mol biol 3,4.ppt
- Количество слайдов: 78
Molecular biology 3, 4 Transcription and translation 1
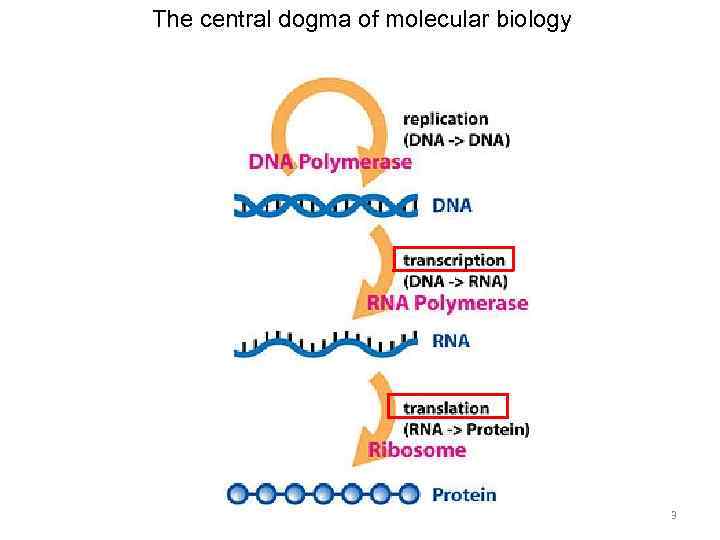
The central dogma of molecular biology 2
The central dogma of molecular biology 3
4
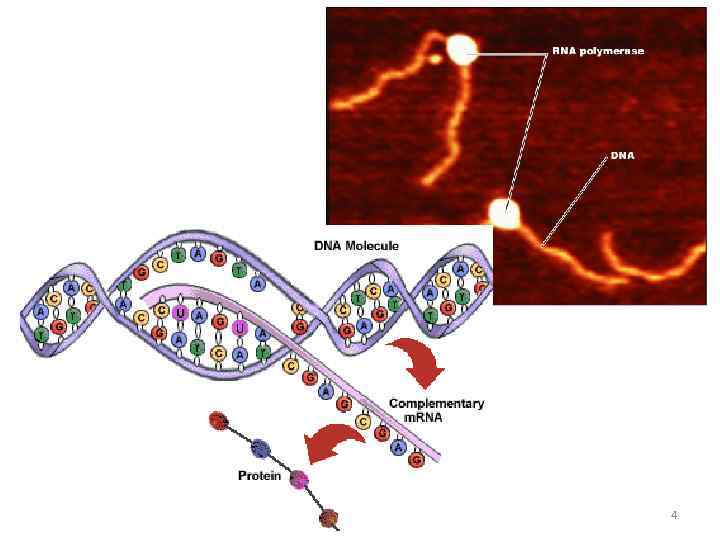
DNA transcription a process of constructing RNA molecule using DNA molecule as a template with resulting transfer of genetic information to the RNA The principles: • complementarity of parent and daughter chains • unipolarity (from 5’ to 3’) • without priming • anti parallel arrangement of parent and daughter chains 5
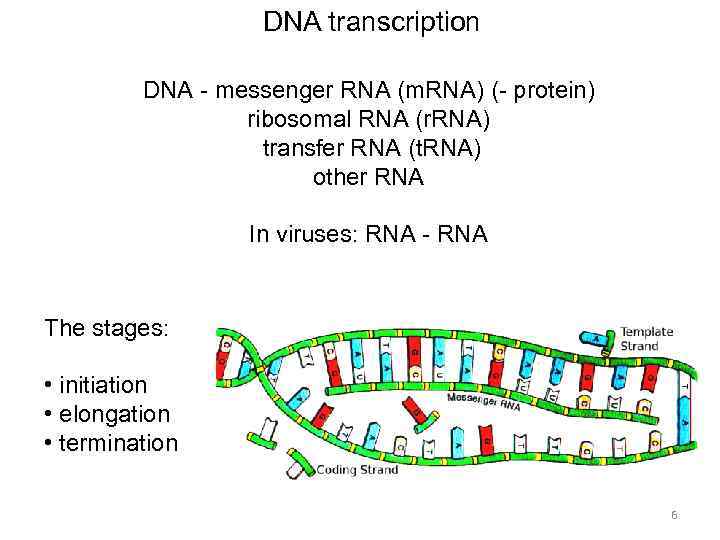
DNA transcription DNA - messenger RNA (m. RNA) (- protein) ribosomal RNA (r. RNA) transfer RNA (t. RNA) other RNA In viruses: RNA - RNA The stages: • initiation • elongation • termination 6
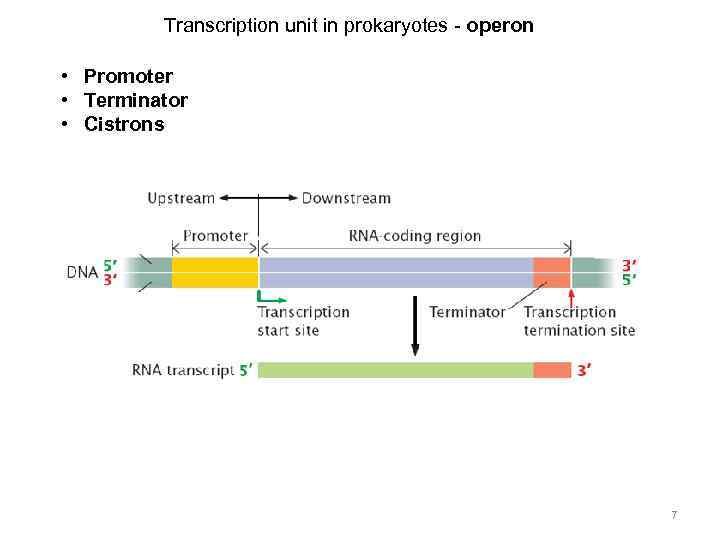
Transcription unit in prokaryotes - operon • Promoter • Terminator • Cistrons 7
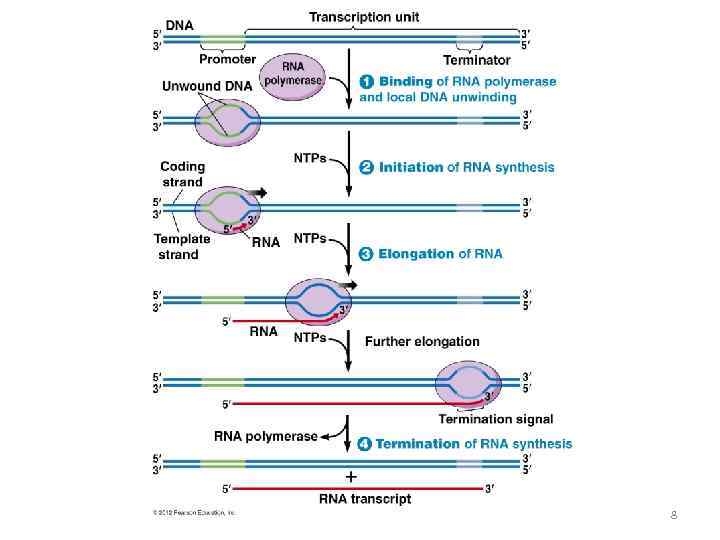
8
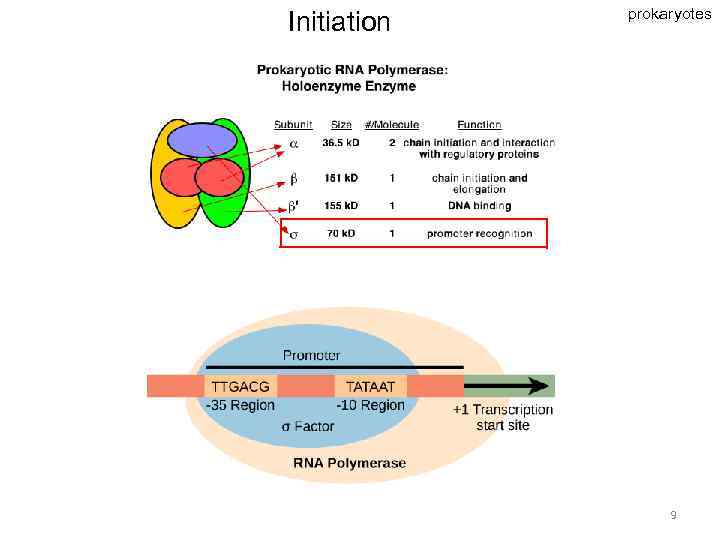
Initiation prokaryotes 9
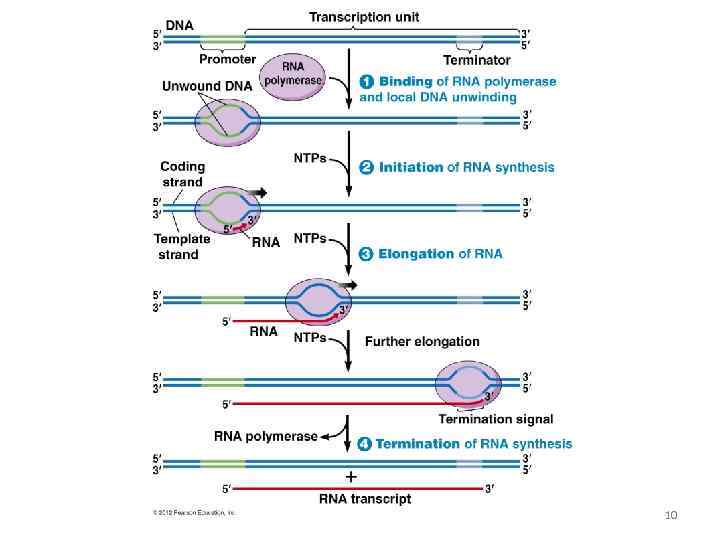
10
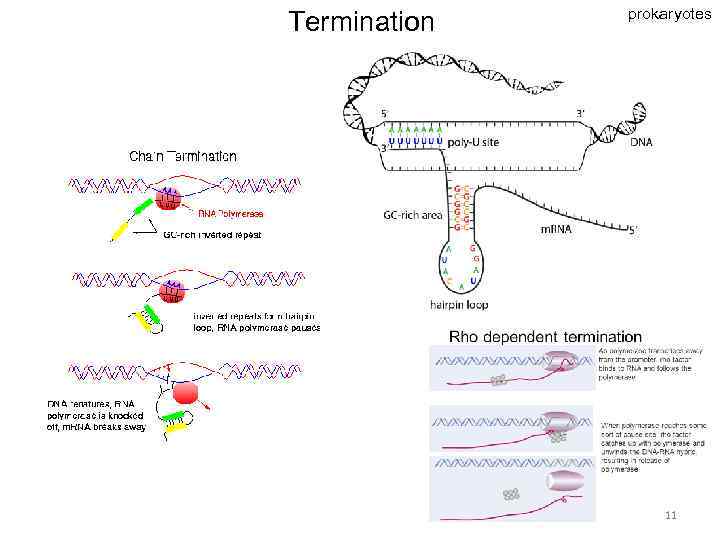
Termination prokaryotes 11
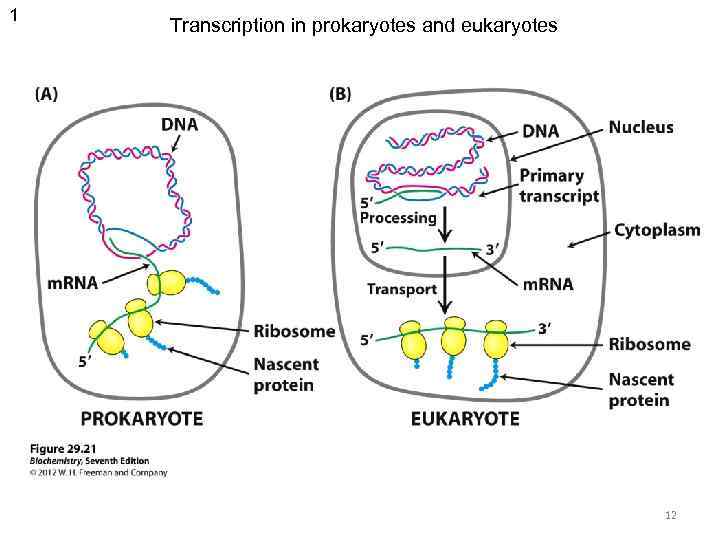
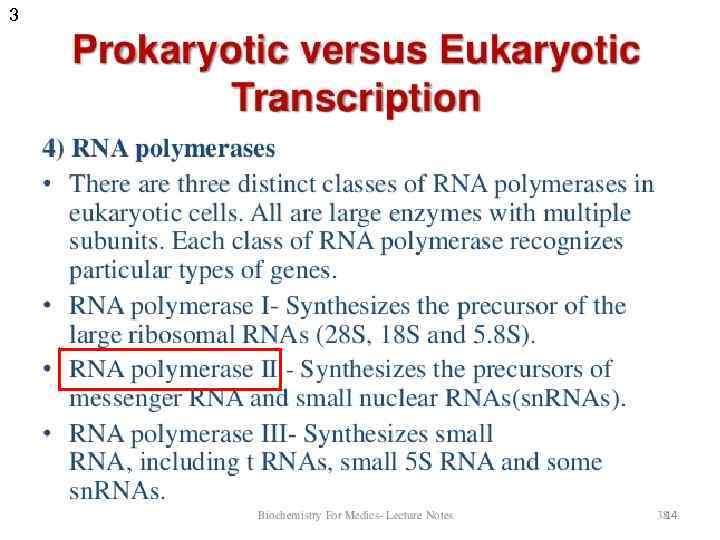
1 Transcription in prokaryotes and eukaryotes 12
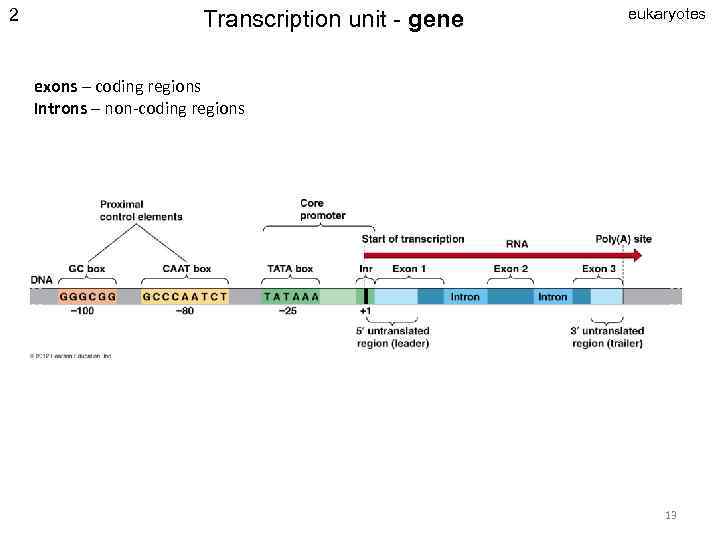
2 Transcription unit - gene eukaryotes exons – coding regions Introns – non-coding regions 13
3 14
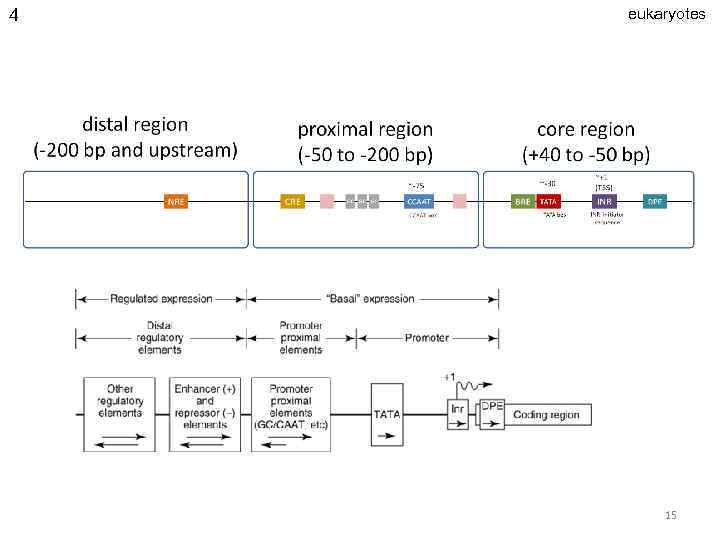
