f394ee107e814e86e1f3f9b19fdc8ec5.ppt
- Количество слайдов: 31

Monday, September 27, 4: 17: 36 PM Outline for today I. Characteristics of a good primer • Degenerated primers • Targeting multiple genes at once. • PCR-RFLP combination of two techniques • Restriction endonucleases II. III. Lec 03 Type of libraries Quantatative PCR and chemistry Slide 72
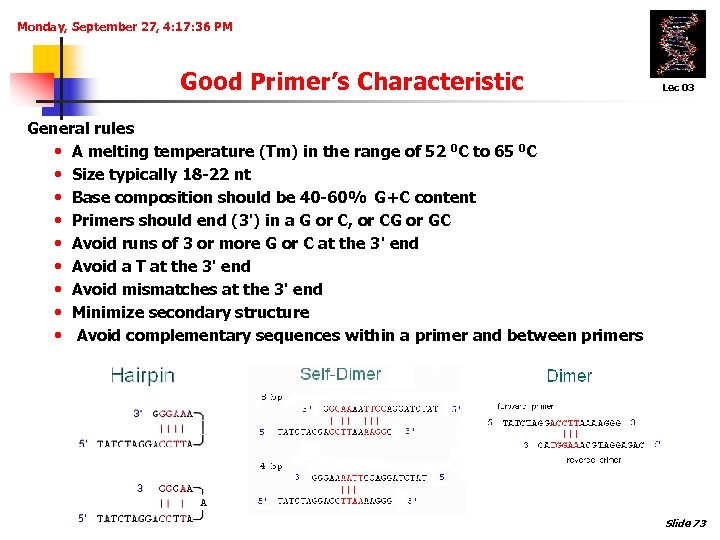
Monday, September 27, 4: 17: 36 PM Good Primer’s Characteristic Lec 03 General rules • A melting temperature (Tm) in the range of 52 0 C to 65 0 C • Size typically 18 -22 nt • Base composition should be 40 -60% G+C content • Primers should end (3') in a G or C, or CG or GC • Avoid runs of 3 or more G or C at the 3' end • Avoid a T at the 3' end • Avoid mismatches at the 3' end • Minimize secondary structure • Avoid complementary sequences within a primer and between primers Slide 73
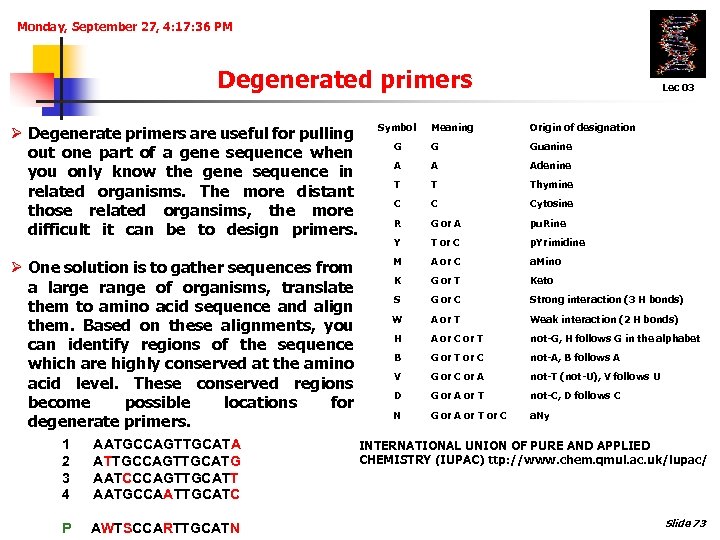
Monday, September 27, 4: 17: 36 PM Degenerated primers Ø Degenerate primers are useful for pulling out one part of a gene sequence when you only know the gene sequence in related organisms. The more distant those related organsims, the more difficult it can be to design primers. Symbol Ø One solution is to gather sequences from a large range of organisms, translate them to amino acid sequence and align them. Based on these alignments, you can identify regions of the sequence which are highly conserved at the amino acid level. These conserved regions become possible locations for degenerate primers. Lec 03 Meaning Origin of designation G G Guanine A A Adenine T T Thymine C C Cytosine R G or A pu. Rine Y T or C p. Yrimidine M A or C a. Mino K G or T Keto S G or C Strong interaction (3 H bonds) W A or T Weak interaction (2 H bonds) H A or C or T not-G, H follows G in the alphabet B G or T or C not-A, B follows A V G or C or A not-T (not-U), V follows U D G or A or T not-C, D follows C N G or A or T or C a. Ny 1 2 3 4 AATGCCAGTTGCATA ATTGCCAGTTGCATG AATCCCAGTTGCATT AATGCCAATTGCATC INTERNATIONAL UNION OF PURE AND APPLIED CHEMISTRY (IUPAC) ttp: //www. chem. qmul. ac. uk/iupac/ P AWTSCCARTTGCATN Slide 73
Monday, September 27, 4: 17: 36 PM Degenerated primers Lec 03 Ø CODEHOP program: Consensus Degenerate Hybrid Oligonucleotide Primers • You can get a multiple alignment from a group of related protein sequences using the Block Maker or other automated methods (such as Clustal. W). http: //blocks. fhcrc. org/blocks/make_blocks. html Slide 73
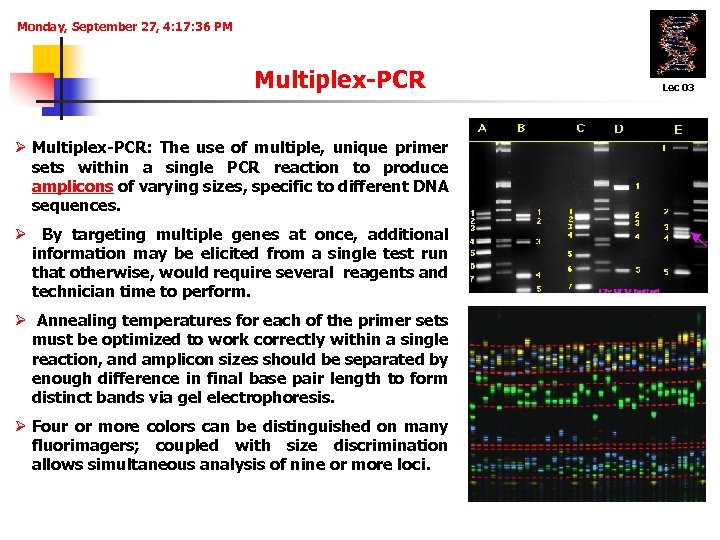
