906692139758ed71d8905407dd9d53e7.ppt
- Количество слайдов: 22

Modified Allele Dosage (MAD) Genotyping by Hi-Res Melting of Whole Amplicons Jason T. Mc. Kinney Scientist Human Genetics Applications

Modified what? … Who’s MAD? • Modified Allele Dosage = Pre-PCR DNA mixing • “MAD”? Well, … PHENCODE=ENCODE+Gen. Phen, Hu. GENet, LSDB’s, … – Seems as though “MAD” is a MUST • Genotype by Hi-Res Melting (HRM) of whole amplicons • Detection of heteroduplexes – “Core” application of Hi-Res Melting • Force heteroduplex formation in homozygous samples by mixing DNA pre-PCR – Typically done 1: 1 volume: volume

Stealing from Peter? • Must not compromise ability to distinguish “true” heterozygous samples • Homozygous “heteroduplexes” = – “intermediate” quantity of heteroduplex molecules relative to “true” heterozygous samples and wild type samples
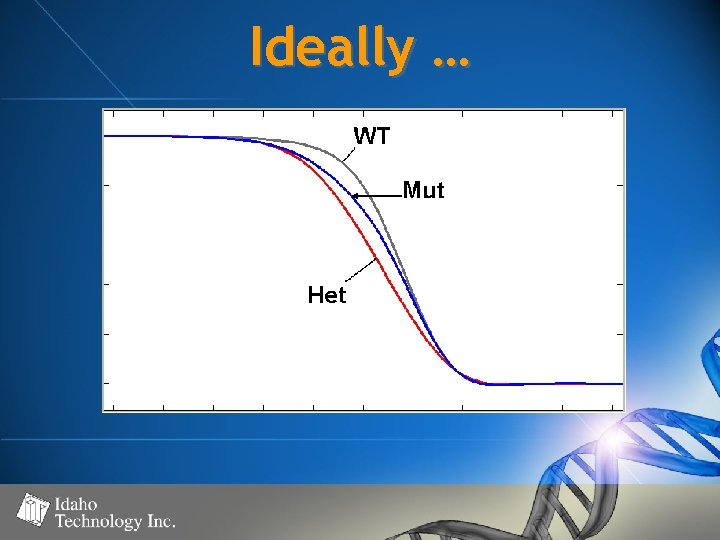
Ideally …
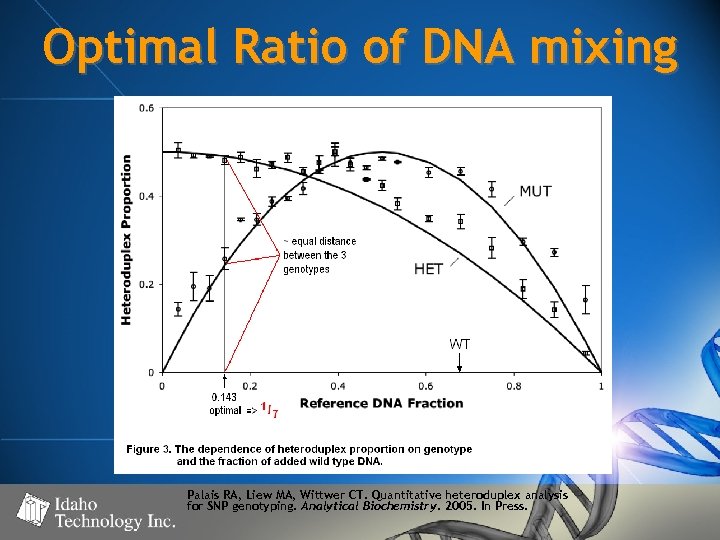
Optimal Ratio of DNA mixing Palais RA, Liew MA, Wittwer CT. Quantitative heteroduplex analysis for SNP genotyping. Analytical Biochemistry. 2005. In Press.