4 eukaryotes 15
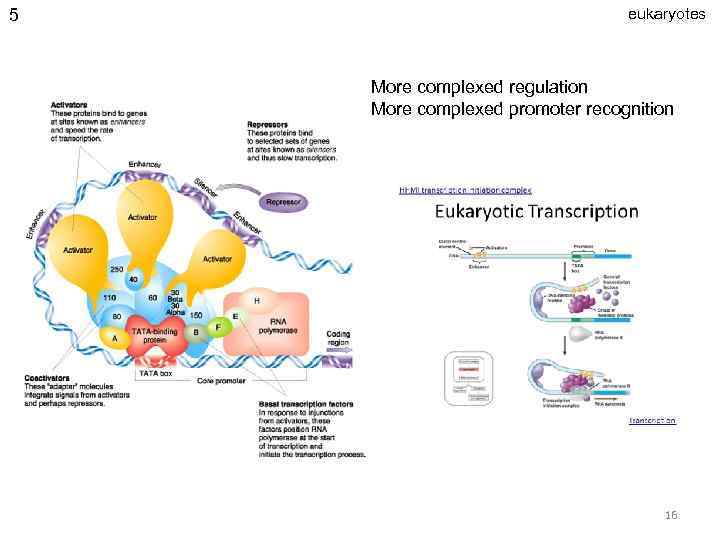
5 eukaryotes More complexed regulation More complexed promoter recognition 16
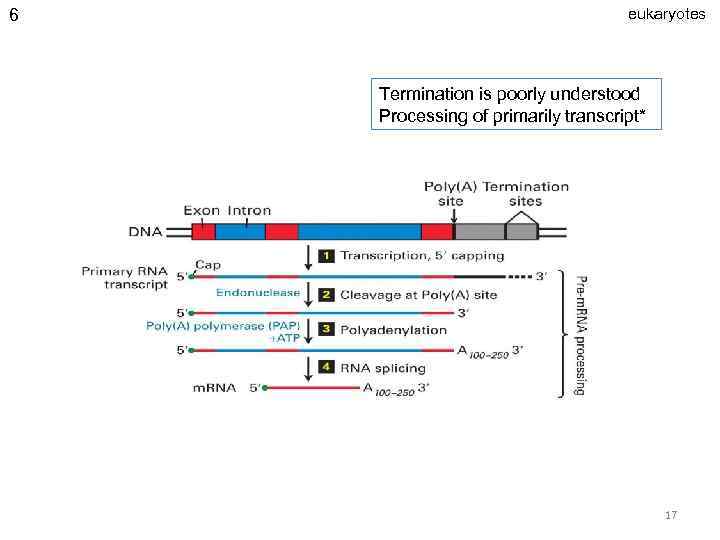
6 eukaryotes Termination is poorly understood Processing of primarily transcript* 17
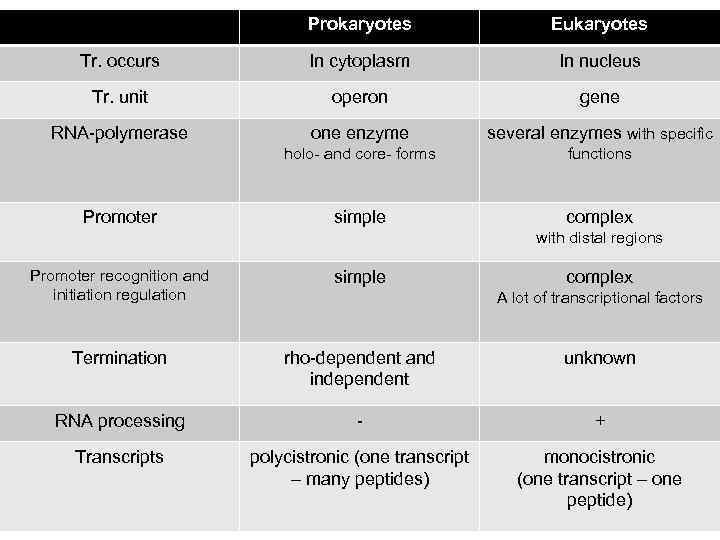
Prokaryotes Eukaryotes Tr. occurs In cytoplasm In nucleus Tr. unit operon gene RNA-polymerase one enzyme several enzymes with specific holo- and core- forms functions simple complex Promoter with distal regions Promoter recognition and initiation regulation simple Termination rho-dependent and independent unknown RNA processing - + Transcripts polycistronic (one transcript – many peptides) monocistronic (one transcript – one peptide) complex A lot of transcriptional factors 18
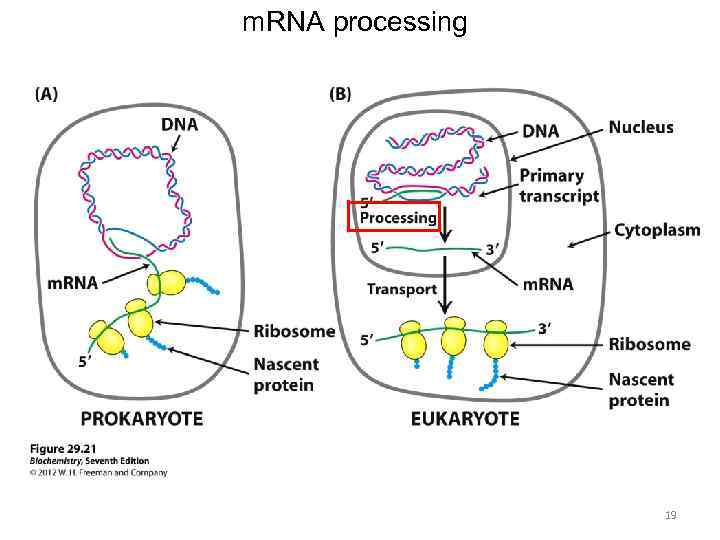
m. RNA processing 19
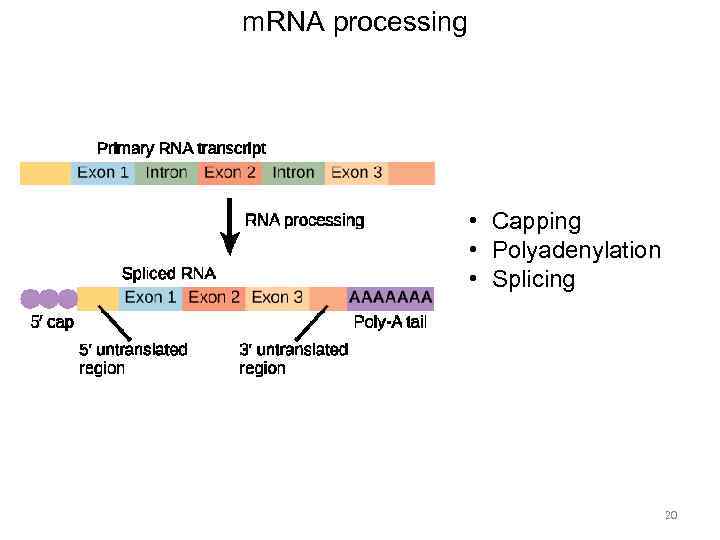
m. RNA processing • Capping • Polyadenylation • Splicing 20
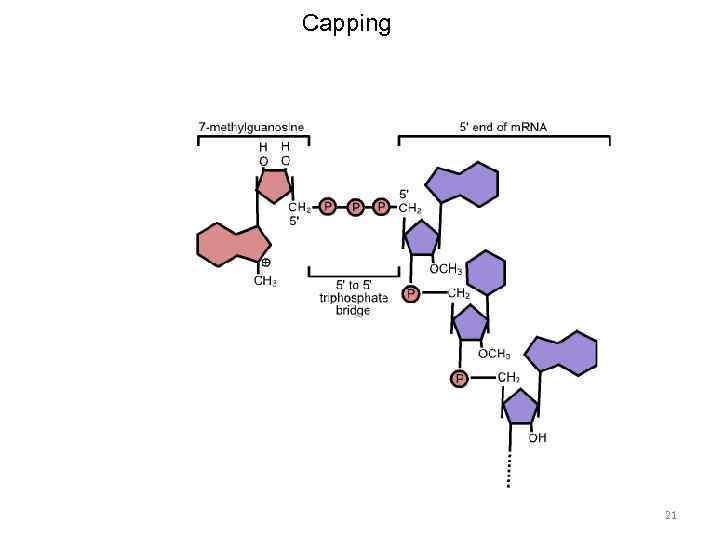
Capping 21
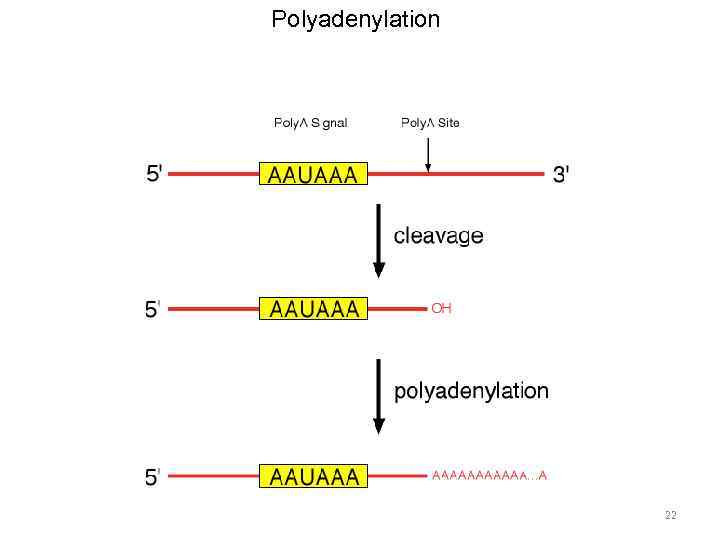
Polyadenylation 22
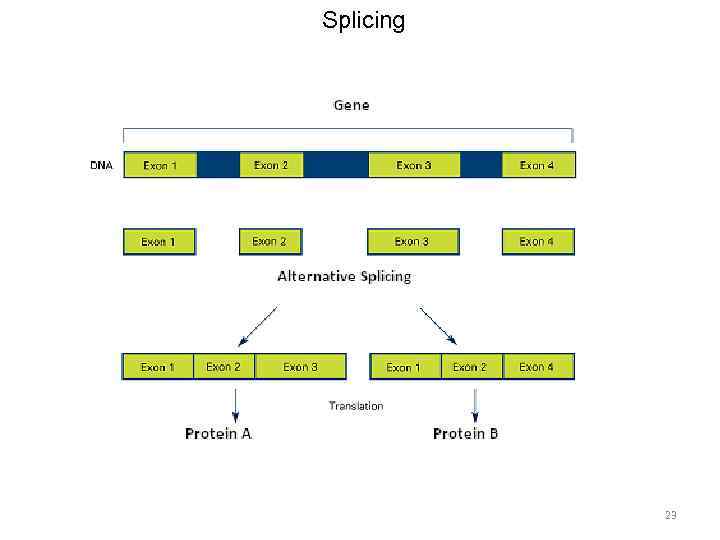
Splicing 23
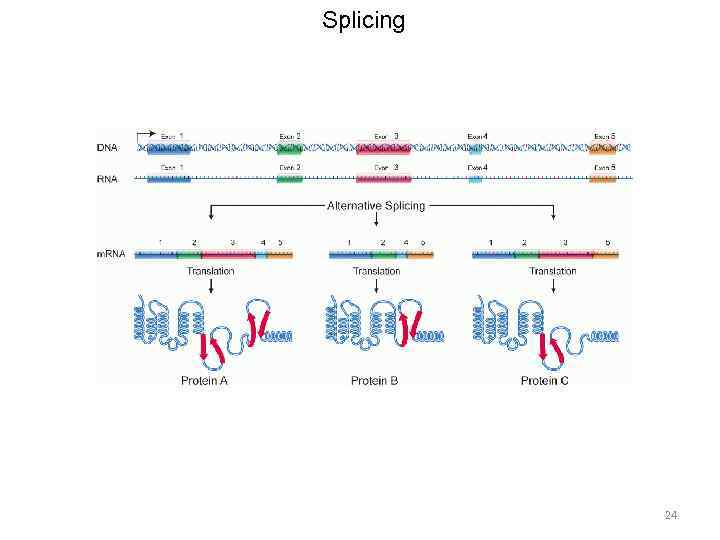
Splicing 24
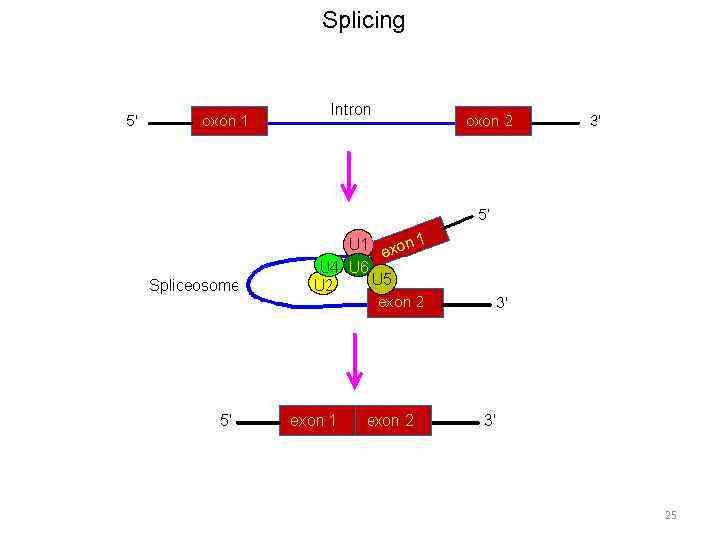
Splicing 25
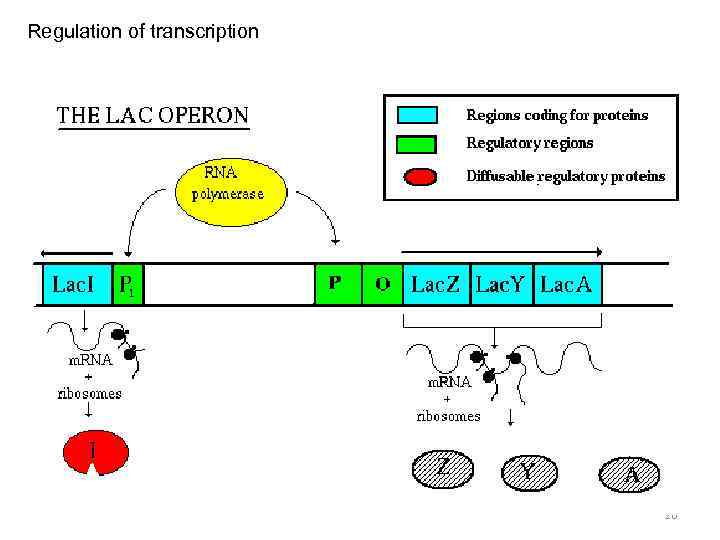
Regulation of transcription 26
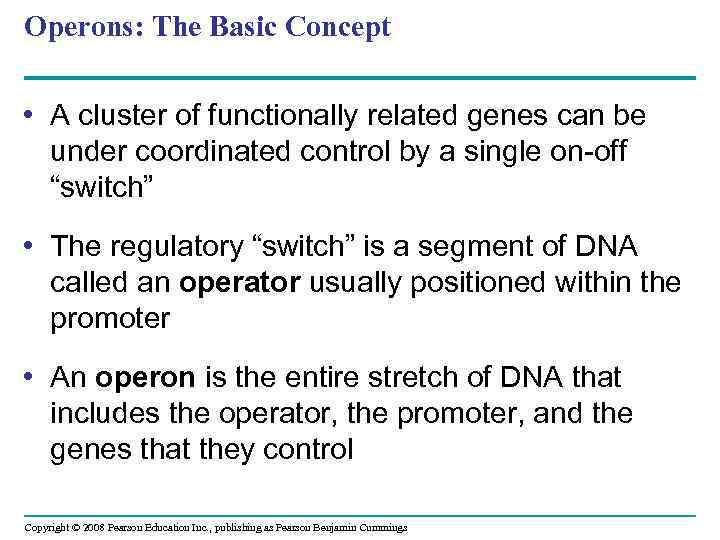
