b48aa524b7bcf54116166d2b4ce9c647.ppt
- Количество слайдов: 33
 Modelling proteomes Ram Samudrala University of Washington
Modelling proteomes Ram Samudrala University of Washington
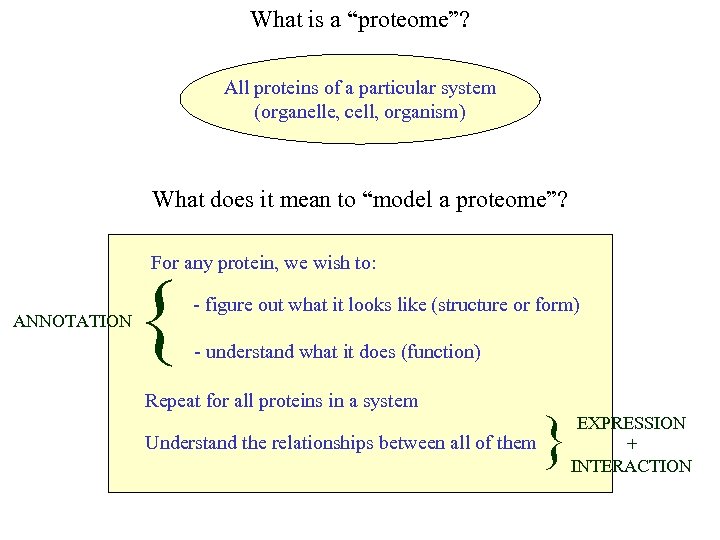 What is a “proteome”? All proteins of a particular system (organelle, cell, organism) What does it mean to “model a proteome”? For any protein, we wish to: ANNOTATION { - figure out what it looks like (structure or form) - understand what it does (function) Repeat for all proteins in a system Understand the relationships between all of them } EXPRESSION + INTERACTION
What is a “proteome”? All proteins of a particular system (organelle, cell, organism) What does it mean to “model a proteome”? For any protein, we wish to: ANNOTATION { - figure out what it looks like (structure or form) - understand what it does (function) Repeat for all proteins in a system Understand the relationships between all of them } EXPRESSION + INTERACTION
 Figuring out protein structure Experimental methods: - X-ray crystallography, NMR, electron diffraction - Accurate but slow Predictive methods: - De novo prediction, comparative modelling - Automated and relatively fast; not as accurate experimental methods
