Find out which parts “do the same thing” – Similar genes are conserved across widely divergent species, often performing similar functions • Structure prediction – Use knowledge of structure of one or more members of a protein MSA to predict structure of other members – Structure is more conserved than sequence • Create “profiles” for protein families – Allow us to search for other members of the family • Genome assembly: Automated reconstruction of “contig” maps of genomic fragments such as ESTs • MSA is the starting point for phylogenetic analysis
Multiple Sequence Alignment: Motivation • Correspondence. Find out which parts “do the same thing” – Similar genes are conserved across widely divergent species, often performing similar functions • Structure prediction – Use knowledge of structure of one or more members of a protein MSA to predict structure of other members – Structure is more conserved than sequence • Create “profiles” for protein families – Allow us to search for other members of the family • Genome assembly: Automated reconstruction of “contig” maps of genomic fragments such as ESTs • MSA is the starting point for phylogenetic analysis
 Multiple Sequence Alignment: Approaches • Optimal Global Alignments -Dynamic programming – Generalization of Needleman-Wunsch – Find alignment that maximizes a score function – Computationally expensive: Time grows as product of sequence lengths • Global Progressive Alignments - Match closelyrelated sequences first using a guide tree • Global Iterative Alignments - Multiple re-building attempts to find best alignment • Local alignments – Profiles, Blocks, Patterns