Operons: The Basic Concept • A cluster of functionally related genes can be under coordinated control by a single on-off “switch” • The regulatory “switch” is a segment of DNA called an operator usually positioned within the promoter • An operon is the entire stretch of DNA that includes the operator, the promoter, and the genes that they control Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings
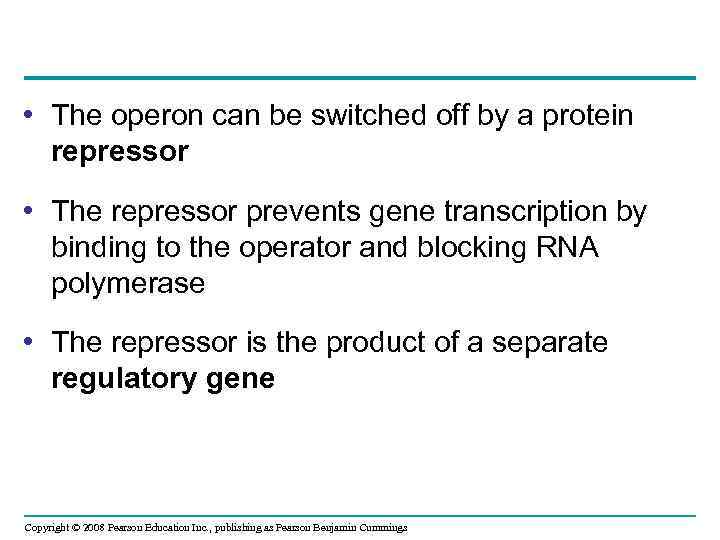
• The operon can be switched off by a protein repressor • The repressor prevents gene transcription by binding to the operator and blocking RNA polymerase • The repressor is the product of a separate regulatory gene Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings
Concept 18. 2: Eukaryotic gene expression can be regulated at any stage • All organisms must regulate which genes are expressed at any given time • In multicellular organisms gene expression is essential for cell specialization Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings
Differential Gene Expression • Almost all the cells in an organism are genetically identical • Differences between cell types result from differential gene expression, the expression of different genes by cells with the same genome • Errors in gene expression can lead to diseases including cancer • Gene expression is regulated at many stages Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings
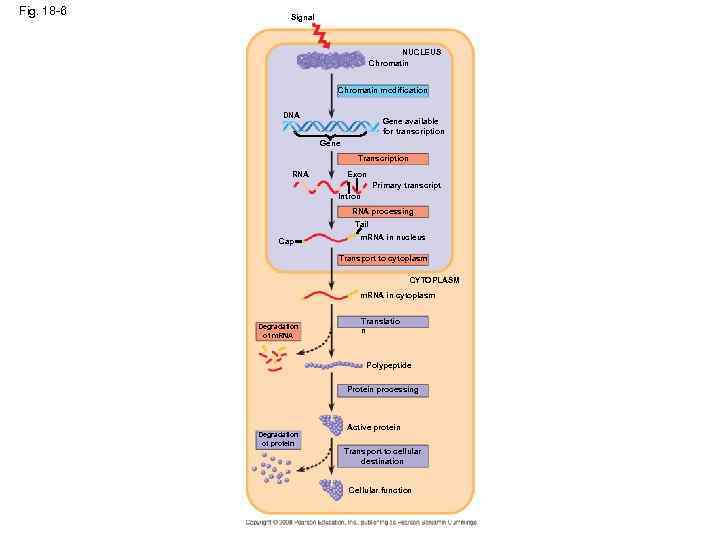