Figuring out protein structure Experimental methods: - X-ray crystallography, NMR, electron diffraction - Accurate but slow Predictive methods: - De novo prediction, comparative modelling - Automated and relatively fast; not as accurate experimental methods
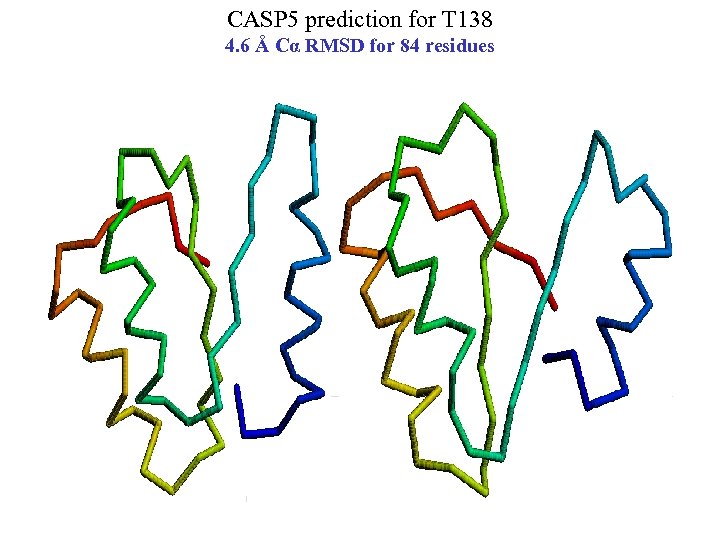 CASP 5 prediction for T 138 4. 6 Å Cα RMSD for 84 residues
CASP 5 prediction for T 138 4. 6 Å Cα RMSD for 84 residues
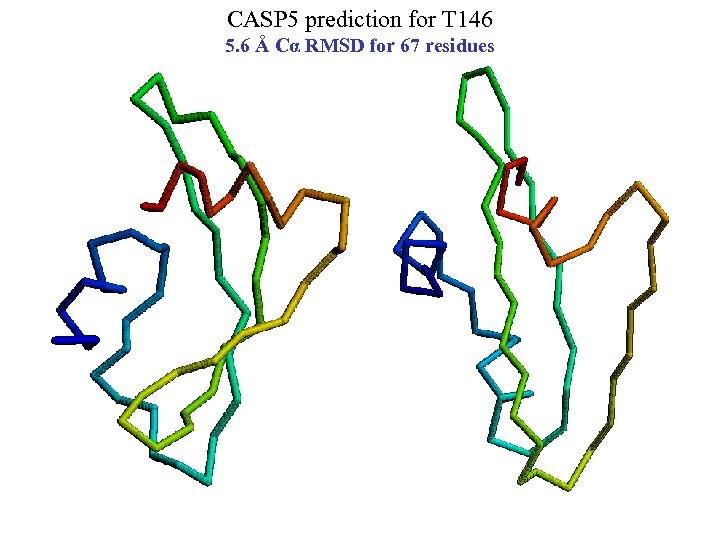 CASP 5 prediction for T 146 5. 6 Å Cα RMSD for 67 residues
CASP 5 prediction for T 146 5. 6 Å Cα RMSD for 67 residues
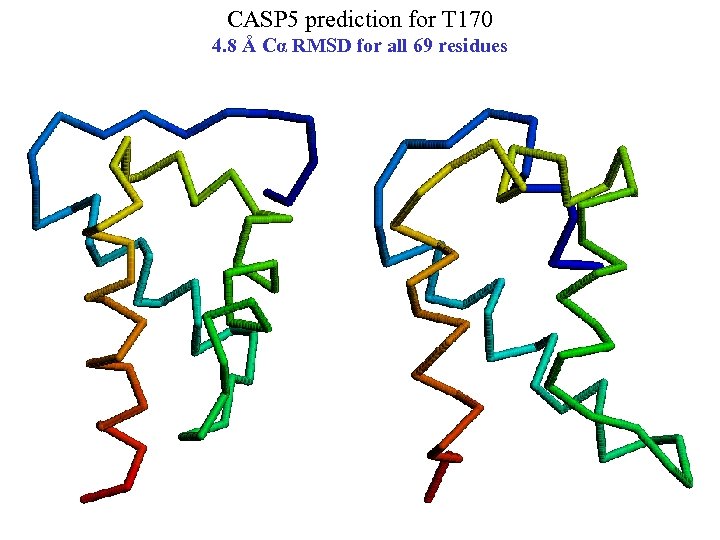 CASP 5 prediction for T 170 4. 8 Å Cα RMSD for all 69 residues
CASP 5 prediction for T 170 4. 8 Å Cα RMSD for all 69 residues
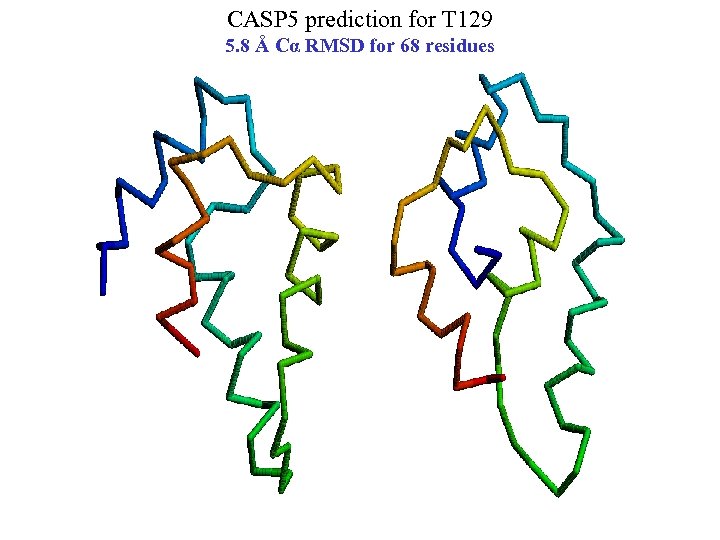 CASP 5 prediction for T 129 5. 8 Å Cα RMSD for 68 residues
CASP 5 prediction for T 129 5. 8 Å Cα RMSD for 68 residues
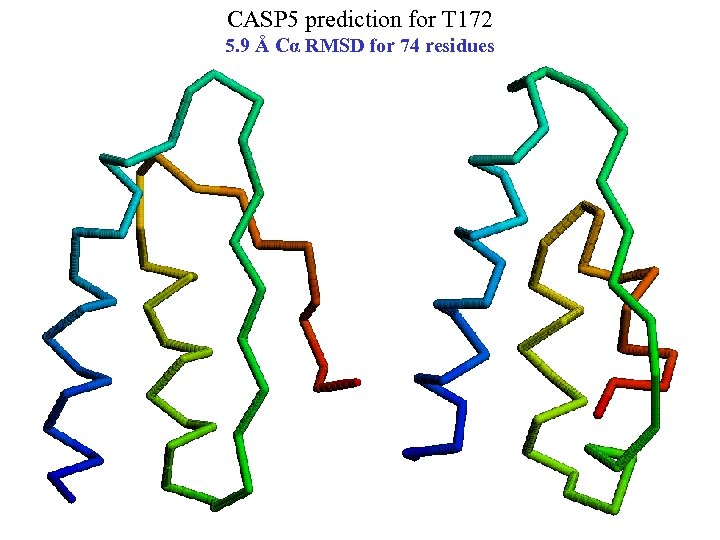 CASP 5 prediction for T 172 5. 9 Å Cα RMSD for 74 residues
CASP 5 prediction for T 172 5. 9 Å Cα RMSD for 74 residues
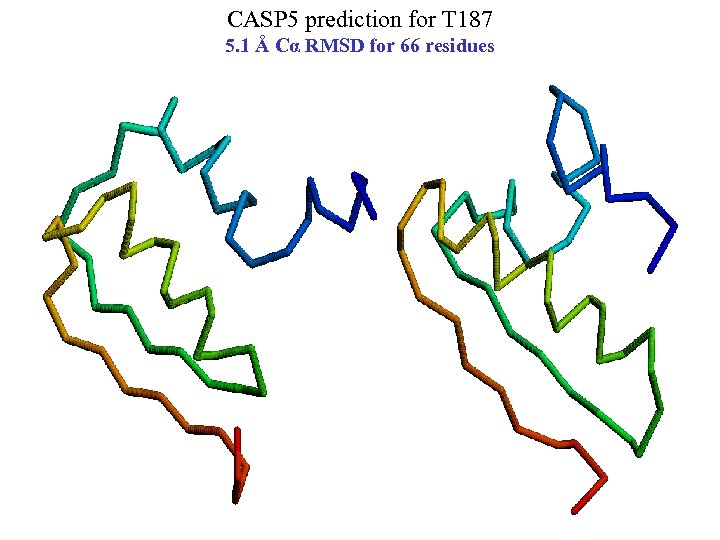 CASP 5 prediction for T 187 5. 1 Å Cα RMSD for 66 residues
CASP 5 prediction for T 187 5. 1 Å Cα RMSD for 66 residues
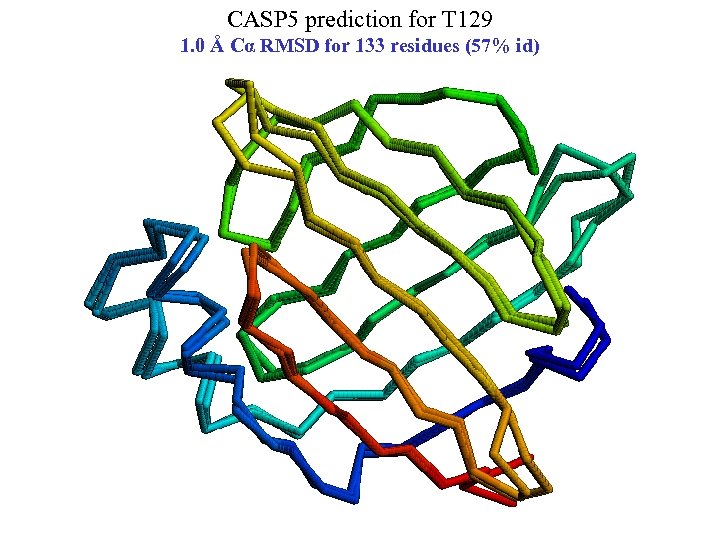 CASP 5 prediction for T 129 1. 0 Å Cα RMSD for 133 residues (57% id)