Multiple Sequence Alignment: Approaches • Optimal Global Alignments -Dynamic programming – Generalization of Needleman-Wunsch – Find alignment that maximizes a score function – Computationally expensive: Time grows as product of sequence lengths • Global Progressive Alignments - Match closelyrelated sequences first using a guide tree • Global Iterative Alignments - Multiple re-building attempts to find best alignment • Local alignments – Profiles, Blocks, Patterns
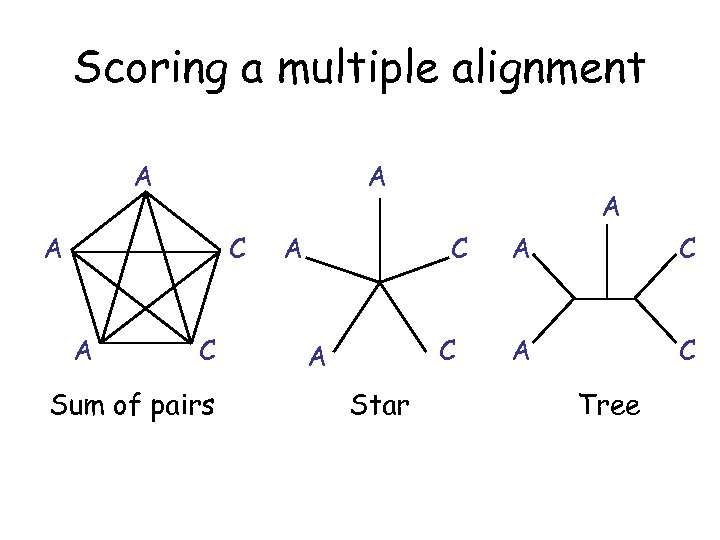 Scoring a multiple alignment A A A C Sum of pairs A A C C A Star A C Tree
Scoring a multiple alignment A A A C Sum of pairs A A C C A Star A C Tree
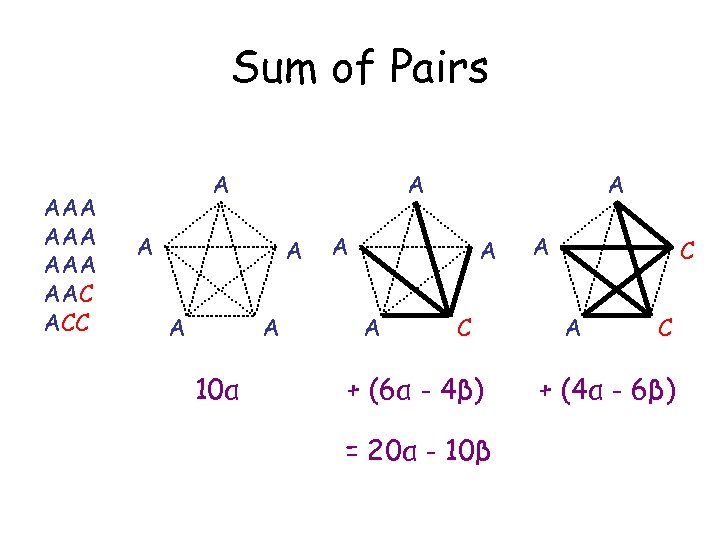 Sum of Pairs AAA AAA AAC ACC A A A 10α A A C + (6α - 4β) = 20α - 10β A C + (4α - 6β)
Sum of Pairs AAA AAA AAC ACC A A A 10α A A C + (6α - 4β) = 20α - 10β A C + (4α - 6β)
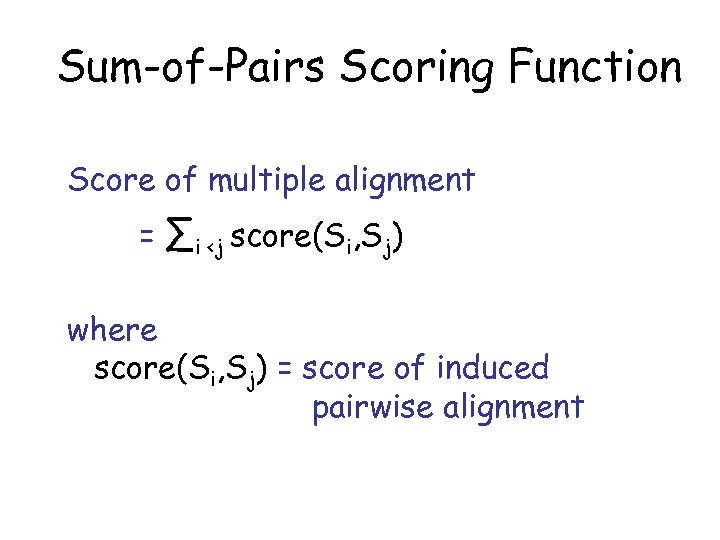 Sum-of-Pairs Scoring Function Score of multiple alignment = ∑i
Sum-of-Pairs Scoring Function Score of multiple alignment = ∑i
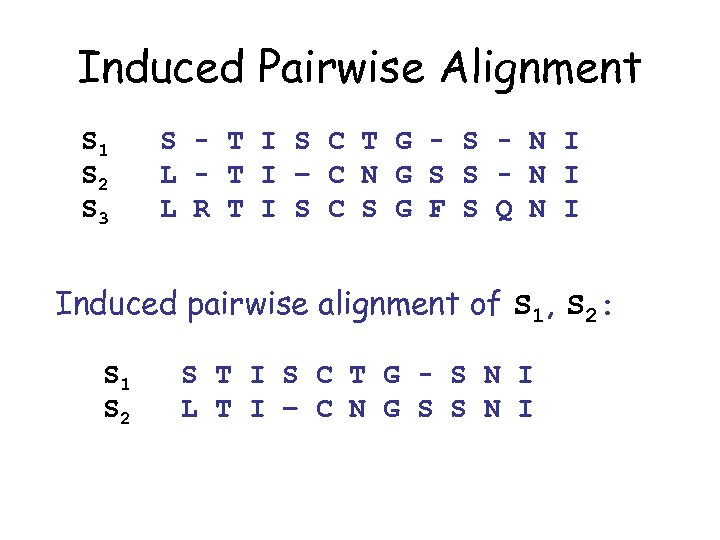 Induced Pairwise Alignment S 1 S 2 S 3 S - T I S C T G - S - N I L - T I – C N G S S - N I L R T I S C S G F S Q N I Induced pairwise alignment of S 1, S 2: S 1 S 2 S T I S C T G - S N I L T I – C N G S S N I
Induced Pairwise Alignment S 1 S 2 S 3 S - T I S C T G - S - N I L - T I – C N G S S - N I L R T I S C S G F S Q N I Induced pairwise alignment of S 1, S 2: S 1 S 2 S T I S C T G - S N I L T I – C N G S S N I
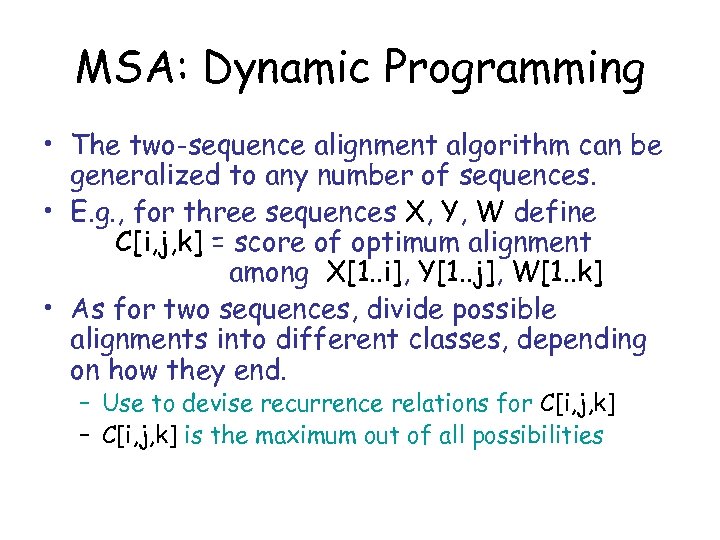 MSA: Dynamic Programming • The two-sequence alignment algorithm can be generalized to any number of sequences. • E. g. , for three sequences X, Y, W define C[i, j, k] = score of optimum alignment among X[1. . i], Y[1. . j], W[1. . k] • As for two sequences, divide possible alignments into different classes, depending on how they end. – Use to devise recurrence relations for C[i, j, k] – C[i, j, k] is the maximum out of all possibilities