Monday, September 27, 4: 17: 36 PM Multiplex-PCR Ø Multiplex-PCR: The use of multiple, unique primer sets within a single PCR reaction to produce amplicons of varying sizes, specific to different DNA sequences. Ø By targeting multiple genes at once, additional information may be elicited from a single test run that otherwise, would require several reagents and technician time to perform. Ø Annealing temperatures for each of the primer sets must be optimized to work correctly within a single reaction, and amplicon sizes should be separated by enough difference in final base pair length to form distinct bands via gel electrophoresis. Ø Four or more colors can be distinguished on many fluorimagers; coupled with size discrimination allows simultaneous analysis of nine or more loci. Lec 03
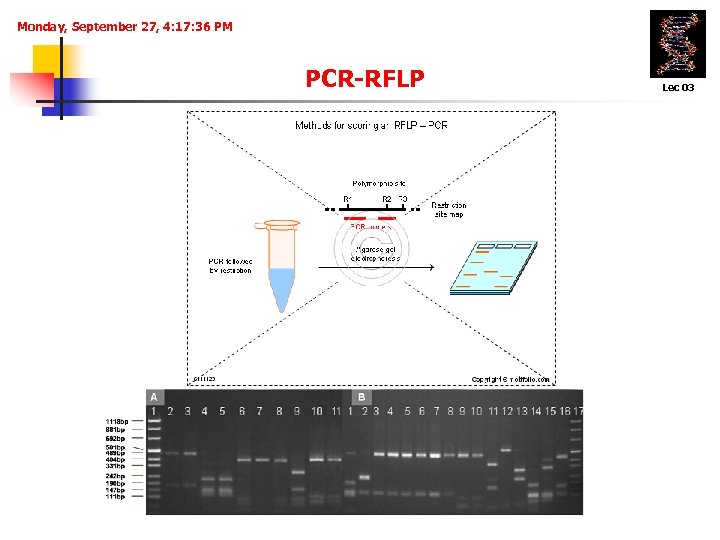
Monday, September 27, 4: 17: 36 PM PCR-RFLP Lec 03
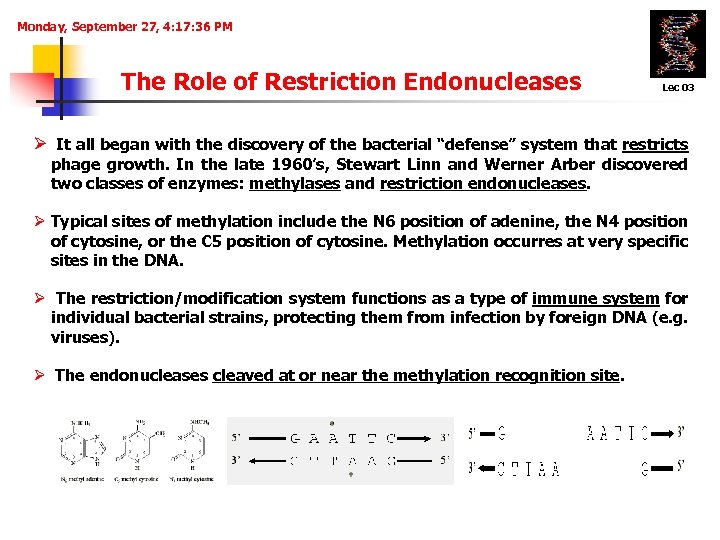
Monday, September 27, 4: 17: 36 PM The Role of Restriction Endonucleases Lec 03 Ø It all began with the discovery of the bacterial “defense” system that restricts phage growth. In the late 1960’s, Stewart Linn and Werner Arber discovered two classes of enzymes: methylases and restriction endonucleases. Ø Typical sites of methylation include the N 6 position of adenine, the N 4 position of cytosine, or the C 5 position of cytosine. Methylation occurres at very specific sites in the DNA. Ø The restriction/modification system functions as a type of immune system for individual bacterial strains, protecting them from infection by foreign DNA (e. g. viruses). Ø The endonucleases cleaved at or near the methylation recognition site.
Monday, September 27, 4: 17: 36 PM Types of Restriction Endonucleases Lec 03 Type I- multisubunit, endonuclease and methylase activities, cleave at random up to 1000 bp from recognition sequence. Type II- cleave DNA within recognition sequence, require no ATP, most monomers. Type III- multisubunit, endonuclease and methylase about 25 bp from recognition sequence Ø Restriction endonucleases are named using the 1 st three letters of their name from the Latin name of their source microorganism Hind III. • First letter is from the genus H from Haemophilus. • Next two letters are the 1 st two letters of the species name in from influenzae. • If microorganism produces only 1 restriction enzyme, end the name with Roman numeral I Hind I. • If more than one restriction enzyme is produced, the others are numbered sequentially II, IV, etc.
