16bdf61efeae576f15ec5b35dce9409f.ppt
- Количество слайдов: 42
Molecular Characterization of Infectious Bronchitis Virus and Newcastle Disease Virus in Layer and Broiler Flocks in different parts of Jordan Ziad W Jaradat 1 and Mustafa M Ababneh 2 1 Fatima College Of Health Sciences, Al Ain, UAE; 2 Jordan University of Science and Technology, Irbid, Jordan
Avian infectious bronchitis virus (IBV) It is a highly contagious viral respiratory pathogen of chicken mainly. Produce severe economic losses due to mortality, decrease in egg production and affect both internal and external egg quality. The disease occur worldwide. Transmission: Airborne, Direct contact, Indirect by contaminated equipment's. Incubation period: 24 -48 hrs (average = 36 hrs) IBV affects mainly 3 systems in chicken: Respiratory Kidneys Oviduct
Avian Infectious Bronchitis Virus Classification Genus: Coronavirus Family: Coronaviridae Order: Nidovirales Characteristics of Coronaviruses: ü Enveloped ü Non-segmented ü Positive-sense ü have Single-stranded RNA genome
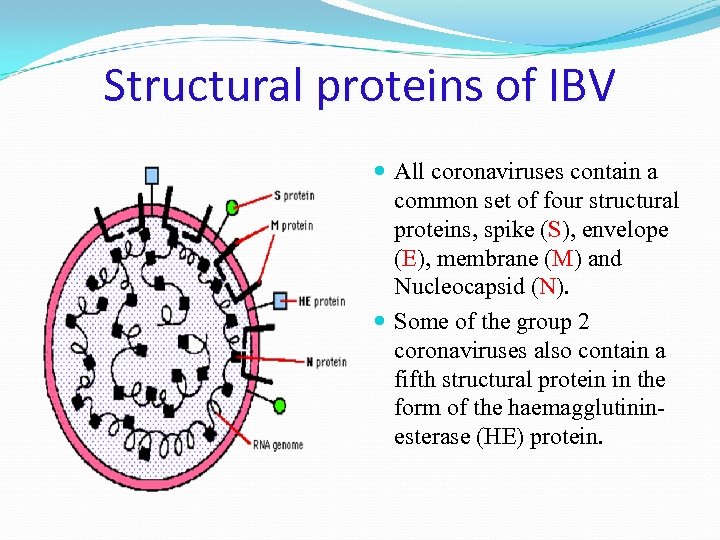
Structural proteins of IBV All coronaviruses contain a common set of four structural proteins, spike (S), envelope (E), membrane (M) and Nucleocapsid (N). Some of the group 2 coronaviruses also contain a fifth structural protein in the form of the haemagglutininesterase (HE) protein.
Newcastle disease Virus (NDV) Newcastle disease (ND) is a highly contagious viral disease and is a continuous threat to the poultry industry worldwide Newcastle disease virus, is a negative-sense, singlestranded RNA virus. NDV strains can be classified as velogenic, mesogenic or lentogenic Phylogenetic classification Order: Mononegavirales Family: Paramyxoviridae Genus: Avulavirus
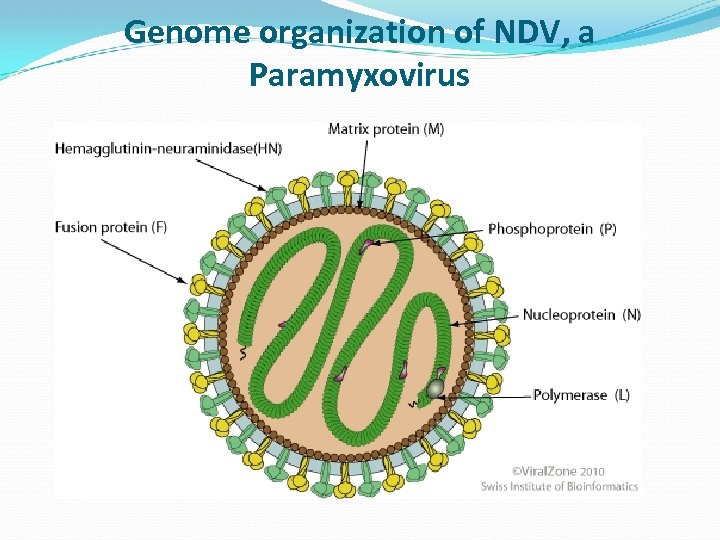
Genome organization of NDV, a Paramyxovirus
Diagnosis of IBV and NDV diagnosis is performed based on: Clinical history, Signs and lesions. Serology (ELISA, Hemagglutination [HA] and Hemagglutinatioin inhibition [HI], Virus Neutralization [VN]) Isolation using specific pathogen free (SPF) embryonated chicken Detection by RT-PCR or Real-Time PCR DNA Sequencing Diagnosis of IBV should include, if possible, identification of the serotype or genotype of the virus because of the great antigenic variation exhibited by IBV strains and the availability of vaccines designed for different serotypes.