MSA: Dynamic Programming • The two-sequence alignment algorithm can be generalized to any number of sequences. • E. g. , for three sequences X, Y, W define C[i, j, k] = score of optimum alignment among X[1. . i], Y[1. . j], W[1. . k] • As for two sequences, divide possible alignments into different classes, depending on how they end. – Use to devise recurrence relations for C[i, j, k] – C[i, j, k] is the maximum out of all possibilities
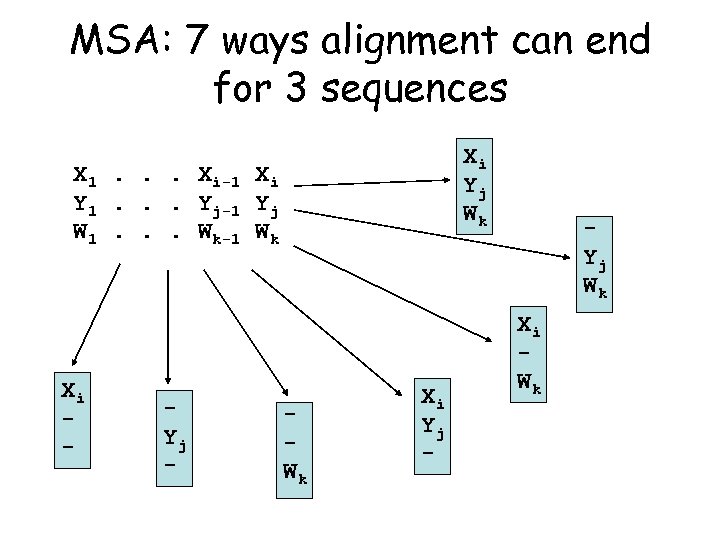 MSA: 7 ways alignment can end for 3 sequences Xi Yj Wk X 1. . . Xi-1 Xi Y 1. . . Yj-1 Yj W 1. . . Wk-1 Wk Xi - Yj - Wk Xi Yj - Yj Wk Xi Wk
MSA: 7 ways alignment can end for 3 sequences Xi Yj Wk X 1. . . Xi-1 Xi Y 1. . . Yj-1 Yj W 1. . . Wk-1 Wk Xi - Yj - Wk Xi Yj - Yj Wk Xi Wk
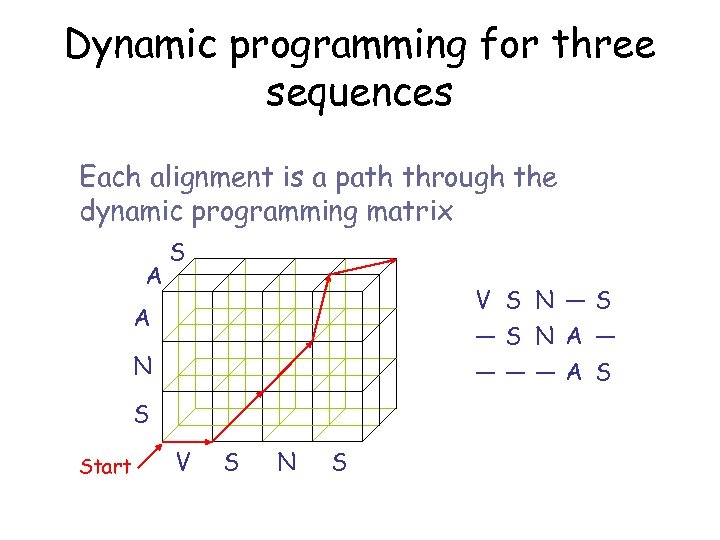 Dynamic programming for three sequences Each alignment is a path through the dynamic programming matrix A S V S N —S —S N A — ———A S A N S Start V S N S
Dynamic programming for three sequences Each alignment is a path through the dynamic programming matrix A S V S N —S —S N A — ———A S A N S Start V S N S
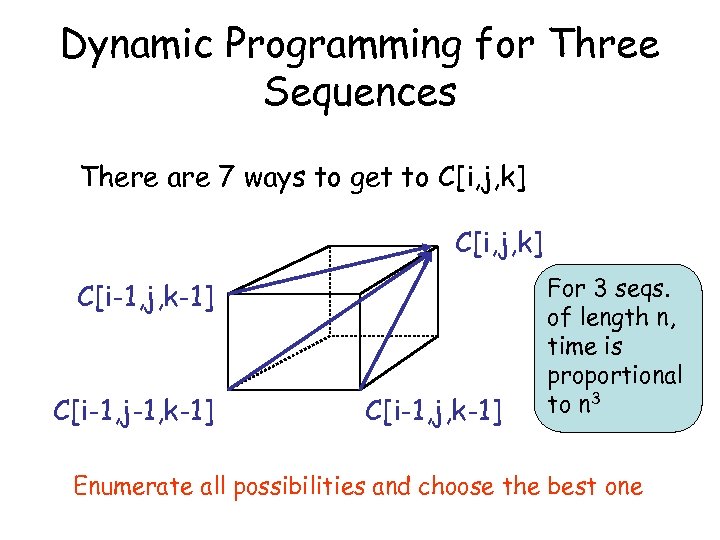 Dynamic Programming for Three Sequences There are 7 ways to get to C[i, j, k] C[i-1, j, k-1] C[i-1, j-1, k-1] C[i-1, j, k-1] For 3 seqs. of length n, time is proportional to n 3 Enumerate all possibilities and choose the best one
Dynamic Programming for Three Sequences There are 7 ways to get to C[i, j, k] C[i-1, j, k-1] C[i-1, j-1, k-1] C[i-1, j, k-1] For 3 seqs. of length n, time is proportional to n 3 Enumerate all possibilities and choose the best one
 Dynamic Programming MSA: General Case • For k sequences of length n, dynamic programming algorithm does (2 k-1) nk operations – Example: 6 sequences of length 100 require 6. 4 X 1013 calculations • Space for table is nk • Implementations (e. g. , Wash. U MSA 2. 1) use tricks and only search subset of dynamic programming table – Even this is expensive. E. g. , Baylor CM Search launcher limits MSA to 8 sequences of 800 characters and 10 minutes processing time