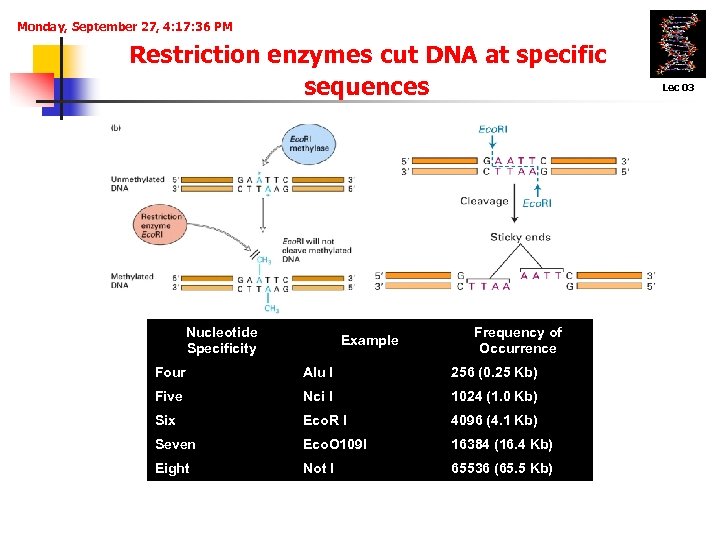
Monday, September 27, 4: 17: 36 PM Restriction enzymes cut DNA at specific sequences Nucleotide Specificity Example Frequency of Occurrence Four Alu I 256 (0. 25 Kb) Five Nci I 1024 (1. 0 Kb) Six Eco. R I 4096 (4. 1 Kb) Seven Eco. O 109 I 16384 (16. 4 Kb) Eight Not I 65536 (65. 5 Kb) Lec 03
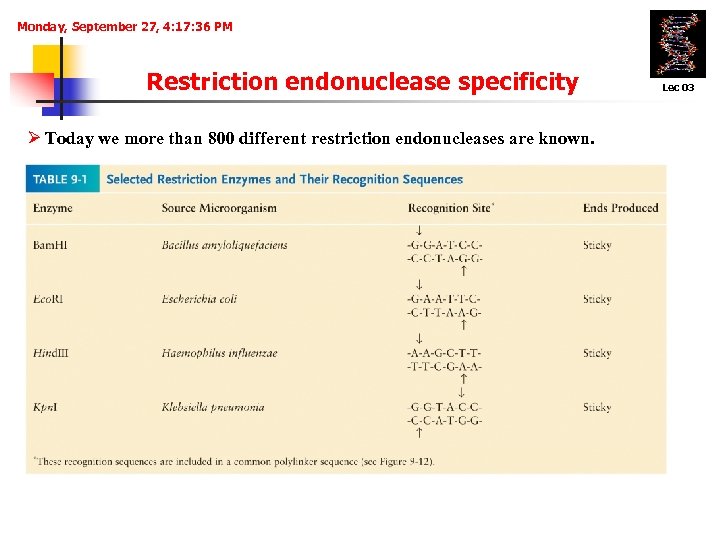
Monday, September 27, 4: 17: 36 PM Restriction endonuclease specificity Ø Today we more than 800 different restriction endonucleases are known. Lec 03

Monday, September 27, 4: 17: 36 PM libraries Lec 03 Ø The term of library represents the collection of all of the vector molecules, each carrying a piece of foriner DNA of the organism, 1. 2. 3. 4. 5. 6. Plasmids Phage Cosmids or Fosmids Bacterial artificial chromosomes (BAC) Yeast artificial chromosomes (YAC). Human artificial chromosomes (HAC)
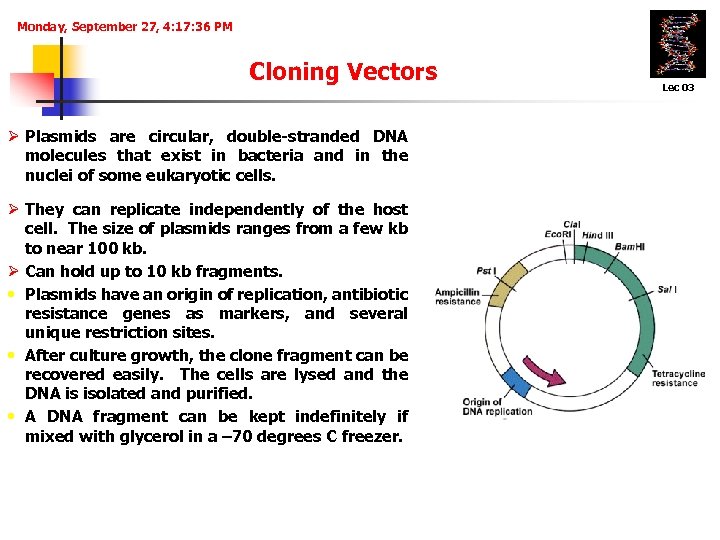
Monday, September 27, 4: 17: 36 PM Cloning Vectors Ø Plasmids are circular, double-stranded DNA molecules that exist in bacteria and in the nuclei of some eukaryotic cells. Ø They can replicate independently of the host cell. The size of plasmids ranges from a few kb to near 100 kb. Ø Can hold up to 10 kb fragments. • Plasmids have an origin of replication, antibiotic resistance genes as markers, and several unique restriction sites. • After culture growth, the clone fragment can be recovered easily. The cells are lysed and the DNA is isolated and purified. • A DNA fragment can be kept indefinitely if mixed with glycerol in a – 70 degrees C freezer. Lec 03