Previous IB studies in Jordan 1. In one study serum samples were collected from 20 chicken flocks with respiratory disease; 14 broiler. 5 broiler breeder and only one layer flock. Samples were tested by ELISA at initial signs and repeated samples at 10 -14 days later. ELISA titer increased in 70% of flocks after 10 -14 days of clinical signs. Results indicated an exposure of some flocks to different serotypes such as Ark, DE-072 and Mass like serotypes (Gharaibeh, 2007). 2. In another study about 175 tracheal swabs were obtained from broiler flocks at acute phase of respiratory disease and tested by RTPCR. Results indicated 60% of samples were positive for IB. Among the positive results, serotype Mass accounted for 35. 2%, serotype 4/91 accounted for 31. 4%, while serotype D 274 accounted for only 8. 6% (Roussan et al, 2008).

Previous studies cont… 3. A third study conducted by Roussan et al, 2009 performed on 51 chicken flocks; 25 broiler, 15 layer and 11 broiler breeder suffering from respiratory disease. Samples were tested for IBV using RT-PCR. About 64% of the positive samples were broilers while 53% and 54% of the layers and the breeders where positive respectively.
Previous studies Cont…. Recently 5 IBV isolates (JOA 2, JOA 4, Saudi-1, Saudi-2 and Iraqi) were detected by Nested RT-PCR. Strain identification was characterized by sequencing and phylogenetic analysis of the amplified hypervariable region of the spike 1 (S 1) gene. The 5 isolates were found CK/CH/LDL/971. Similarity between the 5 isolates ranged from 96. 9% - 99. 7%, and between these isolates and the CK/CH/ LDL/971 strain was ranged from 96. 6 to 99. 1%. The sequenced fragment of the S 1 gene of the CK/CH/ LDL/971 had less than 80% nucleotide identity to IBV vaccine strains commonly used in the Middle East (M 41 and H 120). (Ababneh, et al. , 2012).

NDV in Jordan NDV is endemic in Jordan with multiple outbreaks in different parts of Jordan. In 2011, an outbreak of NDV was characterized and partial sequence of fusion gene was published. The causative virus found to be of lineage 5 d and closely related to the Chinese strain of NDV.

Objectives of this study: ØTo survey all operating layer farms in Northern Jordan for the presence of IBV. ØTo characterize the isolated IBV strains using RT-PCR and sequencing assays targeting spike and nucleocapsid genes. ØTo characterize the NDV isolates from broiler farms in Jordan.
Materials and Methods This study was done in 40 operating layer farms for the IBV and on 40 broiler farms for the NDV. From each flock 10 birds were randomly selected and euthanized, 10 tracheas were collected under minimal contamination conditions. Tracheas where stored at (– 70°C) for virus isolation until used.
Tissue Samples and Viral RNA Extraction: Viral RNA was isolated from Tracheal samples which were stored at − 70◦C in RNAlater® (Qiagen, Germany). Homogenized tissues were subjected to viral RNA extraction using the Viral Gene-spin Viral DNA/RNA extraction kit (Intron Biotechnology, Korea), as per the manufacturer’s instructions. Extracted viral RNA was stored at − 70◦C until tested by RT-PCR.
RT-PCR (Diagnostic and Phylogenetic). The reverse transcription (RT) step was performed using an RT system (Promega, USA). Briefly, viral RNA was denatured at 70 o. C for 10 minutes, followed by the addition of a reaction mix including 4μL Mg. Cl 2, 2μL reverse transcription 10 x buffer, 2μL d. NTP mixture (10 m. M), 0. 5 μL random primers, 0. 75μL AMV reverse transcriptase enzyme, 1μg RNA, and nuclease-free water to a final volume of 20μL.