Optimal ratio, … in English please? • 1 X WT DNA + 6 X Unknown DNA yields the following heteroduplex ratio’s: • Given HRM detection of ~5 -10% variant DNA, these ratio’s should be easily discriminated
Benefits • Genotype sans probe (labeled or unlabeled) – Cost and technical benefit • No asymmetric amplification – Technical benefit • Use “scanning” primers to genotype • Potential to observe variants other than the SNP of interest – Relative to a probe-based genotyping method
System Requirements • Sequence characterized reference sample (wildtype or variant) • Accurately quantified DNA samples • ds. DNA “saturation” dye (LCGreen Plus) • High-resolution instrumentation for melting curve analysis (HR-1, Light. Scanner) • Well characterized region around SNP is beneficial
Validation of MAD genotyping • Risk burden of HDL haplotypes – 6 genes regulating HDL cholesterol metabolism 1. Primer design a. Area around SNP’s interrogated in 100 chromosomes b. Primers designed to avoid observed variants 2. PCR quality a. Primer concentration across annealing temperature gradient b. Definitive criteria for Hi-Res Melting success
Methods • Quantify DNA samples (~10 ng/ l) • Add reference DNA (wildtype) to master mix at 1/7 total DNA • Add unknown DNA at 6/7 total • Amplification – 96 well thermal block • HRM on Light. Scanner • Automated calling of genotypes
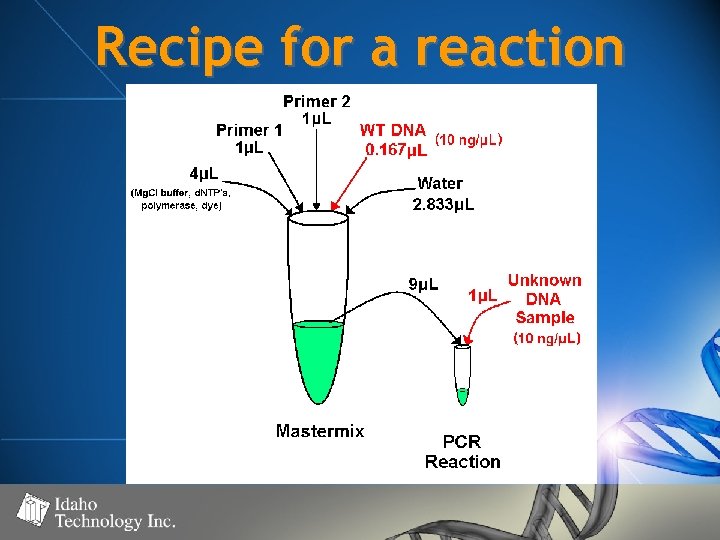
Recipe for a reaction
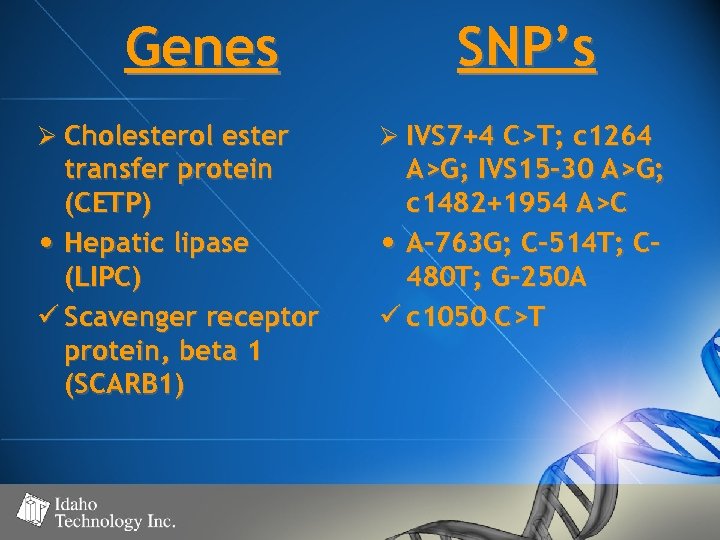
Genes Ø Cholesterol ester transfer protein (CETP) • Hepatic lipase (LIPC) ü Scavenger receptor protein, beta 1 (SCARB 1) SNP’s Ø IVS 7+4 C>T; c 1264 A>G; IVS 15 -30 A>G; c 1482+1954 A>C • A-763 G; C-514 T; C 480 T; G-250 A ü c 1050 C>T

Show me the Data!