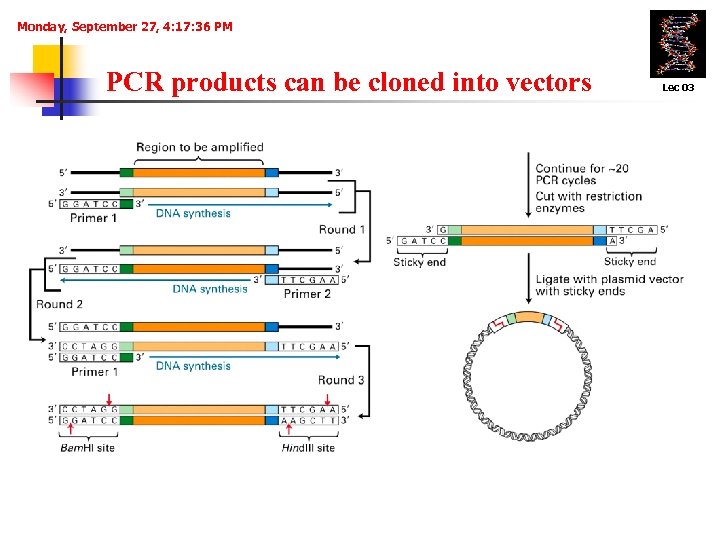
Monday, September 27, 4: 17: 36 PM PCR products can be cloned into vectors Lec 03
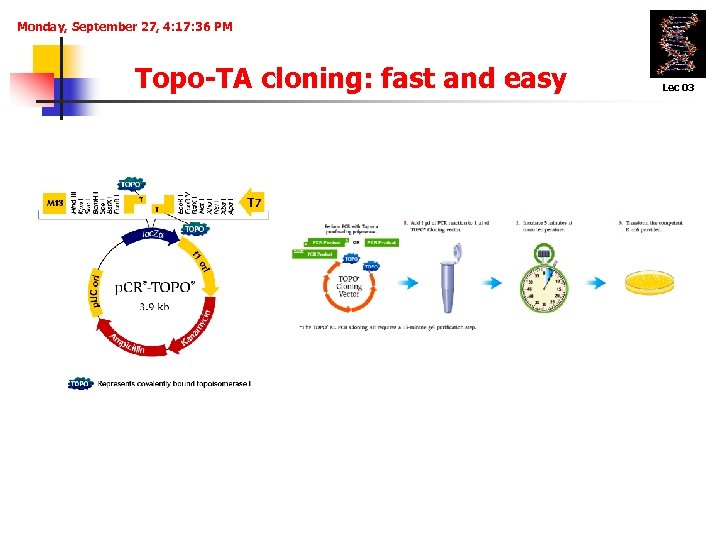
Monday, September 27, 4: 17: 36 PM Topo-TA cloning: fast and easy Lec 03
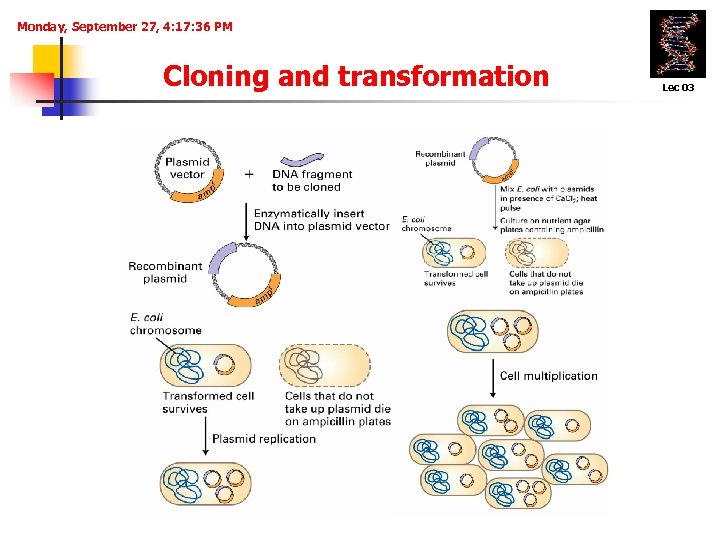
Monday, September 27, 4: 17: 36 PM Cloning and transformation Lec 03

Monday, September 27, 4: 17: 36 PM Phages As Vectors • Bacteriophages Lec 03 are natural vectors that transduce bacterial DNA from one cell to another. Ø Phage vectors infect cells much more efficiently than plasmids transform cells. Ø Clones are not colonies of cells using phage vectors, but rather plaques, a clearing of the bacterial lawn due to phage killing the bacteria in that area. Ø Accommodate more foreign DNA (~ 20 kb).
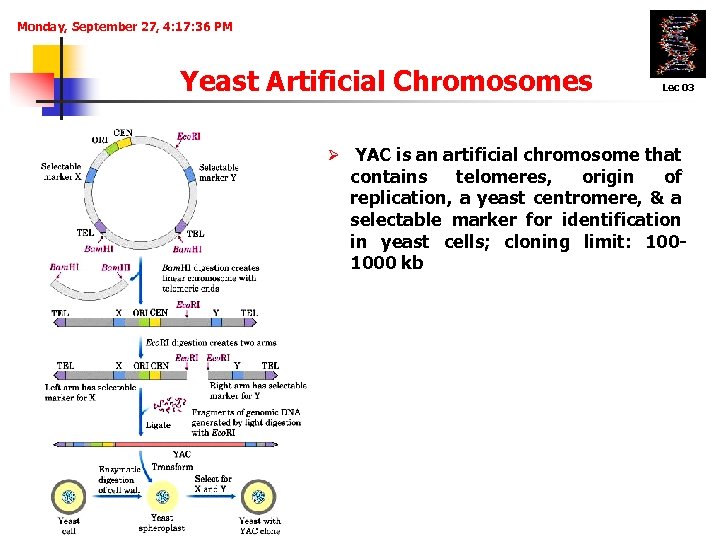
Monday, September 27, 4: 17: 36 PM Yeast Artificial Chromosomes Lec 03 Ø YAC is an artificial chromosome that contains telomeres, origin of replication, a yeast centromere, & a selectable marker for identification in yeast cells; cloning limit: 1001000 kb
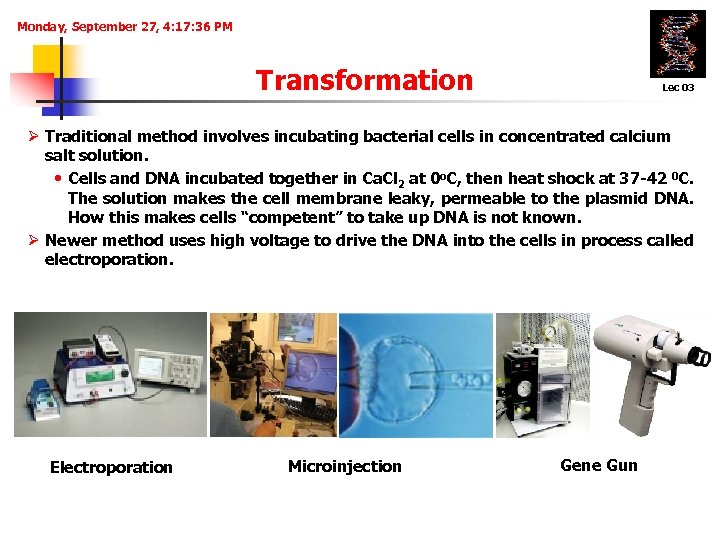
Monday, September 27, 4: 17: 36 PM Transformation Lec 03 Ø Traditional method involves incubating bacterial cells in concentrated calcium salt solution. • Cells and DNA incubated together in Ca. Cl 2 at 0 o. C, then heat shock at 37 -42 0 C. The solution makes the cell membrane leaky, permeable to the plasmid DNA. How this makes cells “competent” to take up DNA is not known. Ø Newer method uses high voltage to drive the DNA into the cells in process called electroporation. Electroporation Microinjection Gene Gun