Then, the reaction was incubated at 42 o. C for 60 minutes followed by 94 o. C for 5 minutes. The c. DNA was diluted up to 100 μL with nucleasefree water for PCR amplification.
RT-PCR Assay Two PCR assays were used for IBV. 1. A diagnostic-nested RT-PCR assay based on the amplification of the neucleocapsid (N) gene 2. A phylogenetic-nested assay for spike (S) gene. IBV-specific primers for the N gene were obtained according to published primers sequences (Farsang, et al 2002). The phylogenetic PCR was performed using nested spike gene primers according as per published protocols (Jones 2005). For NDV , a nested PCR was employed for the partial amplification of Fusion gene following protocol by Nanthakumar, et al (2000).

Results
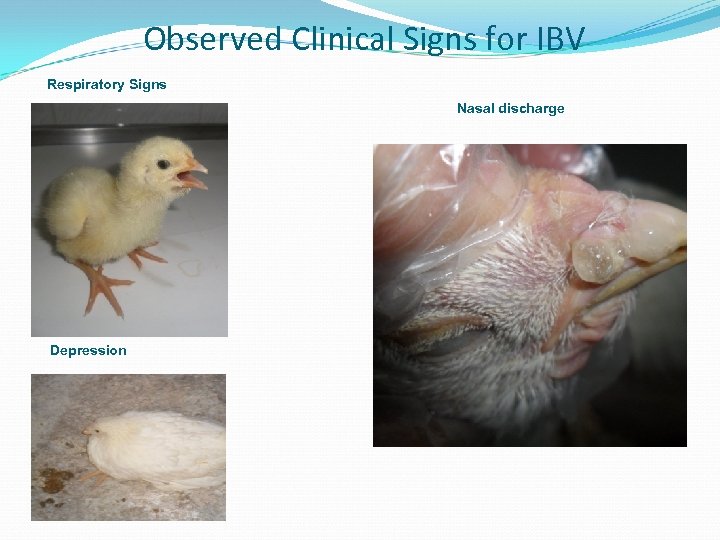