CASP 5 prediction for T 129 1. 0 Å Cα RMSD for 133 residues (57% id)
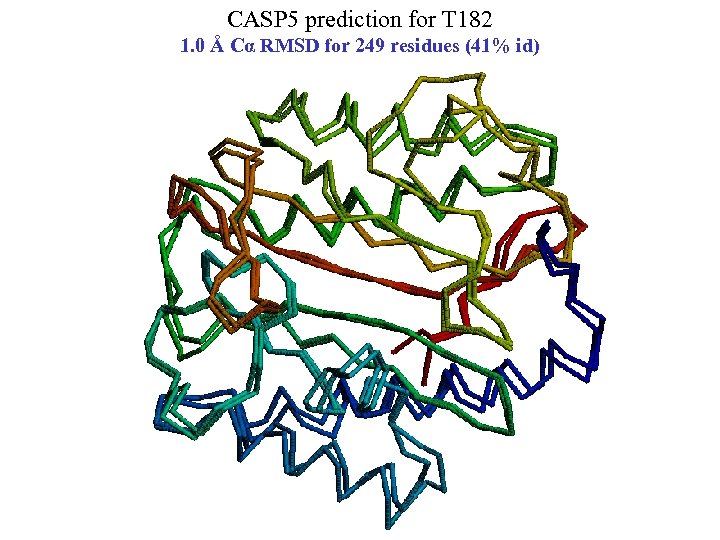 CASP 5 prediction for T 182 1. 0 Å Cα RMSD for 249 residues (41% id)
CASP 5 prediction for T 182 1. 0 Å Cα RMSD for 249 residues (41% id)
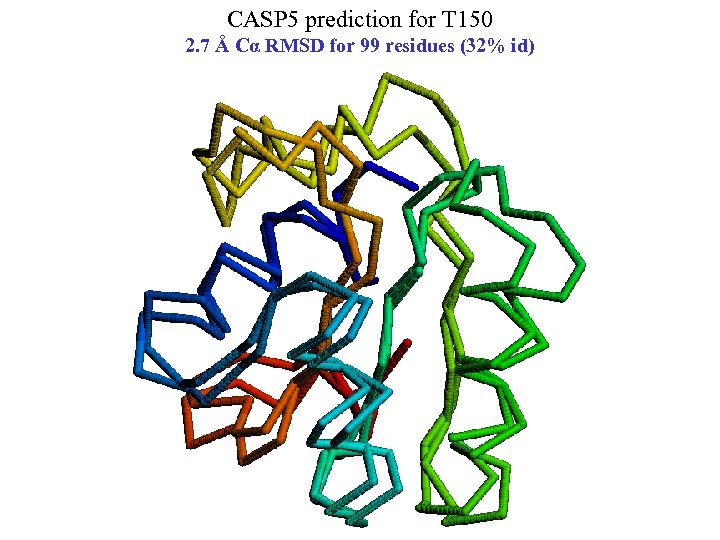 CASP 5 prediction for T 150 2. 7 Å Cα RMSD for 99 residues (32% id)
CASP 5 prediction for T 150 2. 7 Å Cα RMSD for 99 residues (32% id)
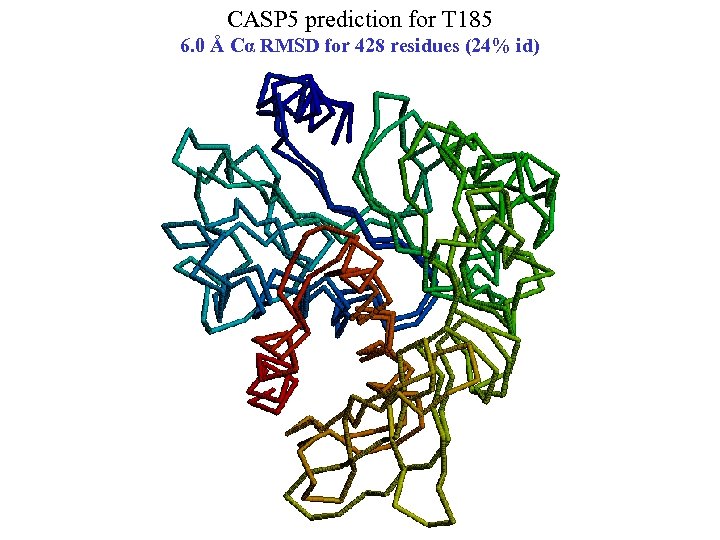 CASP 5 prediction for T 185 6. 0 Å Cα RMSD for 428 residues (24% id)
CASP 5 prediction for T 185 6. 0 Å Cα RMSD for 428 residues (24% id)
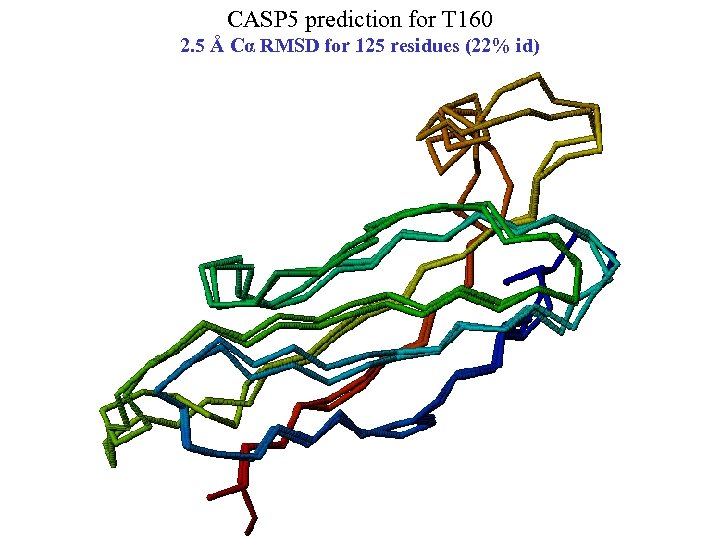 CASP 5 prediction for T 160 2. 5 Å Cα RMSD for 125 residues (22% id)
CASP 5 prediction for T 160 2. 5 Å Cα RMSD for 125 residues (22% id)
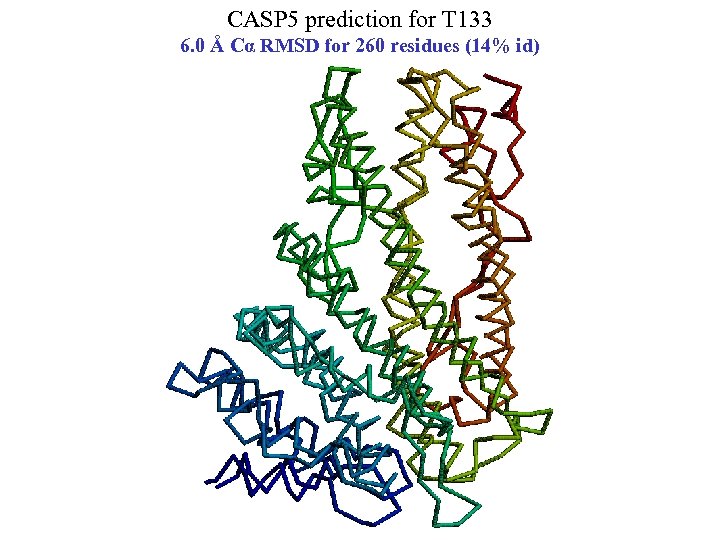 CASP 5 prediction for T 133 6. 0 Å Cα RMSD for 260 residues (14% id)
CASP 5 prediction for T 133 6. 0 Å Cα RMSD for 260 residues (14% id)
 Figuring out protein function Experimental methods: - A variety of single molecule and genomic/proteomic techniques - Usually not complete; slow Predictive methods: - Structure and sequence based methods using experimental knowledge of known functions - Automated and relatively fast; not as accurate as experimental methods; may be comprehensive