Dynamic Programming MSA: General Case • For k sequences of length n, dynamic programming algorithm does (2 k-1) nk operations – Example: 6 sequences of length 100 require 6. 4 X 1013 calculations • Space for table is nk • Implementations (e. g. , Wash. U MSA 2. 1) use tricks and only search subset of dynamic programming table – Even this is expensive. E. g. , Baylor CM Search launcher limits MSA to 8 sequences of 800 characters and 10 minutes processing time
 Problems with SP scoring • Pair-wise comparisons can over-score evolutionarily distant pairs. • Reason: For 3 or more sequences, SP scoring does not correspond to any evolutionary tree But not:
Problems with SP scoring • Pair-wise comparisons can over-score evolutionarily distant pairs. • Reason: For 3 or more sequences, SP scoring does not correspond to any evolutionary tree But not:
 Overcoming problems with SP scoring • Use weights to incorporate evolution in sum of pairs scoring: – Some pair-wise alignments are more important than others • E. g. , more important to have a good alignment between mouse and human sequences than mouse and bird – Assign different weights to different pair-wise alignments. • Weight decreases with evolutionary distance. • Use star tree approach – one sequence is assigned as the ancestor and all others are contrasted it.
Overcoming problems with SP scoring • Use weights to incorporate evolution in sum of pairs scoring: – Some pair-wise alignments are more important than others • E. g. , more important to have a good alignment between mouse and human sequences than mouse and bird – Assign different weights to different pair-wise alignments. • Weight decreases with evolutionary distance. • Use star tree approach – one sequence is assigned as the ancestor and all others are contrasted it.