Observed Clinical Signs for IBV Respiratory Signs Nasal discharge Depression

Eggshell deformity

Lesions of IBV
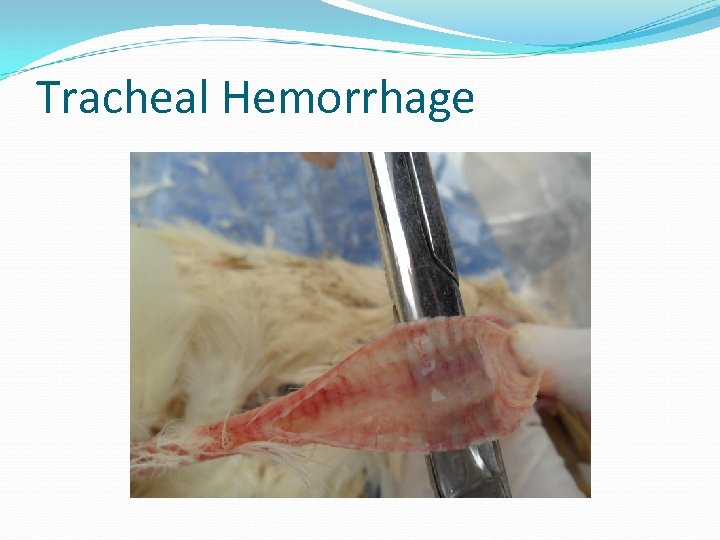
Tracheal Hemorrhage
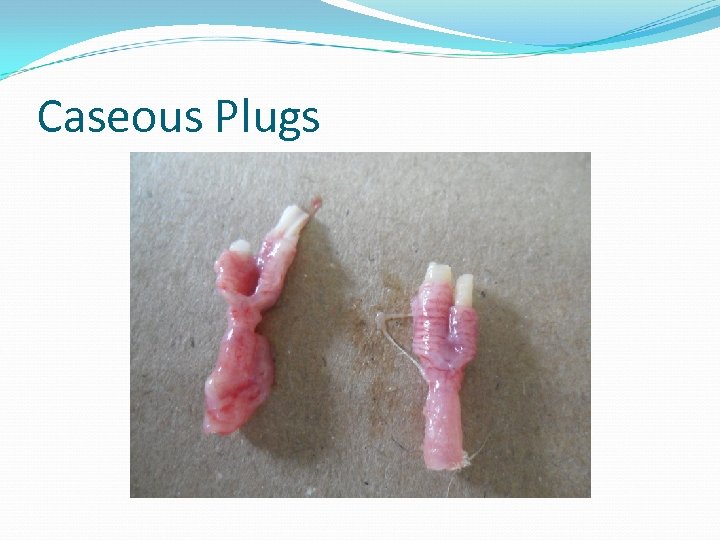
Caseous Plugs
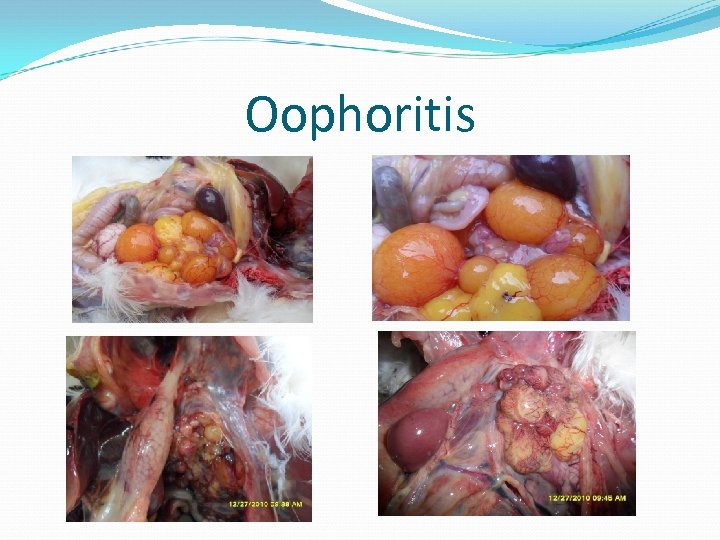
Oophoritis
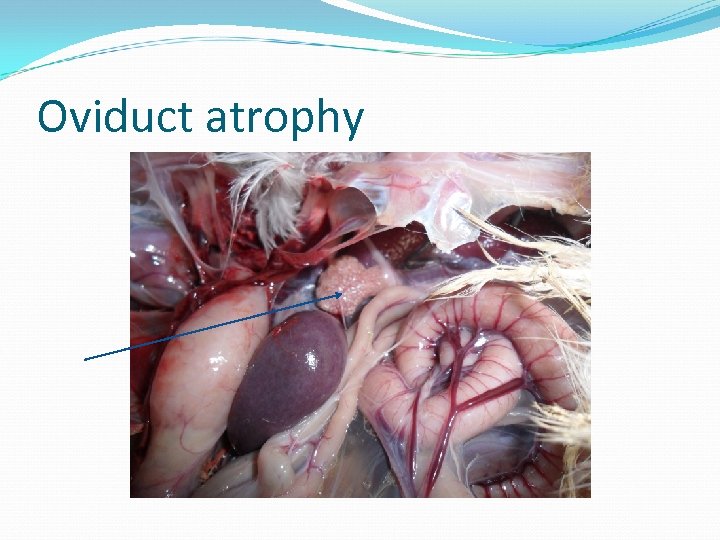
Oviduct atrophy
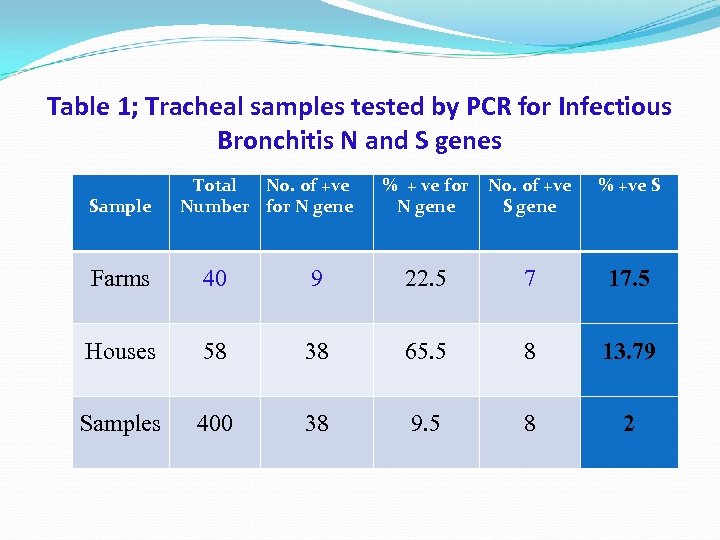
Table 1; Tracheal samples tested by PCR for Infectious Bronchitis N and S genes Sample Total No. of +ve Number for N gene % + ve for N gene No. of +ve S gene % +ve S Farms 40 9 22. 5 7 17. 5 Houses 58 38 65. 5 8 13. 79 Samples 400 38 9. 5 8 2
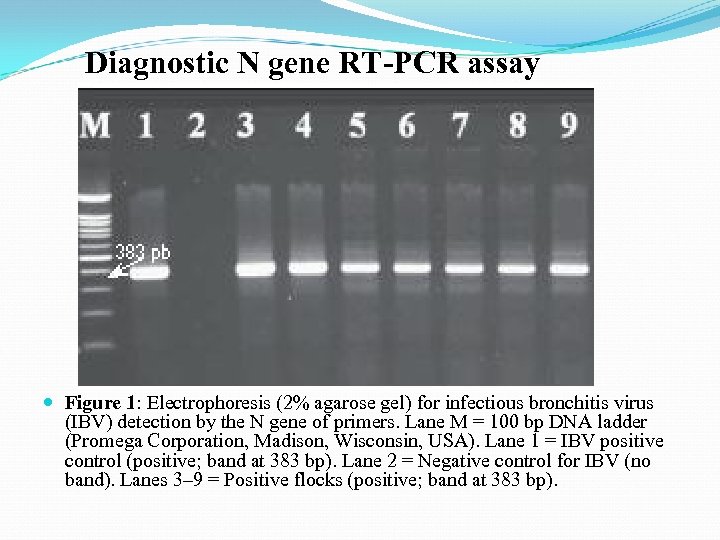