Fig. 18 -6 Signal NUCLEUS Chromatin modification DNA Gene available for transcription Gene Transcription RNA Exon Primary transcript Intron RNA processing Tail Cap m. RNA in nucleus Transport to cytoplasm CYTOPLASM m. RNA in cytoplasm Degradation of m. RNA Translatio n Polypeptide Protein processing Degradation of protein Active protein Transport to cellular destination Cellular function
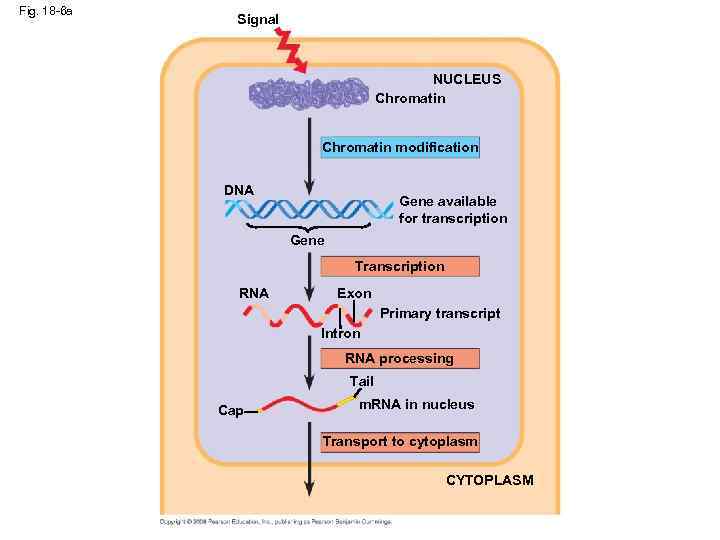
Fig. 18 -6 a Signal NUCLEUS Chromatin modification DNA Gene available for transcription Gene Transcription RNA Exon Primary transcript Intron RNA processing Tail Cap m. RNA in nucleus Transport to cytoplasm CYTOPLASM
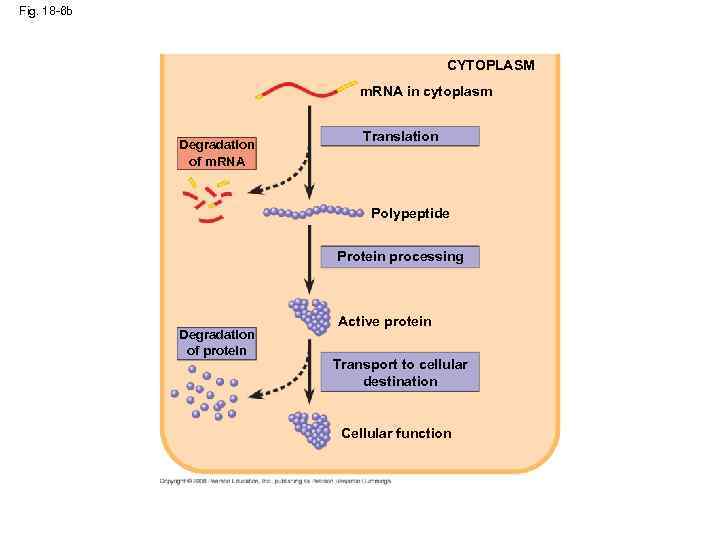
Fig. 18 -6 b CYTOPLASM m. RNA in cytoplasm Degradation of m. RNA Translation Polypeptide Protein processing Degradation of protein Active protein Transport to cellular destination Cellular function
Regulation of Chromatin Structure • Genes within highly packed heterochromatin are usually not expressed • Chemical modifications to histones and DNA of chromatin influence both chromatin structure and gene expression Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings
Histone Modifications • In histone acetylation, acetyl groups are attached to positively charged lysines in histone tails • This process loosens chromatin structure, thereby promoting the initiation of transcription • The addition of methyl groups (methylation) can condense chromatin; the addition of phosphate groups (phosphorylation) next to a methylated amino acid can loosen chromatin Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings
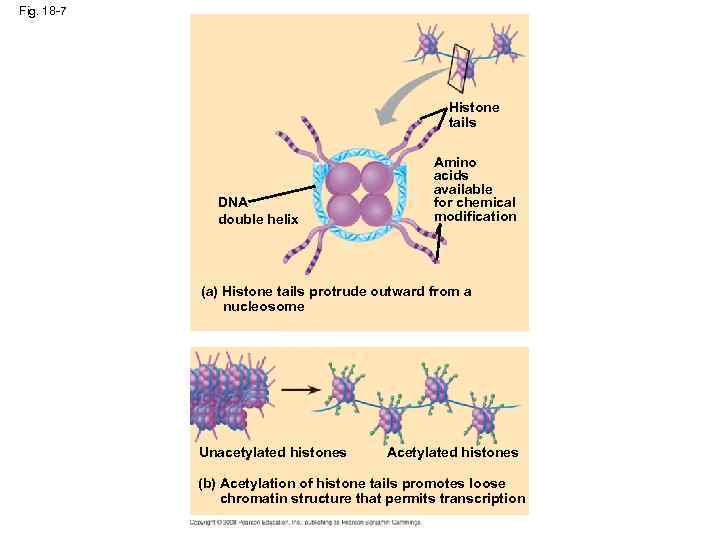
Fig. 18 -7 Histone tails DNA double helix Amino acids available for chemical modification (a) Histone tails protrude outward from a nucleosome Unacetylated histones Acetylated histones (b) Acetylation of histone tails promotes loose chromatin structure that permits transcription
DNA Methylation • DNA methylation, the addition of methyl groups to certain bases in DNA, is associated with reduced transcription in some species • DNA methylation cause long-term inactivation of genes in cellular differentiation Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings
Regulation of Transcription Initiation • Chromatin-modifying enzymes provide initial control of gene expression by making a region of DNA either more or less able to bind the transcription machinery Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings
The Roles of Transcription Factors • To initiate transcription, eukaryotic RNA polymerase requires the assistance of proteins called transcription factors • General transcription factors are essential for the transcription of all protein-coding genes • In eukaryotes, high levels of transcription of particular genes depend on control elements interacting with specific transcription factors Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings
Enhancers and Specific Transcription Factors • Proximal control elements are located close to the promoter • Distal control elements, groups of which are called enhancers, may be far away from a gene or even located in an intron Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

• An activator is a protein that binds to an enhancer and stimulates transcription of a gene • Bound activators cause mediator proteins to interact with proteins at the promoter Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings
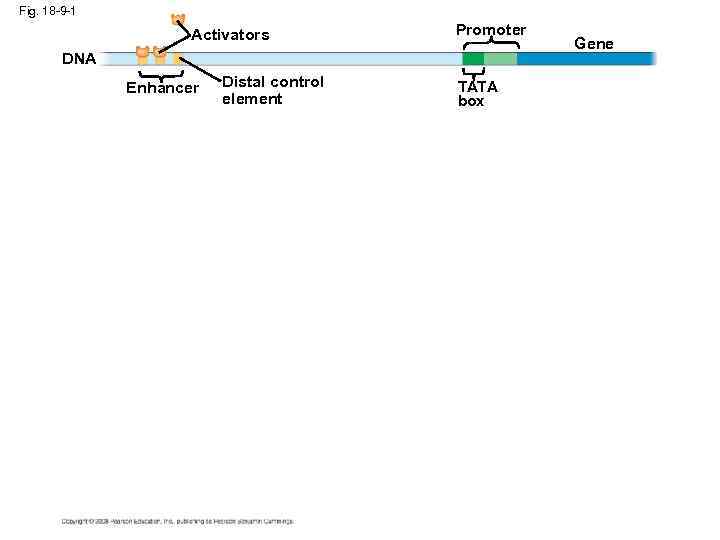
Fig. 18 -9 -1 Activators Promoter DNA Enhancer Distal control element TATA box Gene
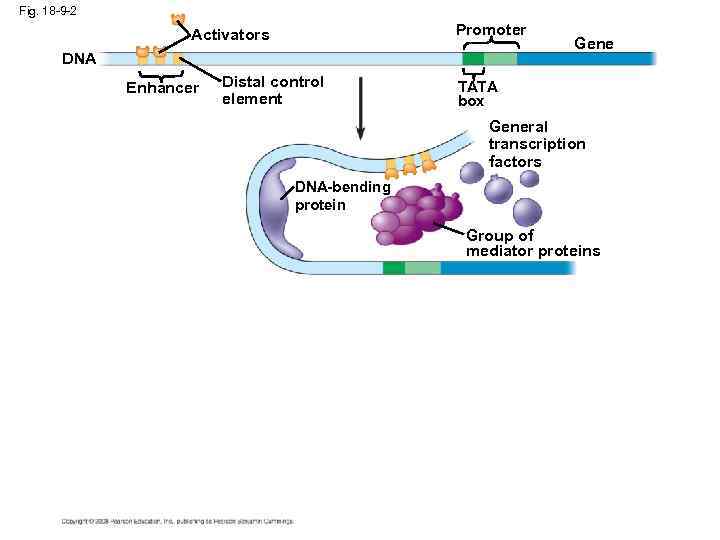
Fig. 18 -9 -2 Promoter Activators DNA Enhancer Distal control element Gene TATA box General transcription factors DNA-bending protein Group of mediator proteins
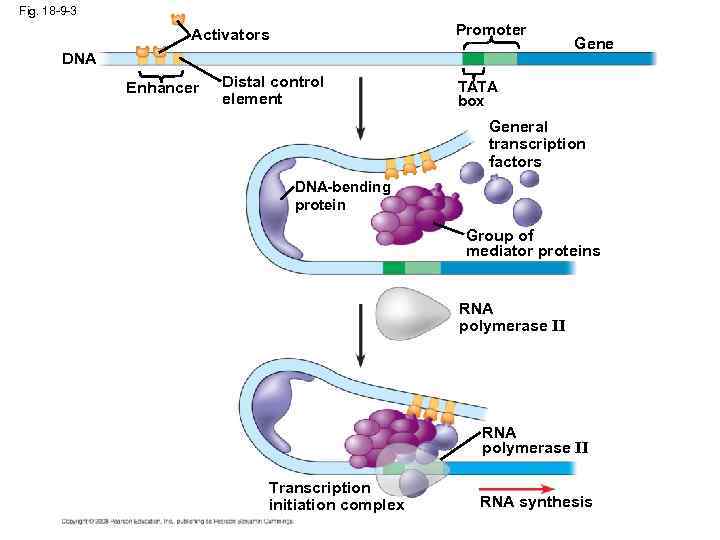
Fig. 18 -9 -3 Promoter Activators DNA Enhancer Distal control element Gene TATA box General transcription factors DNA-bending protein Group of mediator proteins RNA polymerase II Transcription initiation complex RNA synthesis
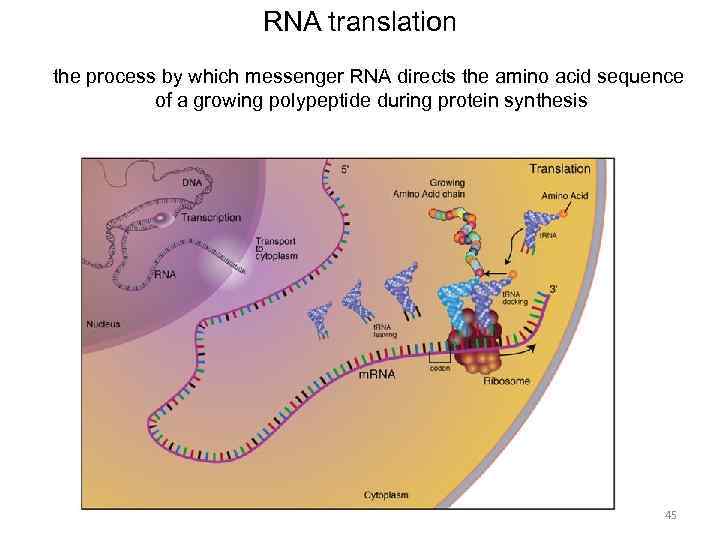
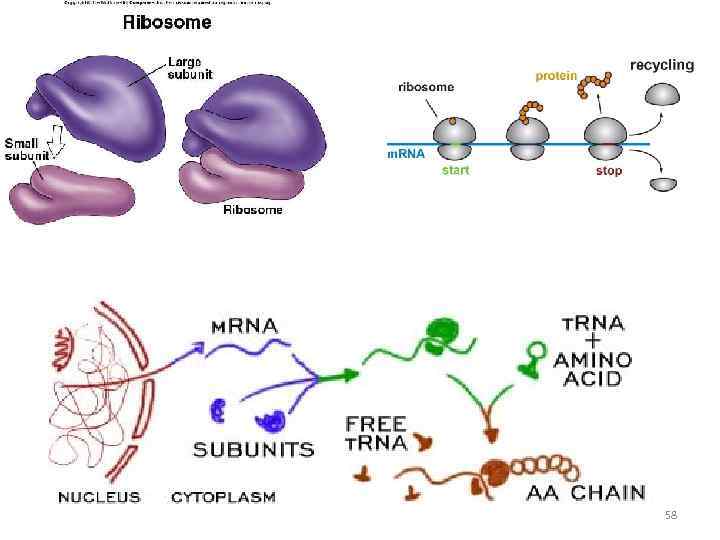
RNA translation the process by which messenger RNA directs the amino acid sequence of a growing polypeptide during protein synthesis 45
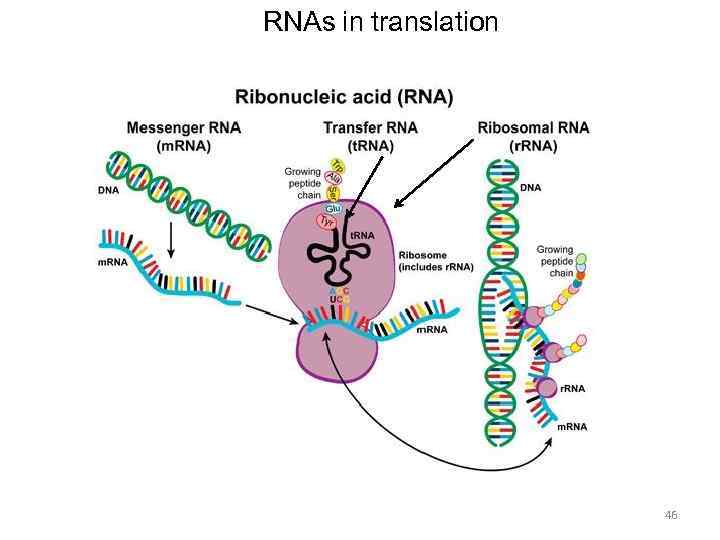
RNAs in translation 46
47
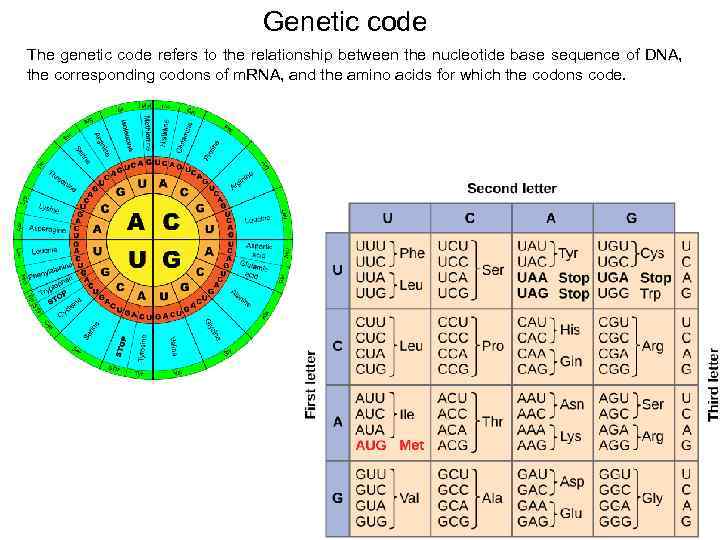