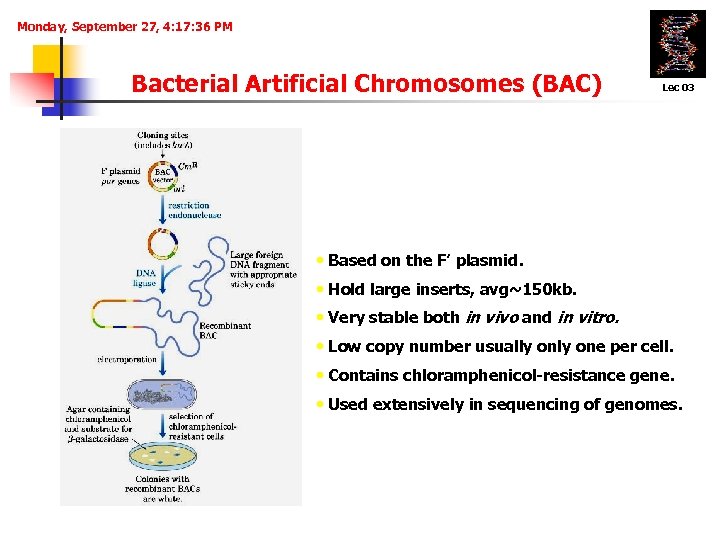
Monday, September 27, 4: 17: 36 PM Bacterial Artificial Chromosomes (BAC) Lec 03 • Based on the F’ plasmid. • Hold large inserts, avg~150 kb. • Very stable both in vivo and in vitro. • Low copy number usually one per cell. • Contains chloramphenicol-resistance gene. • Used extensively in sequencing of genomes.
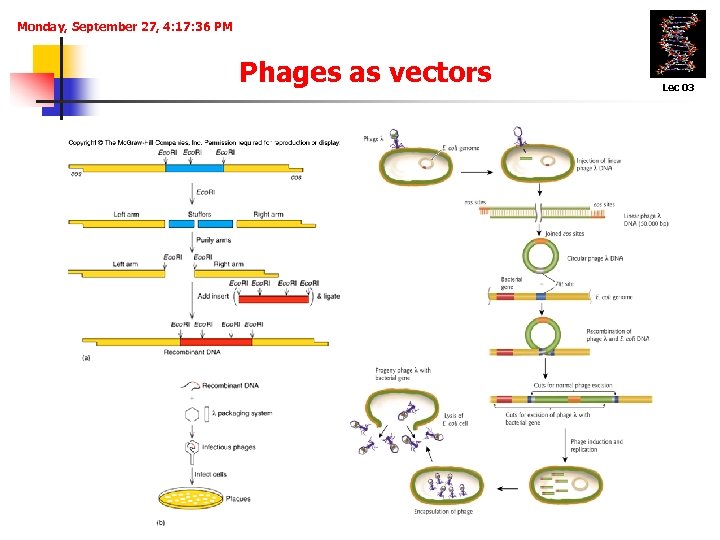
Monday, September 27, 4: 17: 36 PM Phages as vectors Lec 03
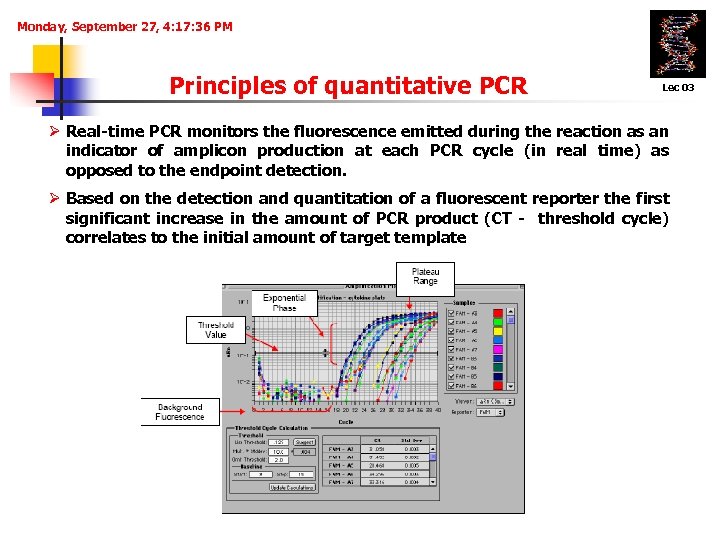
Monday, September 27, 4: 17: 36 PM Principles of quantitative PCR Lec 03 Ø Real-time PCR monitors the fluorescence emitted during the reaction as an indicator of amplicon production at each PCR cycle (in real time) as opposed to the endpoint detection. Ø Based on the detection and quantitation of a fluorescent reporter the first significant increase in the amount of PCR product (CT - threshold cycle) correlates to the initial amount of target template
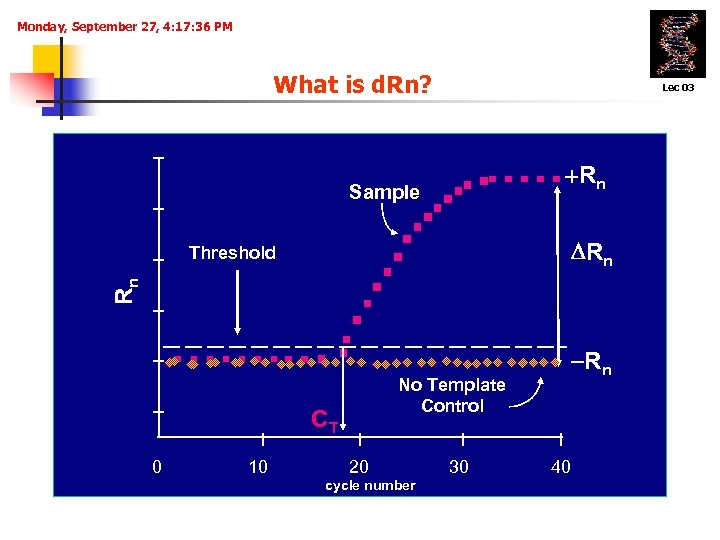
Monday, September 27, 4: 17: 36 PM What is d. Rn? Lec 03 +Rn Sample Rn Rn Threshold No Template Control CT 0 10 -Rn 20 cycle number 30 40
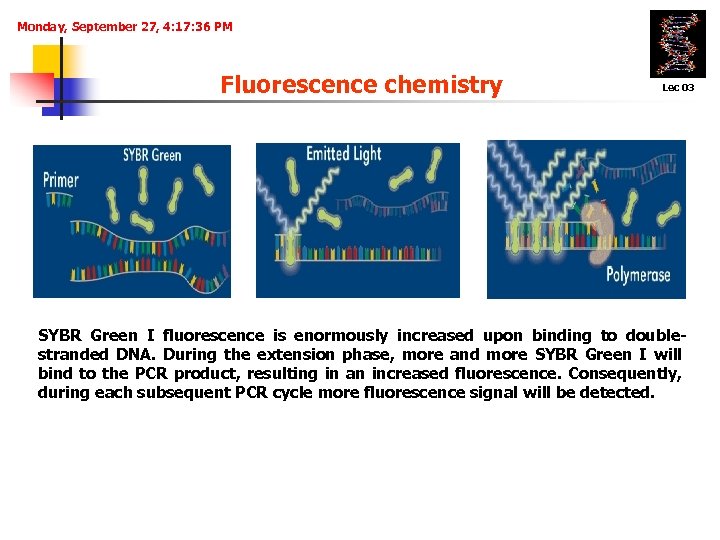
Monday, September 27, 4: 17: 36 PM Fluorescence chemistry Lec 03 1. DNA-binding agents (SYBR I Green technique) Denaturation SYBR Green 95°C Annealing (60°C) enormously Extension (72°C) binding to I fluorescence is increased upon doublestranded DNA. During the extension phase, more and more SYBR Green I will bind to the PCR product, resulting in an increased fluorescence. Consequently, during each subsequent PCR cycle more fluorescence signal will be detected.