Diagnostic N gene RT-PCR assay Figure 1: Electrophoresis (2% agarose gel) for infectious bronchitis virus (IBV) detection by the N gene of primers. Lane M = 100 bp DNA ladder (Promega Corporation, Madison, Wisconsin, USA). Lane 1 = IBV positive control (positive; band at 383 bp). Lane 2 = Negative control for IBV (no band). Lanes 3– 9 = Positive flocks (positive; band at 383 bp).
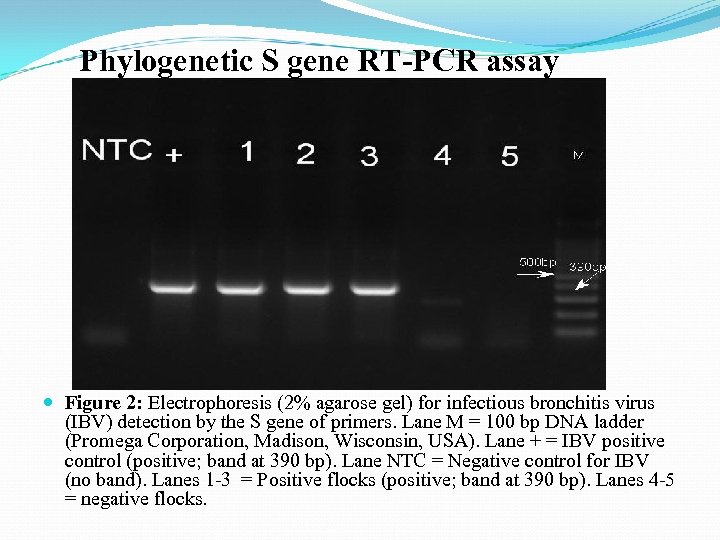
Phylogenetic S gene RT-PCR assay Figure 2: Electrophoresis (2% agarose gel) for infectious bronchitis virus (IBV) detection by the S gene of primers. Lane M = 100 bp DNA ladder (Promega Corporation, Madison, Wisconsin, USA). Lane + = IBV positive control (positive; band at 390 bp). Lane NTC = Negative control for IBV (no band). Lanes 1 -3 = Positive flocks (positive; band at 390 bp). Lanes 4 -5 = negative flocks.