Figuring out protein function Experimental methods: - A variety of single molecule and genomic/proteomic techniques - Usually not complete; slow Predictive methods: - Structure and sequence based methods using experimental knowledge of known functions - Automated and relatively fast; not as accurate as experimental methods; may be comprehensive
 Structure and sequence based functional studies Using structure to rationalise mutagenesis data: - Dr adhesin study with Steve Moseley (Molec. Micro. 2002) - Invb study with Sam Miller Using structure to rationalise putative function: - Bacterial tubulin study with Jim Staley (PNAS 2003) Using structure and sequence to predict inhibitor binding: - Protease inhibitor binding studies using docking (BMC 2003) - Sequence-based prediction using regression models for HIV protease and RT with John Mittler (submitted) Structure and sequence based methods for functional classification: - In progress - General method to assign function from sequence - Analyse binding of 130 K compounds to structures - Build machine learning models of functional families
Structure and sequence based functional studies Using structure to rationalise mutagenesis data: - Dr adhesin study with Steve Moseley (Molec. Micro. 2002) - Invb study with Sam Miller Using structure to rationalise putative function: - Bacterial tubulin study with Jim Staley (PNAS 2003) Using structure and sequence to predict inhibitor binding: - Protease inhibitor binding studies using docking (BMC 2003) - Sequence-based prediction using regression models for HIV protease and RT with John Mittler (submitted) Structure and sequence based methods for functional classification: - In progress - General method to assign function from sequence - Analyse binding of 130 K compounds to structures - Build machine learning models of functional families
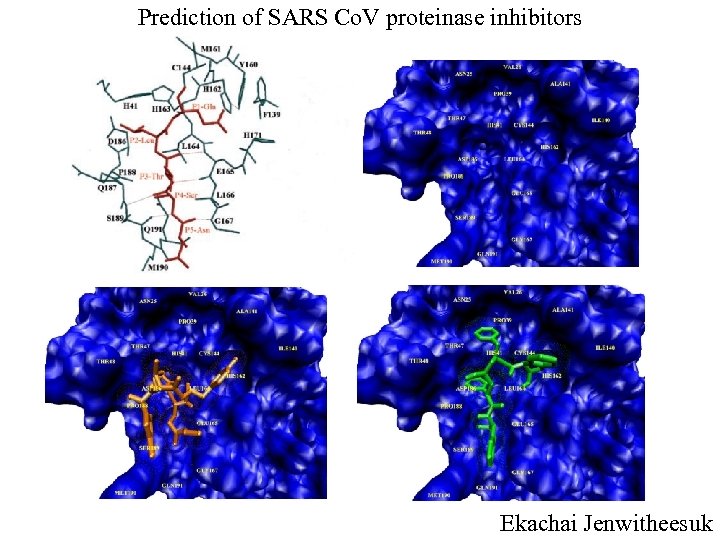 Prediction of SARS Co. V proteinase inhibitors Ekachai Jenwitheesuk
Prediction of SARS Co. V proteinase inhibitors Ekachai Jenwitheesuk
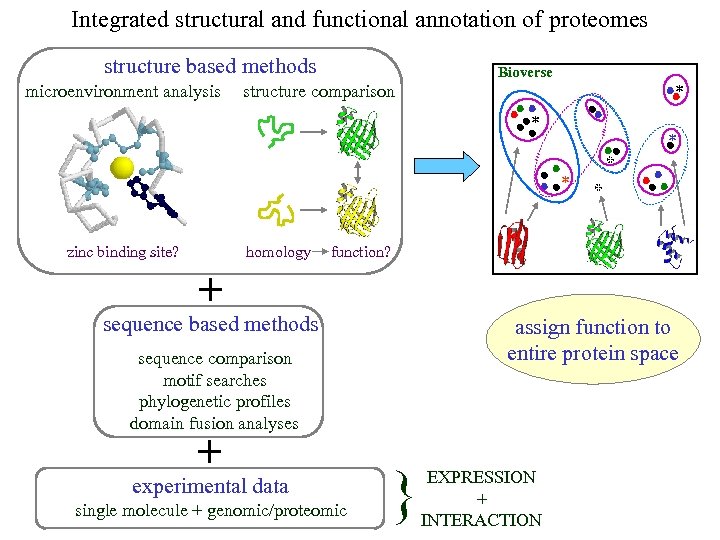 Integrated structural and functional annotation of proteomes structure based methods microenvironment analysis Bioverse structure comparison * * homology zinc binding site? * * function? + sequence based methods assign function to entire protein space sequence comparison motif searches phylogenetic profiles domain fusion analyses + experimental data single molecule + genomic/proteomic } EXPRESSION + INTERACTION