 Star Alignments • Construct multiple alignments using pair-wise alignment relative to a fixed sequence • Out of a set S = {S 1, S 2, . . . , Sr} of sequences, pick sequence Sc that maximizes star_score(c) = ∑ {sim(Sc, Si) : 1 ≤ i ≤ r, i ≠ c} where sim(Si, Sj) is the optimal score of a pair-wise alignment between Si and Sj
Star Alignments • Construct multiple alignments using pair-wise alignment relative to a fixed sequence • Out of a set S = {S 1, S 2, . . . , Sr} of sequences, pick sequence Sc that maximizes star_score(c) = ∑ {sim(Sc, Si) : 1 ≤ i ≤ r, i ≠ c} where sim(Si, Sj) is the optimal score of a pair-wise alignment between Si and Sj
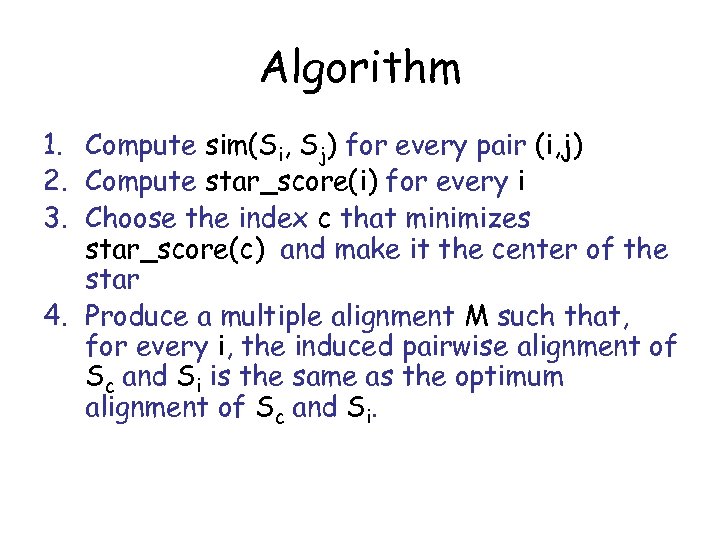 Algorithm 1. Compute sim(Si, Sj) for every pair (i, j) 2. Compute star_score(i) for every i 3. Choose the index c that minimizes star_score(c) and make it the center of the star 4. Produce a multiple alignment M such that, for every i, the induced pairwise alignment of Sc and Si is the same as the optimum alignment of Sc and Si.
Algorithm 1. Compute sim(Si, Sj) for every pair (i, j) 2. Compute star_score(i) for every i 3. Choose the index c that minimizes star_score(c) and make it the center of the star 4. Produce a multiple alignment M such that, for every i, the induced pairwise alignment of Sc and Si is the same as the optimum alignment of Sc and Si.
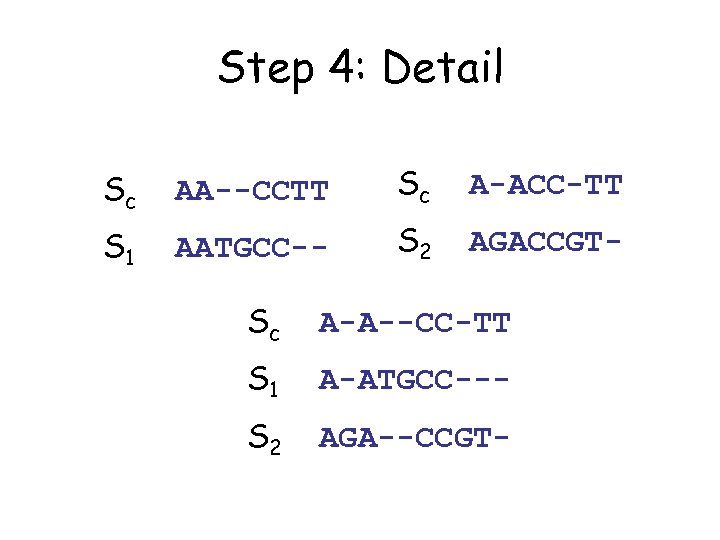 Step 4: Detail Sc AA--CCTT Sc A-ACC-TT S 1 AATGCC-- S 2 AGACCGT- Sc A-A--CC-TT S 1 A-ATGCC--- S 2 AGA--CCGT-
Step 4: Detail Sc AA--CCTT Sc A-ACC-TT S 1 AATGCC-- S 2 AGACCGT- Sc A-A--CC-TT S 1 A-ATGCC--- S 2 AGA--CCGT-