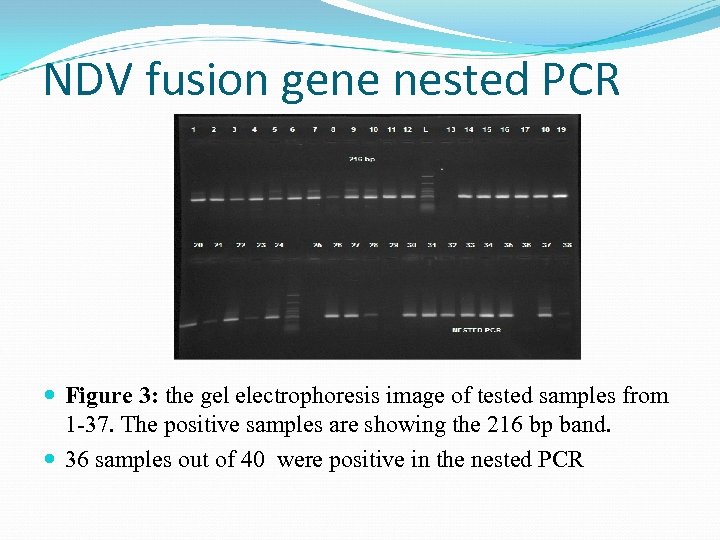
NDV fusion gene nested PCR Figure 3: the gel electrophoresis image of tested samples from 1 -37. The positive samples are showing the 216 bp band. 36 samples out of 40 were positive in the nested PCR
DNA Sequencing Amplified partial PCR products of S gene on gel were sliced then these PCR products were submitted to the Macrogen Co. Seoul, Korea for sequencing using big dye terminator technology. Sequence analysis procedure was carried out using lasergene Software. Sequence alignment was conducted using reference IBV and NDV strains obtained from Gen. Bank.
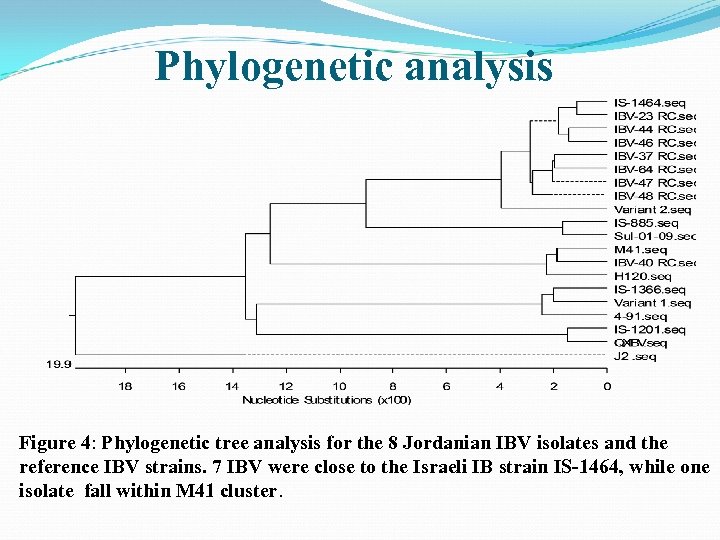
Phylogenetic analysis Figure 4: Phylogenetic tree analysis for the 8 Jordanian IBV isolates and the reference IBV strains. 7 IBV were close to the Israeli IB strain IS-1464, while one isolate fall within M 41 cluster.