Integrated structural and functional annotation of proteomes structure based methods microenvironment analysis Bioverse structure comparison * * homology zinc binding site? * * function? + sequence based methods assign function to entire protein space sequence comparison motif searches phylogenetic profiles domain fusion analyses + experimental data single molecule + genomic/proteomic } EXPRESSION + INTERACTION
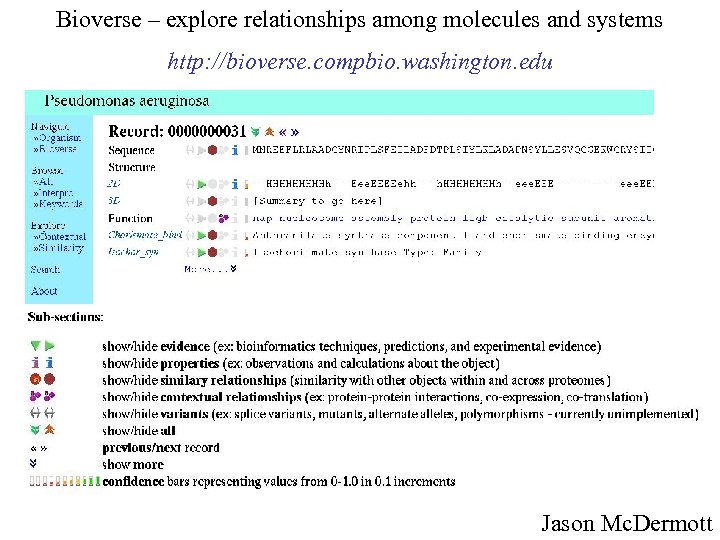 Bioverse – explore relationships among molecules and systems http: //bioverse. compbio. washington. edu Jason Mc. Dermott
Bioverse – explore relationships among molecules and systems http: //bioverse. compbio. washington. edu Jason Mc. Dermott
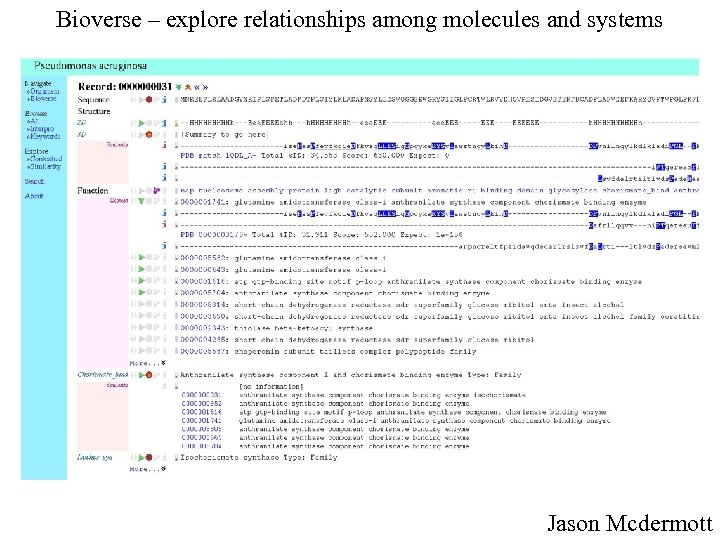 Bioverse – explore relationships among molecules and systems Jason Mcdermott
Bioverse – explore relationships among molecules and systems Jason Mcdermott
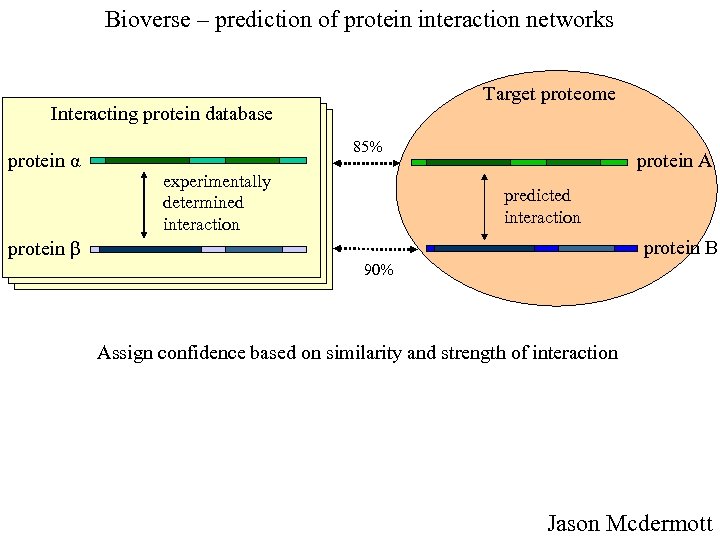 Bioverse – prediction of protein interaction networks Target proteome Interacting protein database protein α 85% experimentally determined interaction protein A predicted interaction protein B protein β 90% Assign confidence based on similarity and strength of interaction Jason Mcdermott