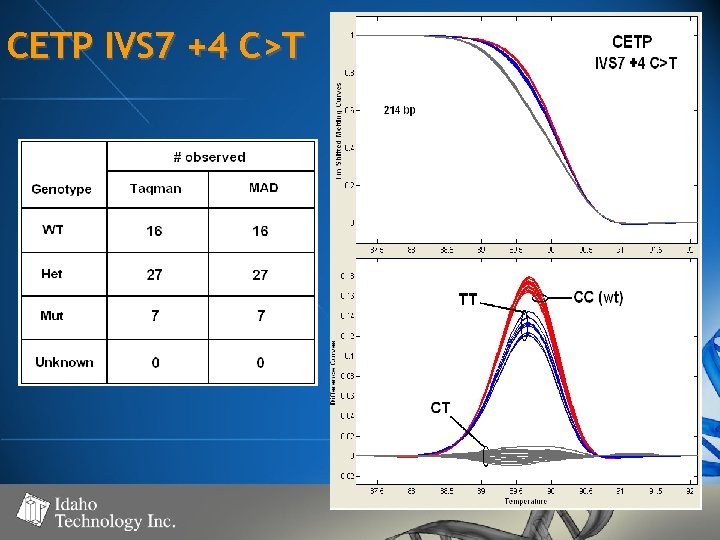
CETP IVS 7 +4 C>T
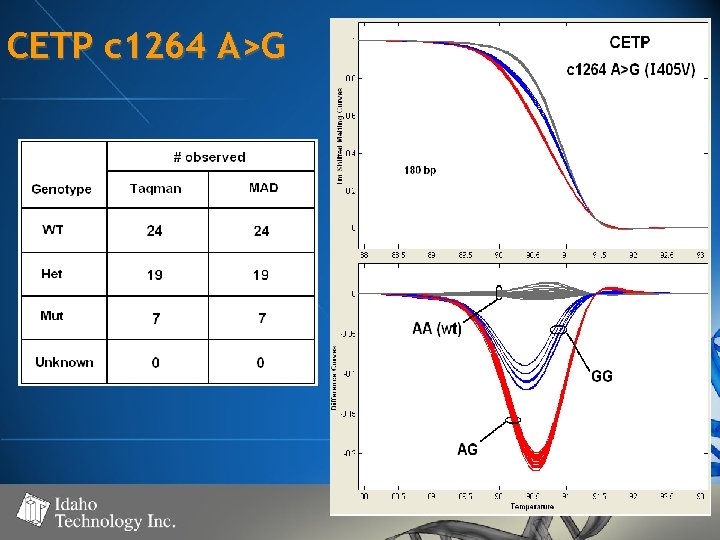
CETP c 1264 A>G
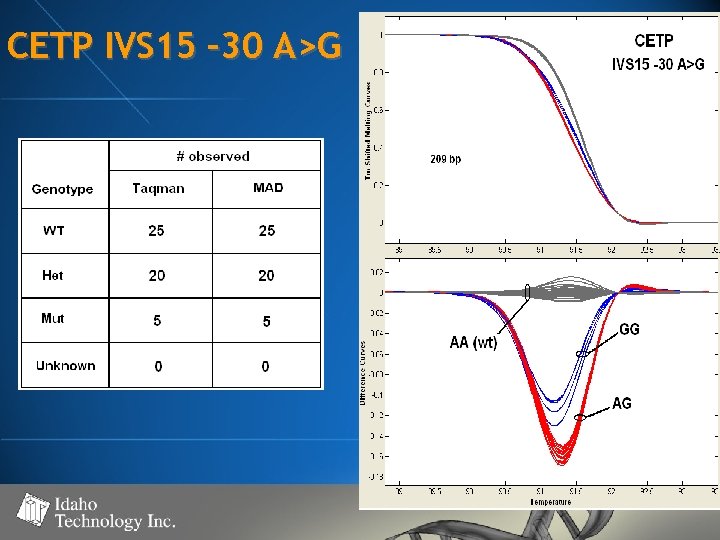
CETP IVS 15 – 30 A>G
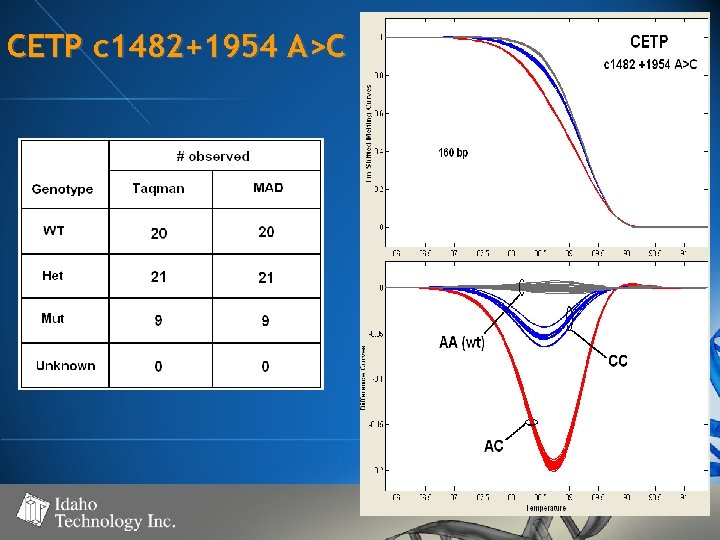
CETP c 1482+1954 A>C
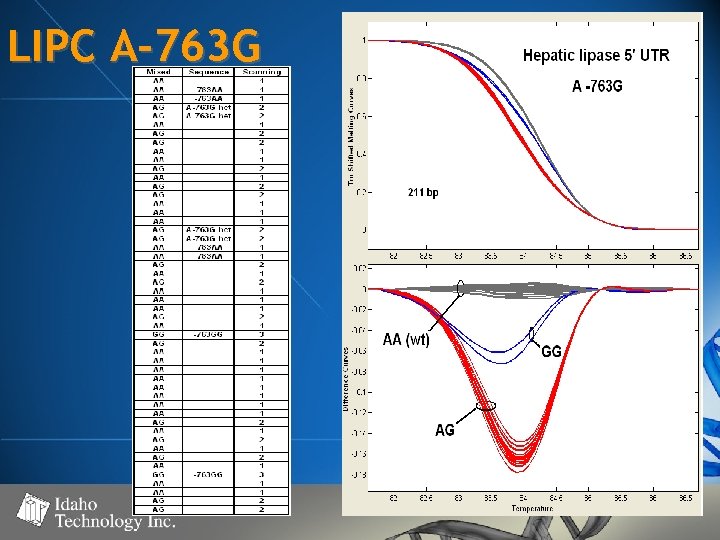