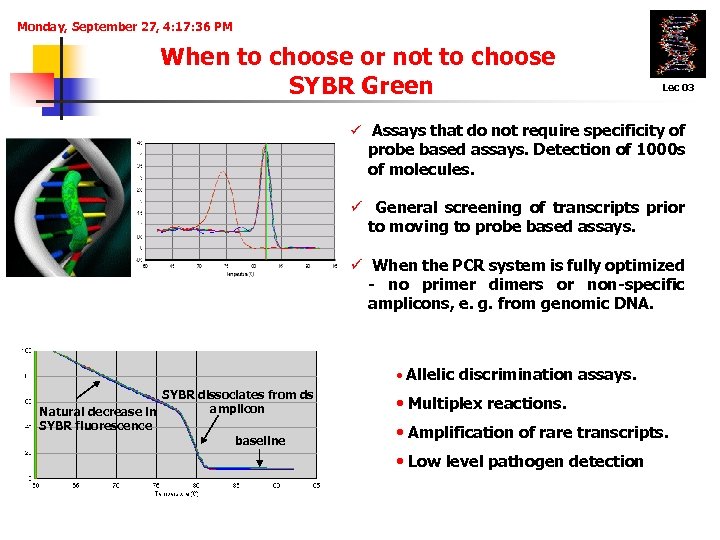
Monday, September 27, 4: 17: 36 PM When to choose or not to choose SYBR Green Lec 03 ü Assays that do not require specificity of probe based assays. Detection of 1000 s of molecules. ü General screening of transcripts prior to moving to probe based assays. ü When the PCR system is fully optimized - no primer dimers or non-specific amplicons, e. g. from genomic DNA. • Allelic discrimination assays. SYBR dissociates from ds amplicon Natural decrease in SYBR fluorescence baseline • Multiplex reactions. • Amplification of rare transcripts. • Low level pathogen detection
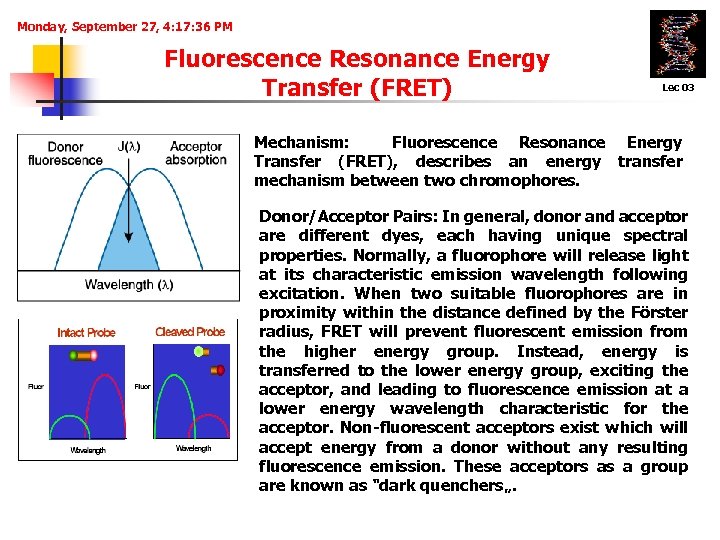
Monday, September 27, 4: 17: 36 PM Fluorescence Resonance Energy Transfer (FRET) Lec 03 Mechanism: Fluorescence Resonance Energy Transfer (FRET), describes an energy transfer mechanism between two chromophores. Donor/Acceptor Pairs: In general, donor and acceptor are different dyes, each having unique spectral properties. Normally, a fluorophore will release light at its characteristic emission wavelength following excitation. When two suitable fluorophores are in proximity within the distance defined by the Förster radius, FRET will prevent fluorescent emission from the higher energy group. Instead, energy is transferred to the lower energy group, exciting the acceptor, and leading to fluorescence emission at a lower energy wavelength characteristic for the acceptor. Non-fluorescent acceptors exist which will accept energy from a donor without any resulting fluorescence emission. These acceptors as a group are known as "dark quenchers„.