Genetic code The genetic code refers to the relationship between the nucleotide base sequence of DNA, the corresponding codons of m. RNA, and the amino acids for which the codons code. 48
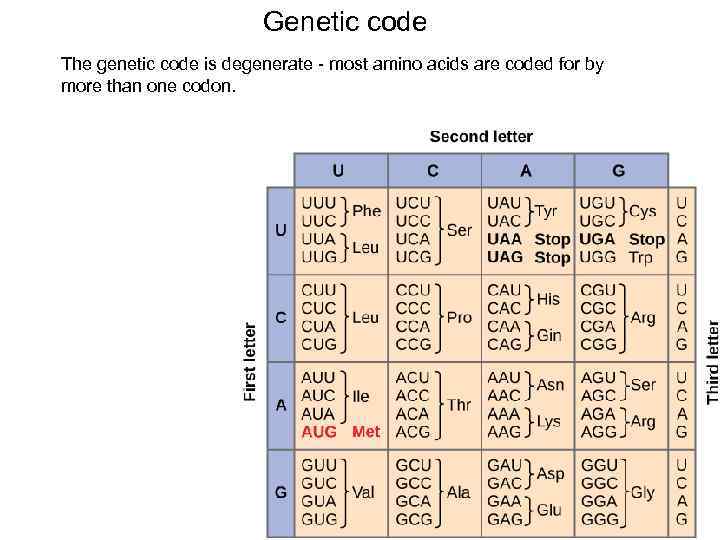
Genetic code The genetic code is degenerate - most amino acids are coded for by more than one codon. 49
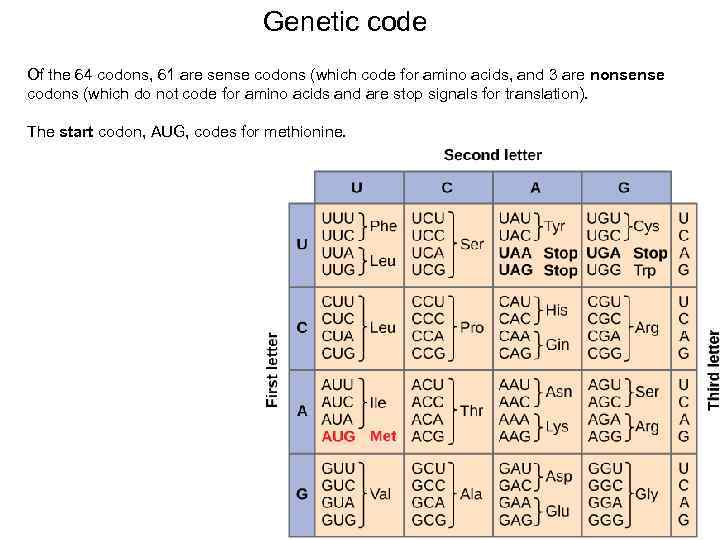
Genetic code Of the 64 codons, 61 are sense codons (which code for amino acids, and 3 are nonsense codons (which do not code for amino acids and are stop signals for translation). The start codon, AUG, codes for methionine. 50
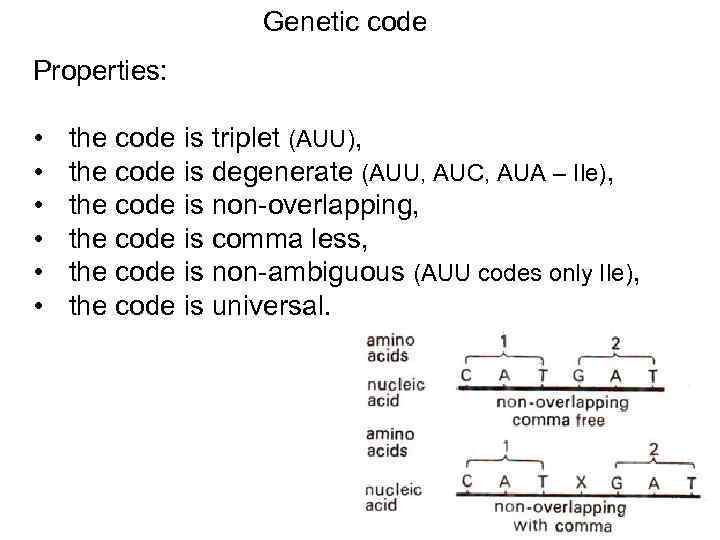
Genetic code Properties: • • • the code is triplet (AUU), the code is degenerate (AUU, AUC, AUA – Ile), the code is non-overlapping, the code is comma less, the code is non-ambiguous (AUU codes only Ile), the code is universal. 51

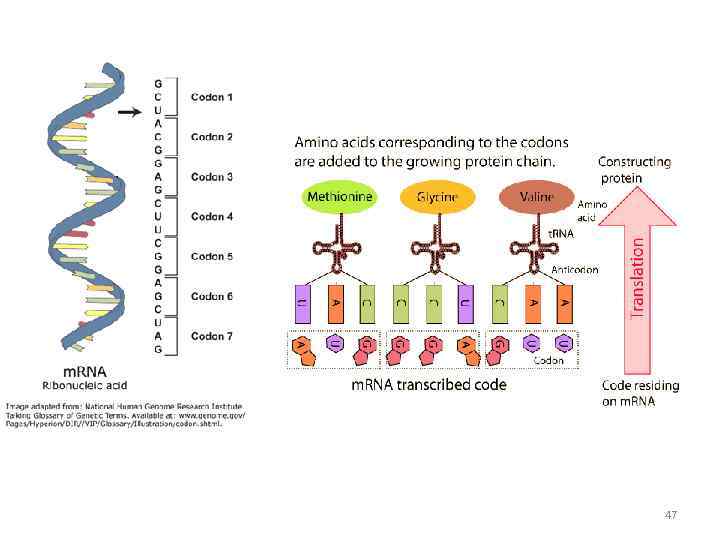
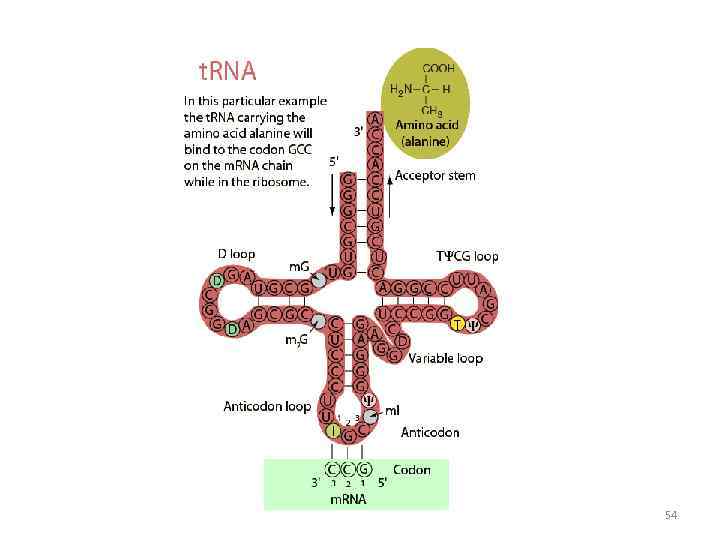
Specific amino acids are attached to molecules of t. RNA. Another portion of the t. RNA has a base triplet called an anticodon. The base pairing of codon and anticodon at the ribosome results in specific amino acids being brought to the site of protein synthesis. The ribosome moves along the m. RNA strand as amino acids are joined to forma growing polypeptide; m. RNA is read in the 5’ to 3’ direction. Translation ends when the ribosome reaches a stop codon on the m. RNA. 52
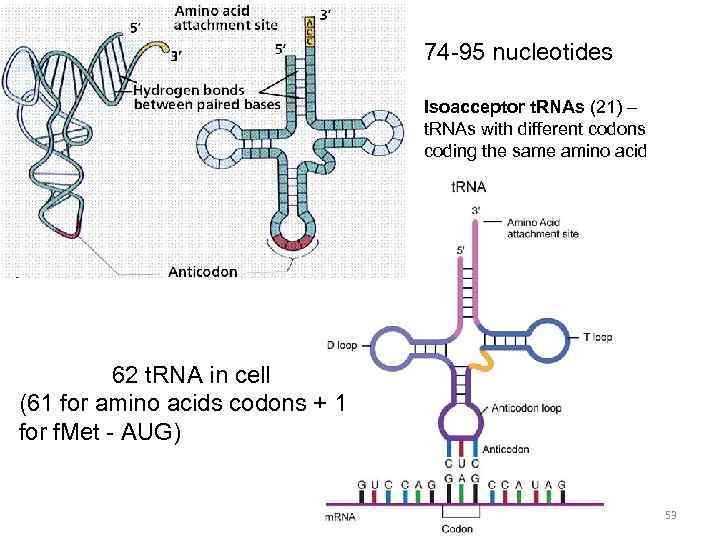