LIPC A-763 G
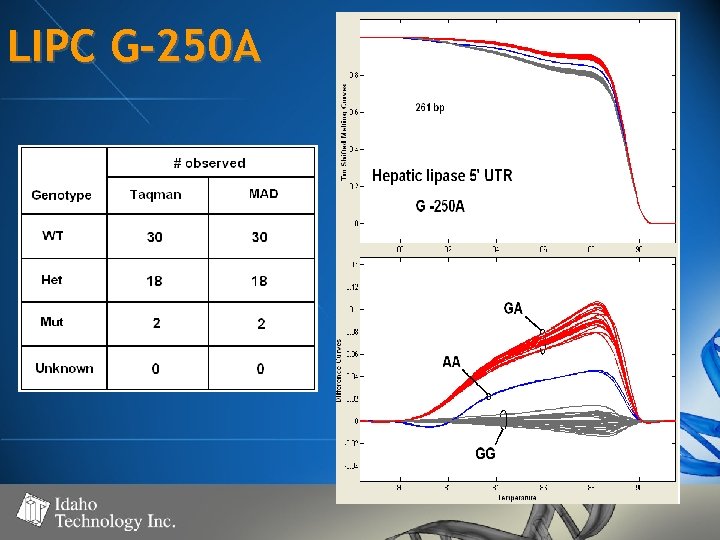
LIPC G-250 A
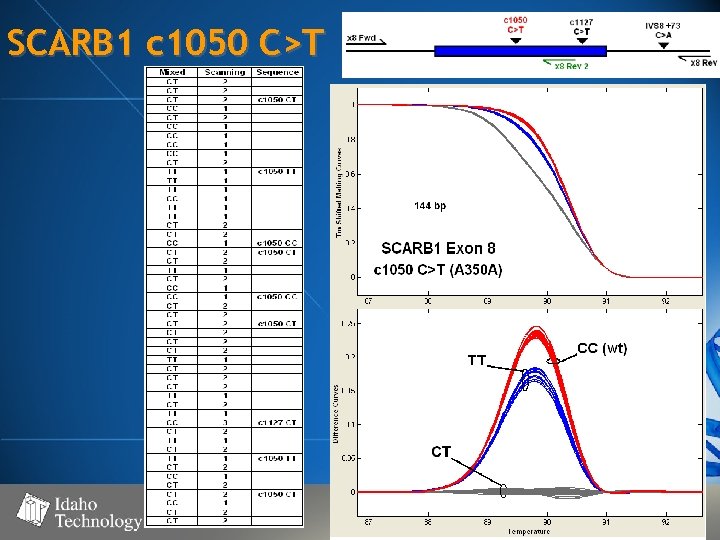
SCARB 1 c 1050 C>T
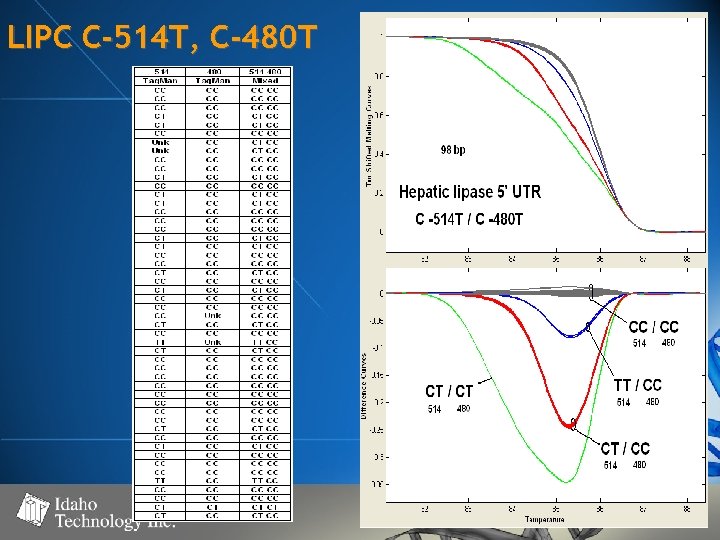
LIPC C-514 T, C-480 T
Summary of Data • SNP’s observed – C>T (4), A>G (3), A>C (1), G>A (1) • Fragment size range - 98 -261 bp – Strong correlation between fragment size and WT – Het difference (r=0. 93) – No correlation between WT – Mut difference and fragment size (r=0. 12) • 50 -60% GC • 100% genotype concordance – Taq. Man (350) – Sequencing (68)
906692139758ed71d8905407dd9d53e7.ppt