Bioverse – prediction of protein interaction networks Target proteome Interacting protein database protein α 85% experimentally determined interaction protein A predicted interaction protein B protein β 90% Assign confidence based on similarity and strength of interaction Jason Mcdermott
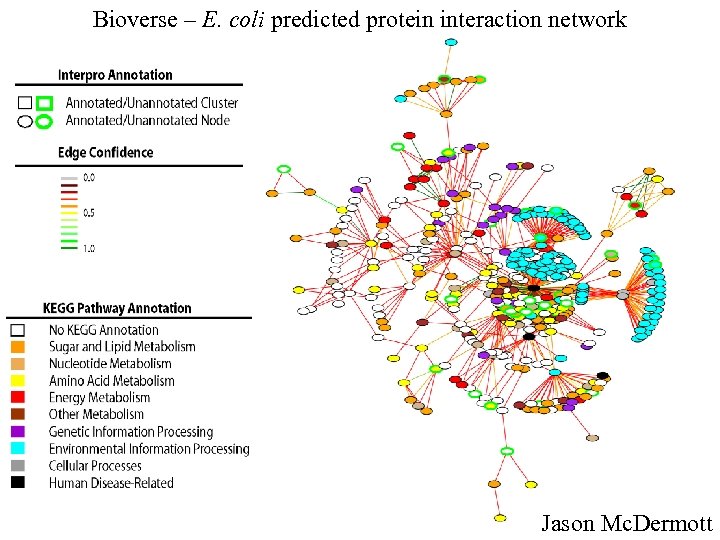 Bioverse – E. coli predicted protein interaction network Jason Mc. Dermott
Bioverse – E. coli predicted protein interaction network Jason Mc. Dermott
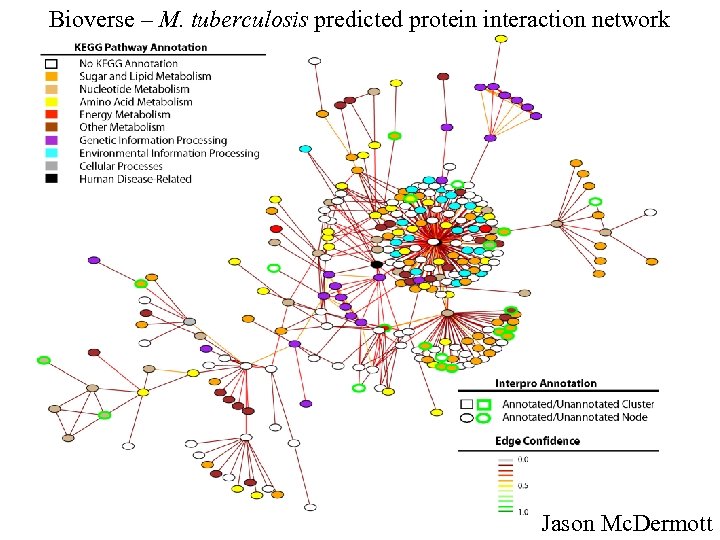 Bioverse – M. tuberculosis predicted protein interaction network Jason Mc. Dermott
Bioverse – M. tuberculosis predicted protein interaction network Jason Mc. Dermott
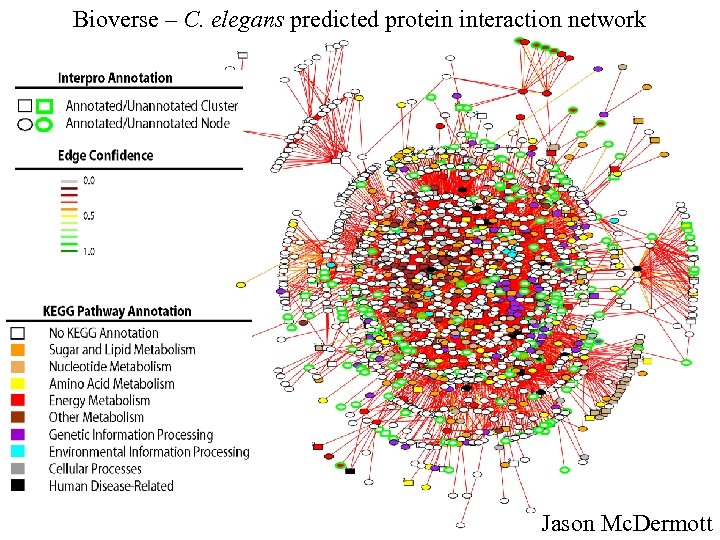 Bioverse – C. elegans predicted protein interaction network Jason Mc. Dermott
Bioverse – C. elegans predicted protein interaction network Jason Mc. Dermott
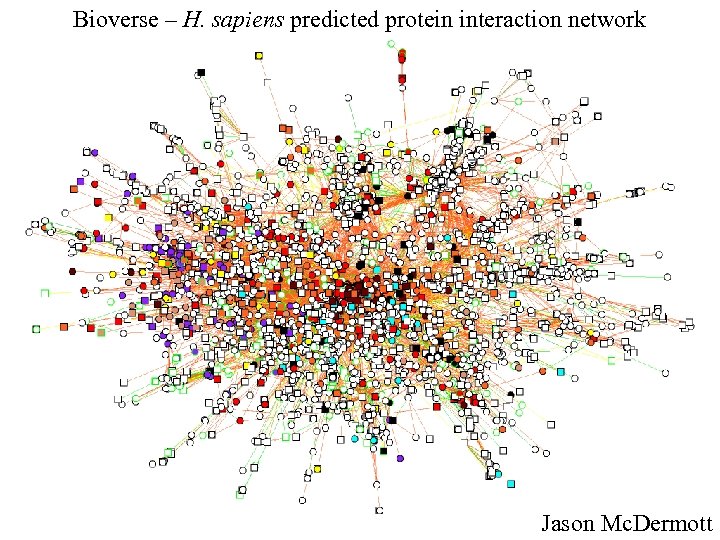 Bioverse – H. sapiens predicted protein interaction network Jason Mc. Dermott
Bioverse – H. sapiens predicted protein interaction network Jason Mc. Dermott