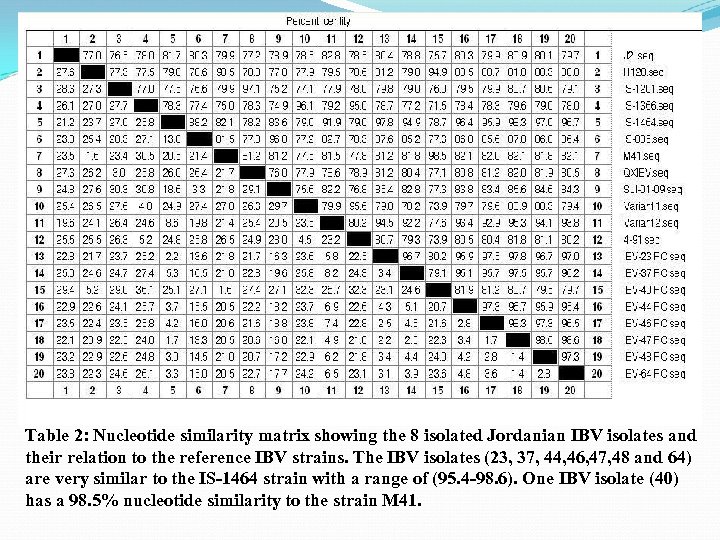
Table 2: Nucleotide similarity matrix showing the 8 isolated Jordanian IBV isolates and their relation to the reference IBV strains. The IBV isolates (23, 37, 44, 46, 47, 48 and 64) are very similar to the IS-1464 strain with a range of (95. 4 -98. 6). One IBV isolate (40) has a 98. 5% nucleotide similarity to the strain M 41.
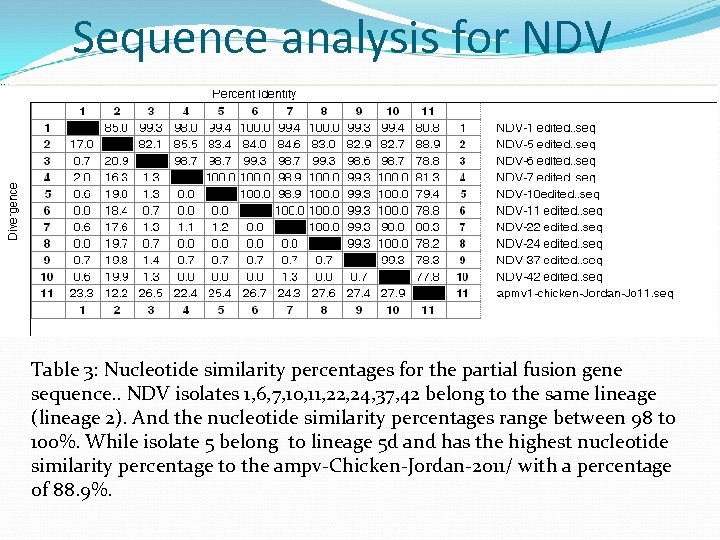
Sequence analysis for NDV Table 3: Nucleotide similarity percentages for the partial fusion gene sequence. . NDV isolates 1, 6, 7, 10, 11, 22, 24, 37, 42 belong to the same lineage (lineage 2). And the nucleotide similarity percentages range between 98 to 100%. While isolate 5 belong to lineage 5 d and has the highest nucleotide similarity percentage to the ampv-Chicken-Jordan-2 o 11/ with a percentage of 88. 9%.
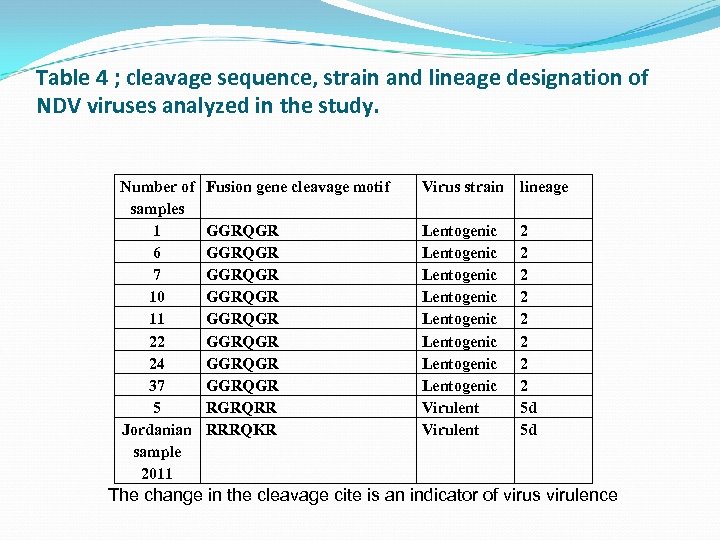
Table 4 ; cleavage sequence, strain and lineage designation of NDV viruses analyzed in the study. Number of samples 1 6 7 10 11 22 24 37 5 Jordanian sample 2011 Fusion gene cleavage motif Virus strain lineage GGRQGR GGRQGR RGRQRR RRRQKR Lentogenic Lentogenic Virulent 2 2 2 2 5 d 5 d The change in the cleavage cite is an indicator of virus virulence
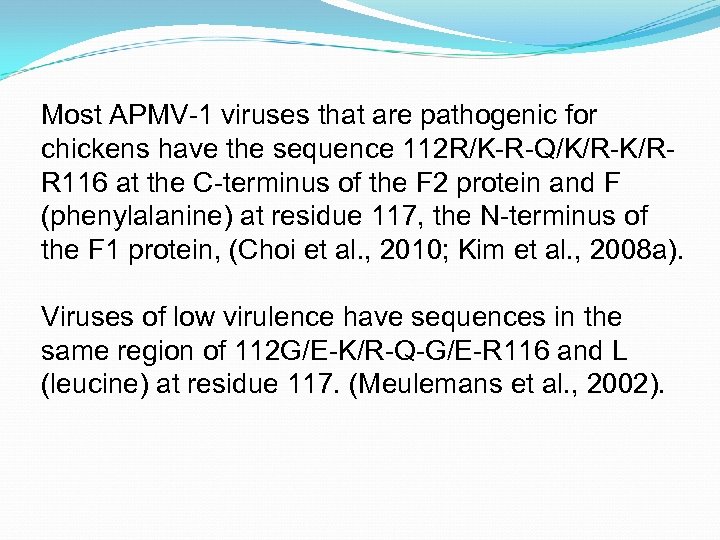
Most APMV-1 viruses that are pathogenic for chickens have the sequence 112 R/K-R-Q/K/R-K/RR 116 at the C-terminus of the F 2 protein and F (phenylalanine) at residue 117, the N-terminus of the F 1 protein, (Choi et al. , 2010; Kim et al. , 2008 a). Viruses of low virulence have sequences in the same region of 112 G/E-K/R-Q-G/E-R 116 and L (leucine) at residue 117. (Meulemans et al. , 2002).

Thus, there appears to be the requirement of at least one pair of basic amino acids at residues 116 and 115 plus a phenylalanine at residue 117 and a basic amino acid (R) at 113 if the virus is to show virulence for chickens.