74 -95 nucleotides Isoacceptor t. RNAs (21) – t. RNAs with different codons coding the same amino acid 62 t. RNA in cell (61 for amino acids codons + 1 for f. Met - AUG) 53
54
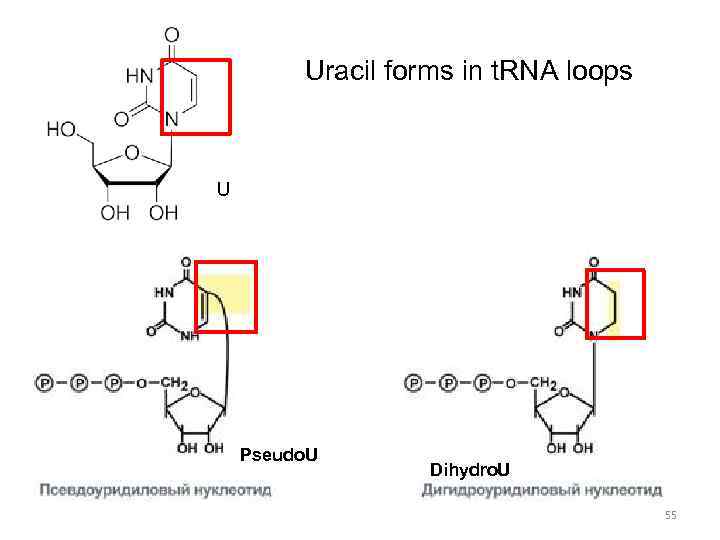
Uracil forms in t. RNA loops U Pseudo. U Dihydro. U 55
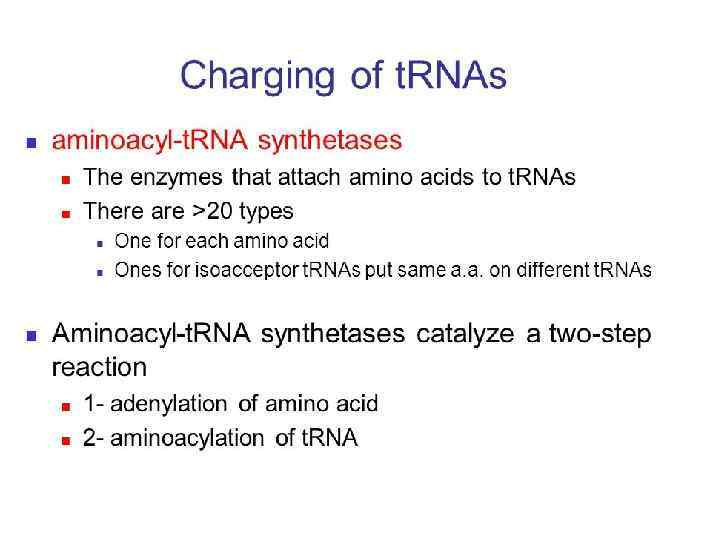
56
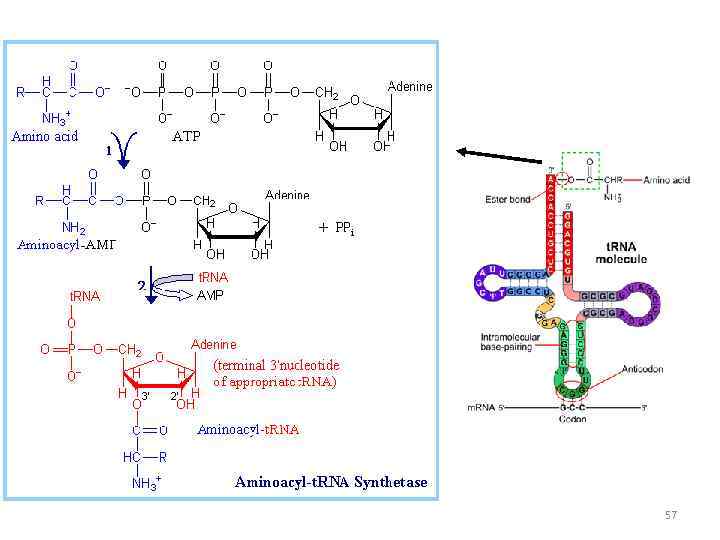
57
58

59
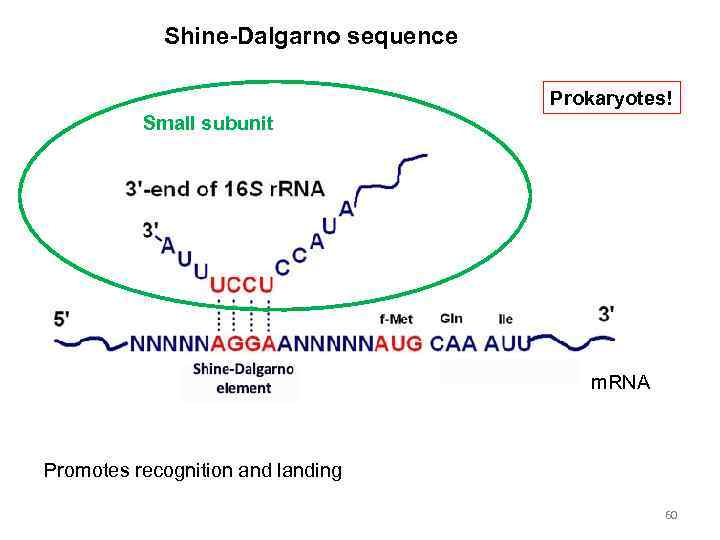
Shine-Dalgarno sequence Prokaryotes! Small subunit m. RNA Promotes recognition and landing 60
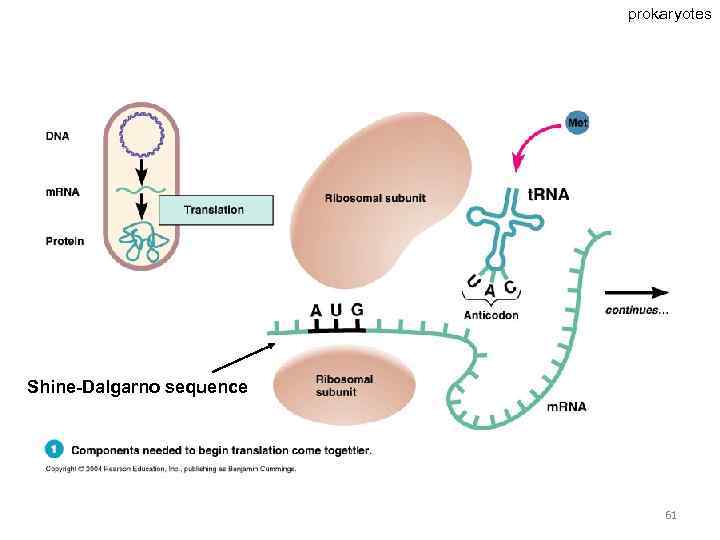
prokaryotes Shine-Dalgarno sequence 61
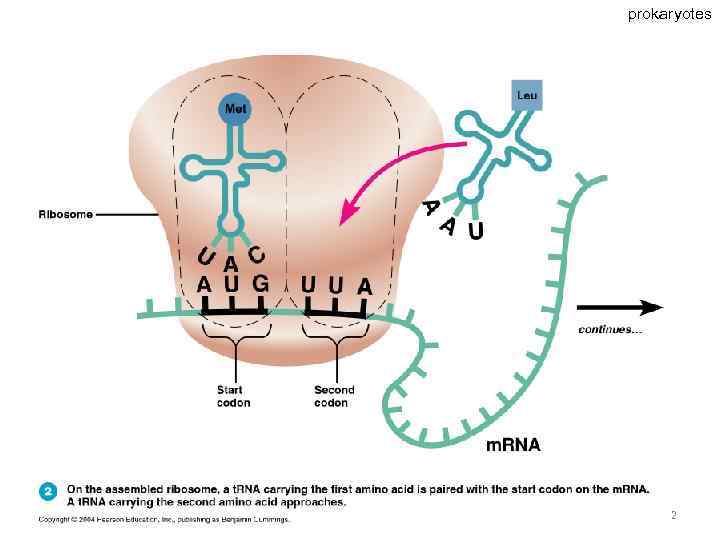
prokaryotes 62
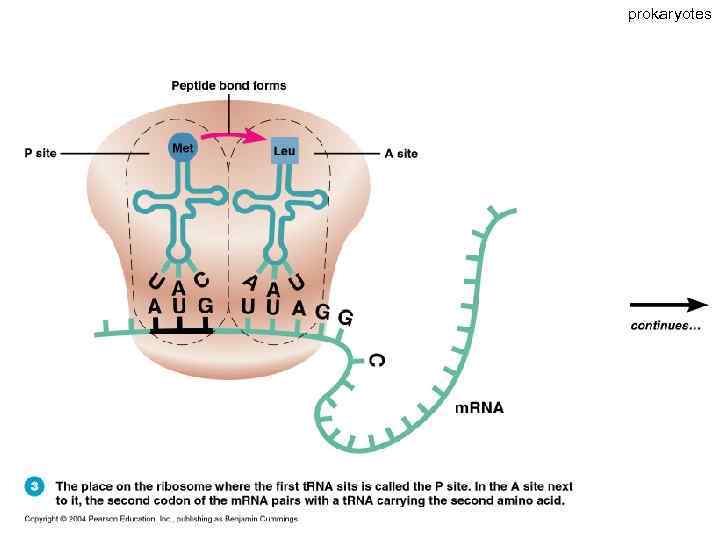
prokaryotes 63
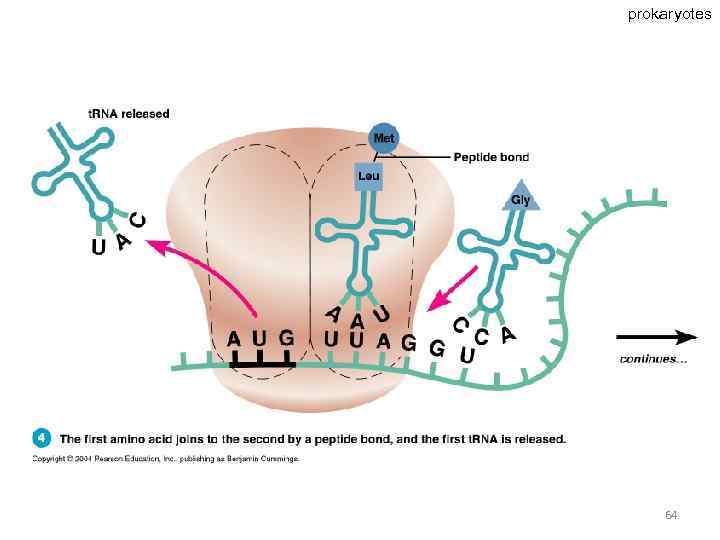
prokaryotes 64
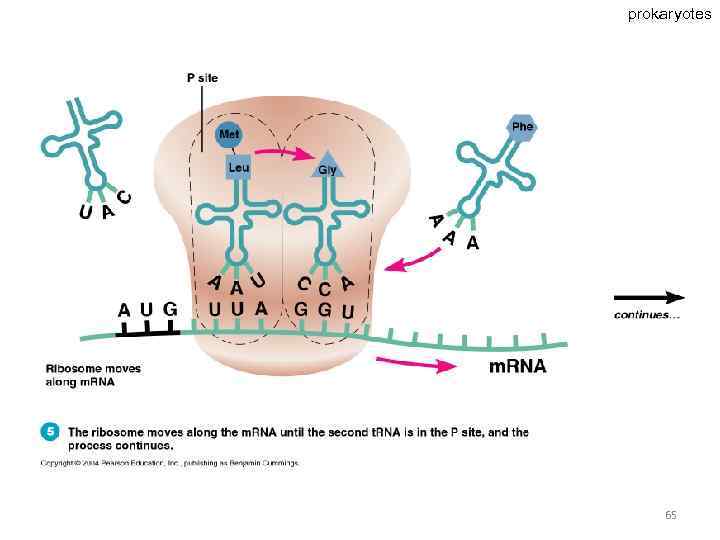
prokaryotes 65
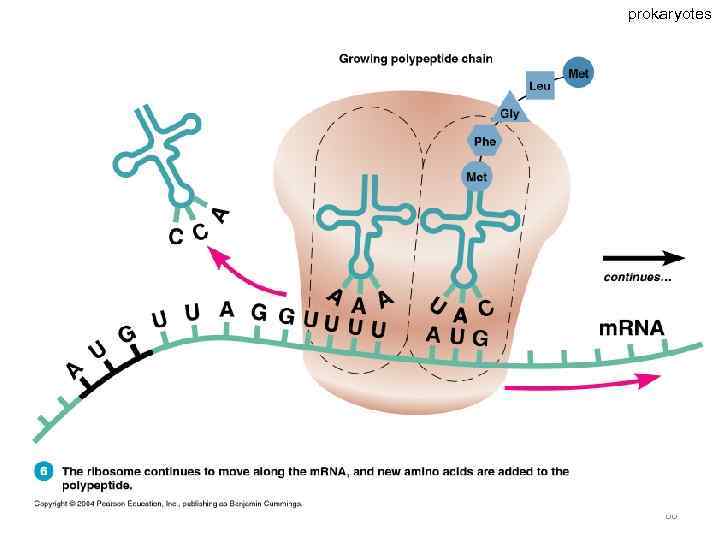
prokaryotes 66
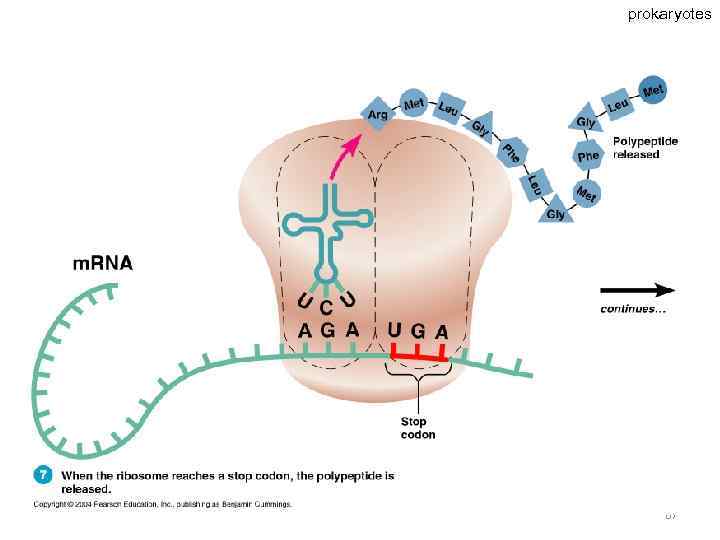
prokaryotes 67
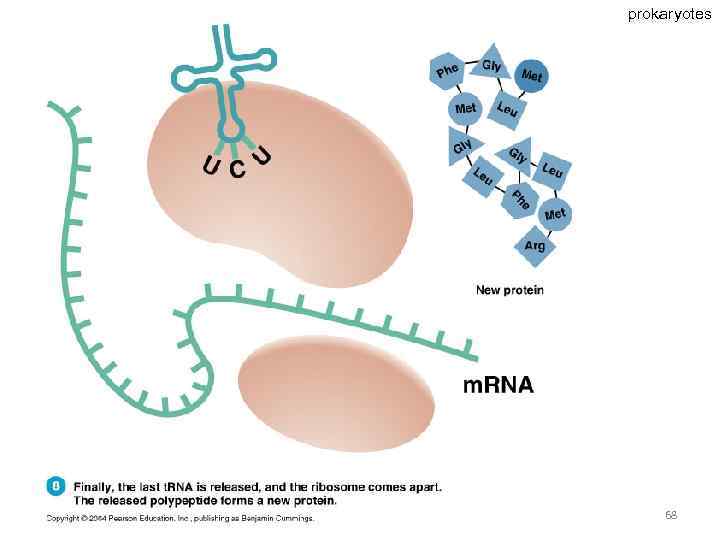
prokaryotes 68
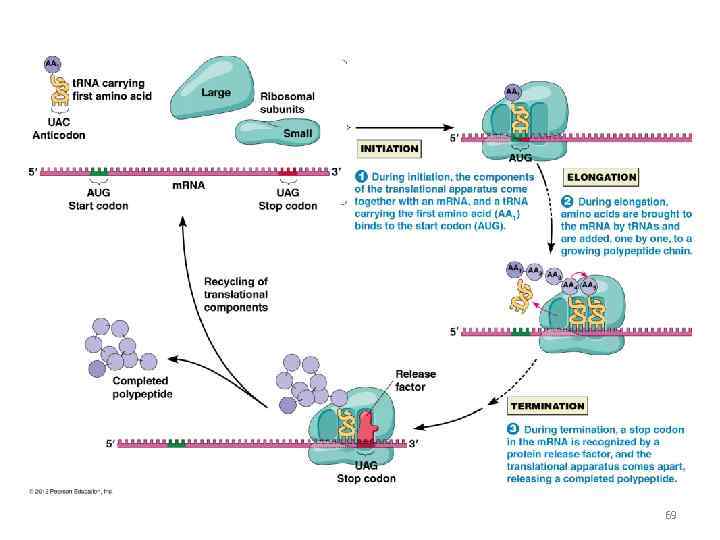
69
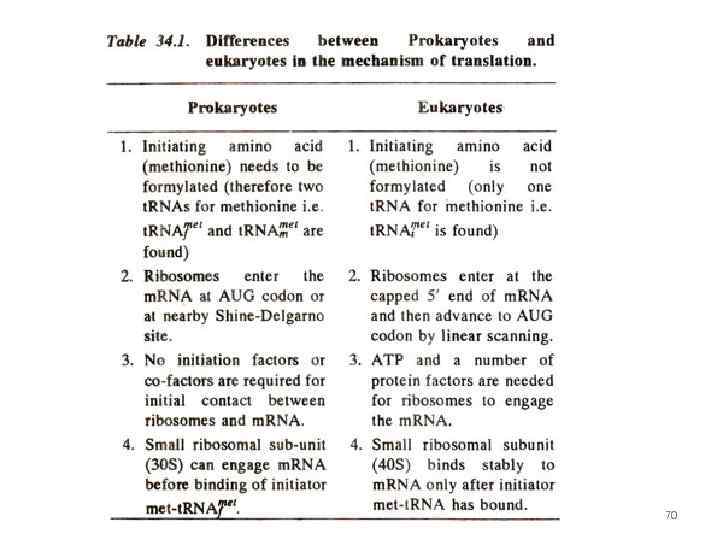
70
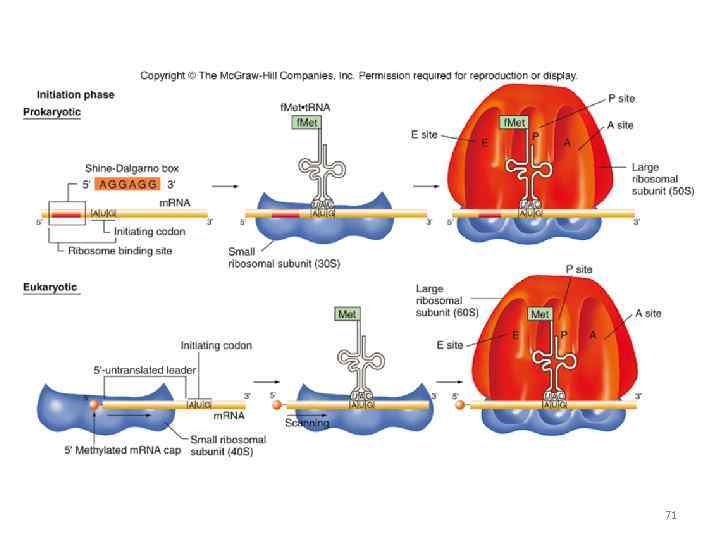
71
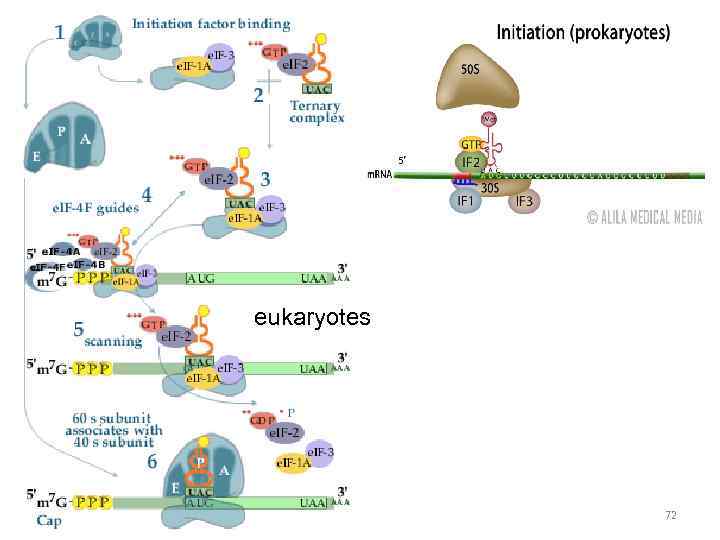
eukaryotes 72
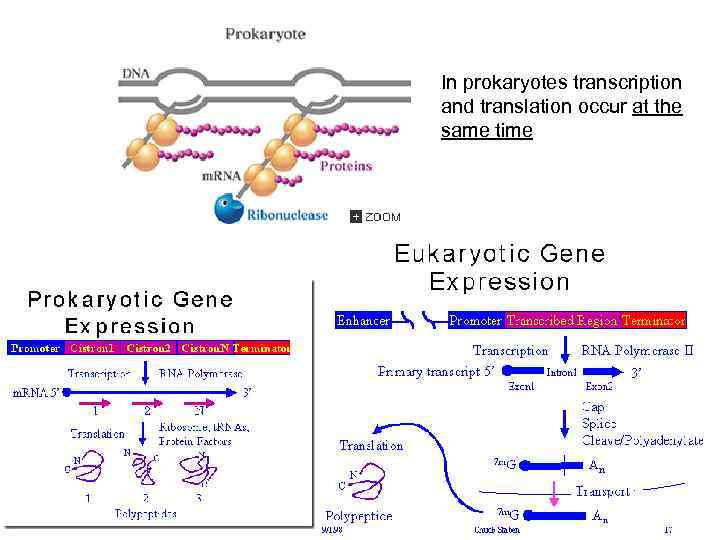
In prokaryotes transcription and translation occur at the same time 73
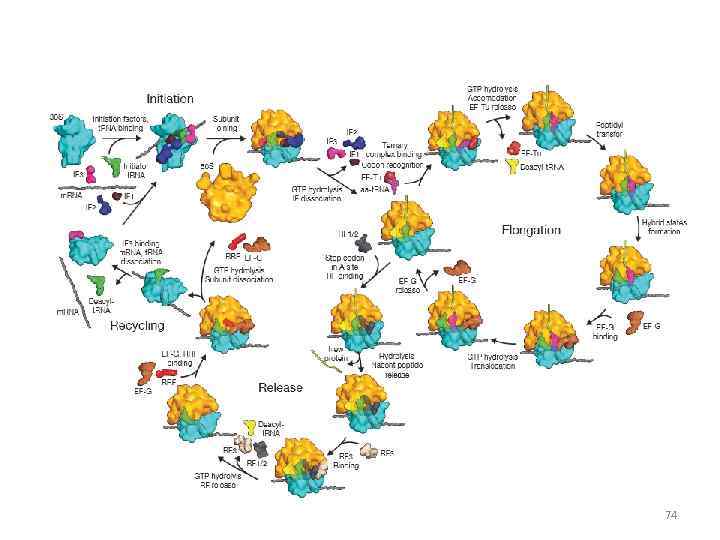
74
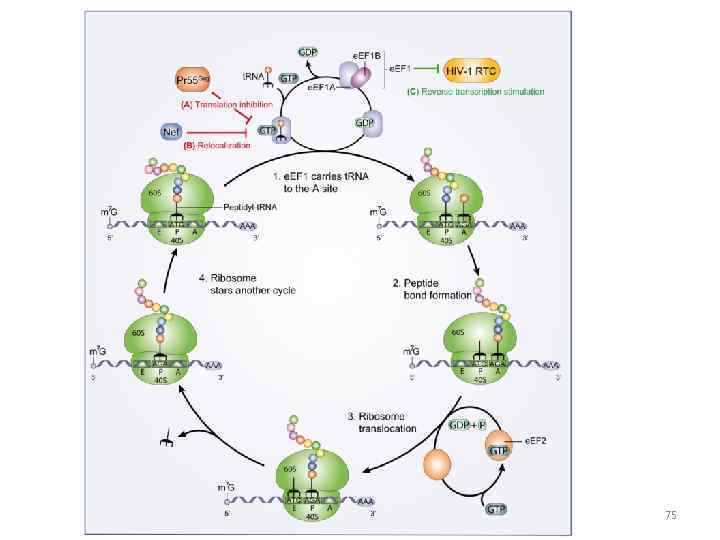
75
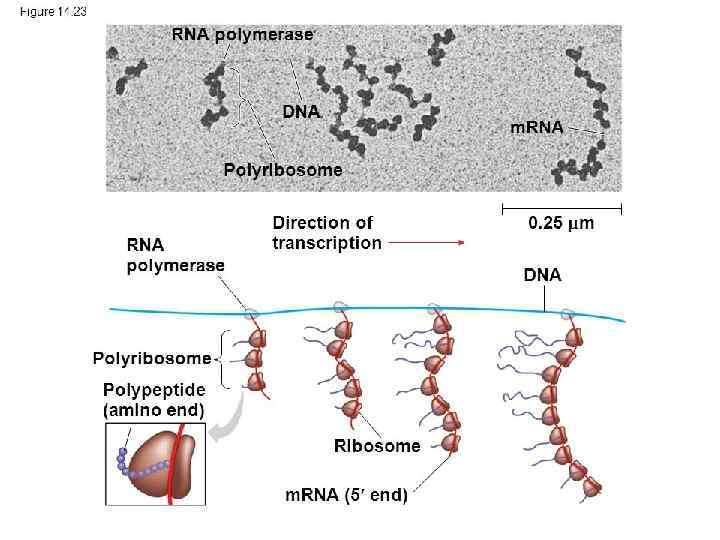
76
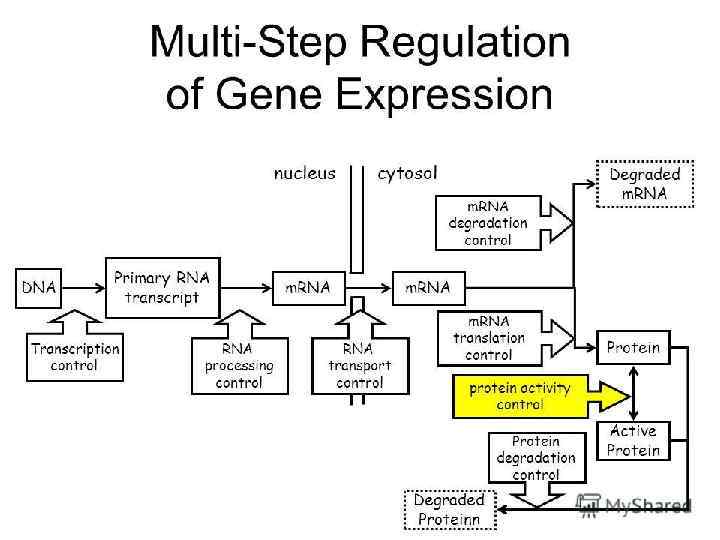
77

Campbell book: p. 333 -354 p. 360 -373 78
mol biol 3,4.ppt