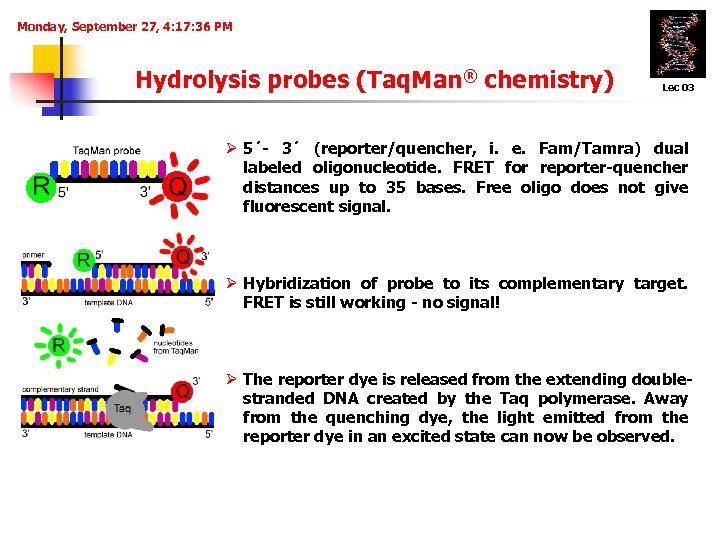
Monday, September 27, 4: 17: 36 PM Hydrolysis probes (Taq. Man® chemistry) 1. Lec 03 Ø 5´- 3´ (reporter/quencher, i. e. Fam/Tamra) dual labeled oligonucleotide. FRET for reporter-quencher distances up to 35 bases. Free oligo does not give fluorescent signal. 2. Ø Hybridization of probe to its complementary target. FRET is still working - no signal! 3. Ø The reporter dye is released from the extending doublestranded DNA created by the Taq polymerase. Away from the quenching dye, the light emitted from the reporter dye in an excited state can now be observed.
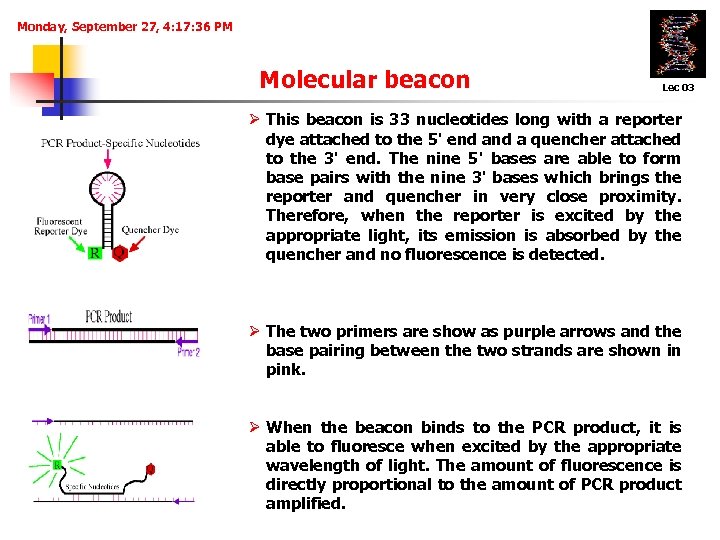
Monday, September 27, 4: 17: 36 PM Molecular beacon Lec 03 Ø This beacon is 33 nucleotides long with a reporter dye attached to the 5' end a quencher attached to the 3' end. The nine 5' bases are able to form base pairs with the nine 3' bases which brings the reporter and quencher in very close proximity. Therefore, when the reporter is excited by the appropriate light, its emission is absorbed by the quencher and no fluorescence is detected. Ø The two primers are show as purple arrows and the base pairing between the two strands are shown in pink. Ø When the beacon binds to the PCR product, it is able to fluoresce when excited by the appropriate wavelength of light. The amount of fluorescence is directly proportional to the amount of PCR product amplified.