Discussion The numbers of positive results in the highly conserved N gene PCR were more than the positive results for the more variable S gene PCR. For Example, 9 out of 40 farms were positive for the N gene (22. 5%), while 7 farms (17. 5%) were positive for S gene). It is known that the N gene is more stable and abundant than the S gene (S. Youn, 2003, J. You et al. , 2005, D. Y. Zhang et al. , 2005).
Eight samples were sequenced, 7 of them (87. 5%) were similar to the IBV strain of IS/1464/06. This strain was reported in: Israel and known as variant II –like strain (Meir, R. and Maharat, O. Unpublished data). Egypt (Mansoura) (El-Mahdy, S. S. , El-Hady, M. M. , Soliman, Y. A. , 2010). Turkey: EU 780077 (IS/ 1464/06) IBV strain that was named as IS/1464/06 in turkey (Kahya, S. , Coven, F. , Temelli, S. , Eyigor, A. , Carli, K. T. 2013).
One sample was found by partial S sequencing to be similar to IBV strain M 41. This pathogenic M 41 strain was isolated in Jordan earlier, Roussan et al, 2009 reported the overall seroprevalence for M 41 (92. 9%), 4/91 (90%), and D 274 (61. 4%), among 40 broiler, 18 layer and 12 broiler breeder. However, they reported (58. 8%) overall PCR positive for IBV without identifying the strain.
For NDV, 36 farms were positive for the presence of NDV virus by nested PCR. Nine samples were selected randomly for sequencing Upon sequencing, 8 samples were identified as lineage 2 and having a sequence of GGRQGR in the cleavage site indicating the lentogenic nature of the virus. While one isolate (No. 5) has a RGRQRR indicating the velogenic nature of this isolate and belong to the genus 5 d.
Conclusion Most of IBV isolates in Jordan are closely related to the IS-1464 strain. Most of NDV isolates are of lineage 2. and one sample was of lineage 5 d.

Thank you
16bdf61efeae576f15ec5b35dce9409f.ppt