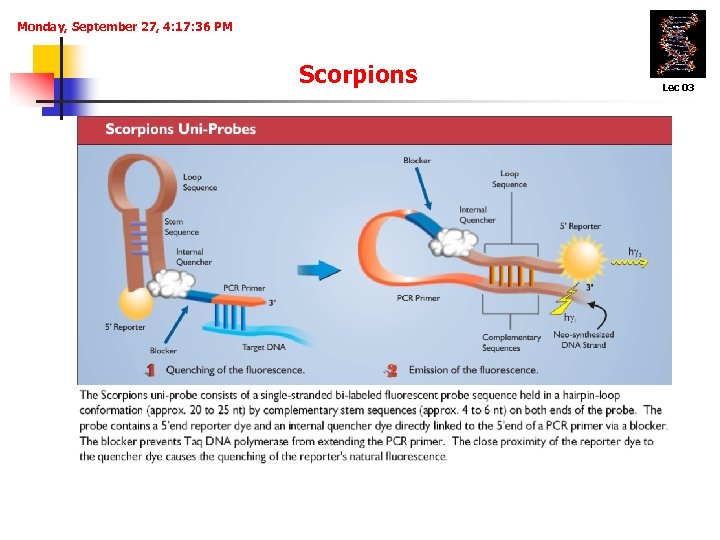
Monday, September 27, 4: 17: 36 PM Scorpions Lec 03
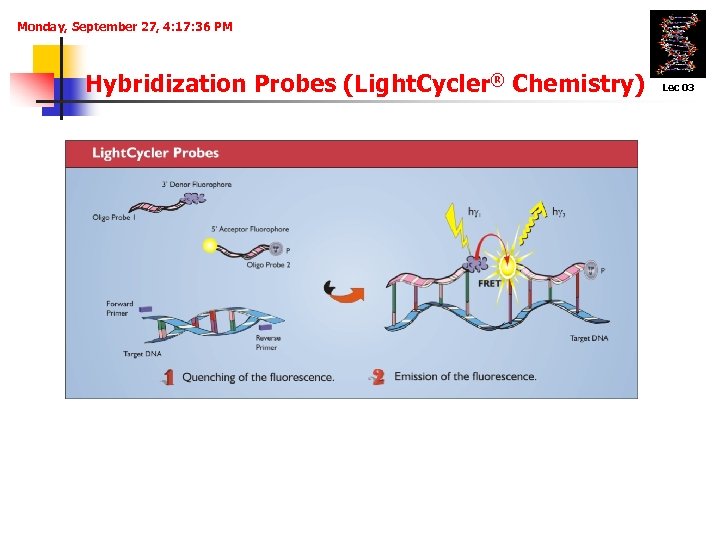
Monday, September 27, 4: 17: 36 PM Hybridization Probes (Light. Cycler® Chemistry) Lec 03 Hybridization of probes to the amplified DNA fragment in a head to tail arrangement. Positioning of the two fluorescence dyes in close proximity to each other. Fluorescence measurement is performed after the annealing step.
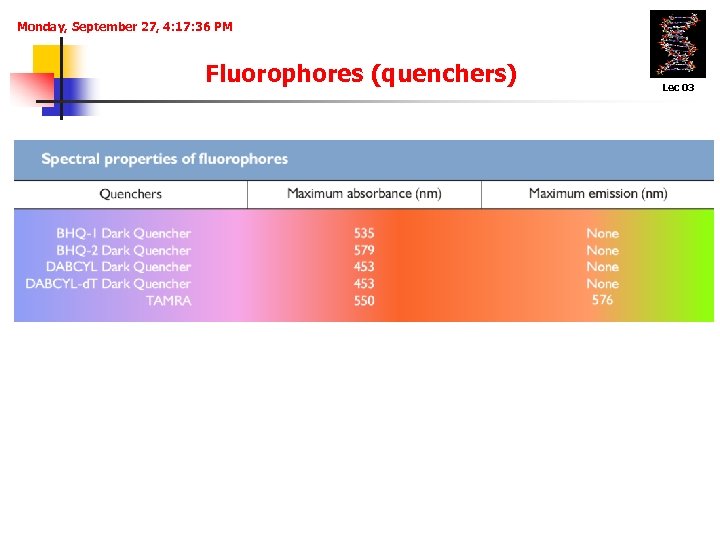
Monday, September 27, 4: 17: 36 PM Fluorophores (quenchers) Lec 03
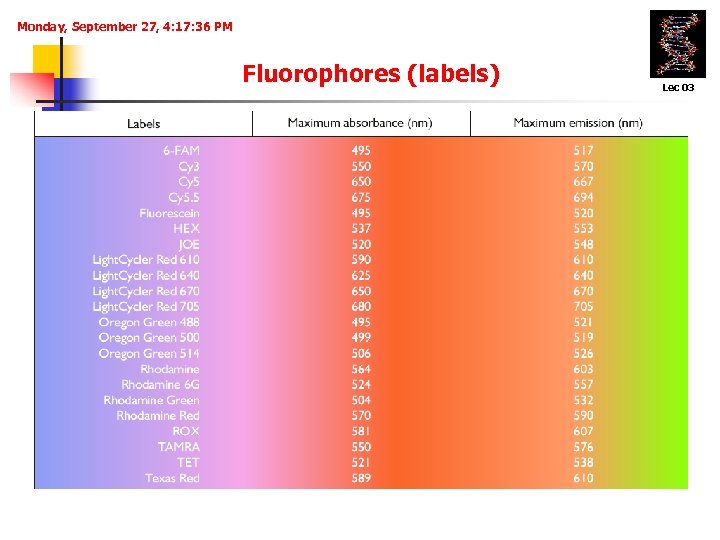
Monday, September 27, 4: 17: 36 PM Fluorophores (labels) Lec 03
f394ee107e814e86e1f3f9b19fdc8ec5.ppt