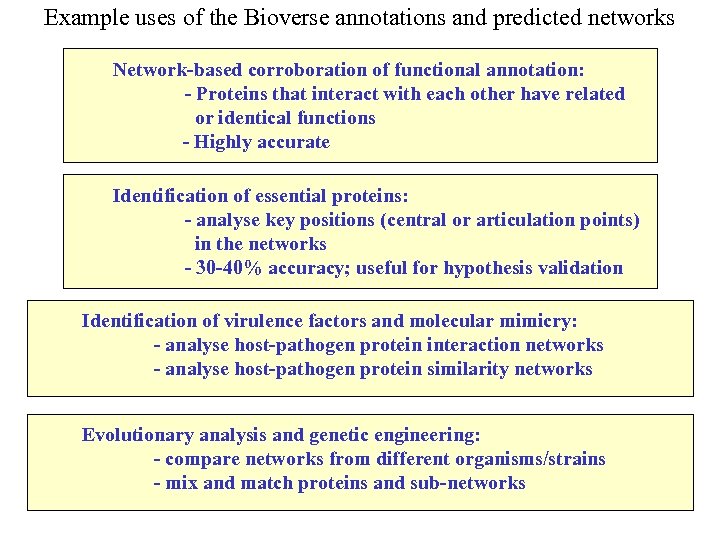 Example uses of the Bioverse annotations and predicted networks Network-based corroboration of functional annotation: - Proteins that interact with each other have related or identical functions - Highly accurate Identification of essential proteins: - analyse key positions (central or articulation points) in the networks - 30 -40% accuracy; useful for hypothesis validation Identification of virulence factors and molecular mimicry: - analyse host-pathogen protein interaction networks - analyse host-pathogen protein similarity networks Evolutionary analysis and genetic engineering: - compare networks from different organisms/strains - mix and match proteins and sub-networks
Example uses of the Bioverse annotations and predicted networks Network-based corroboration of functional annotation: - Proteins that interact with each other have related or identical functions - Highly accurate Identification of essential proteins: - analyse key positions (central or articulation points) in the networks - 30 -40% accuracy; useful for hypothesis validation Identification of virulence factors and molecular mimicry: - analyse host-pathogen protein interaction networks - analyse host-pathogen protein similarity networks Evolutionary analysis and genetic engineering: - compare networks from different organisms/strains - mix and match proteins and sub-networks
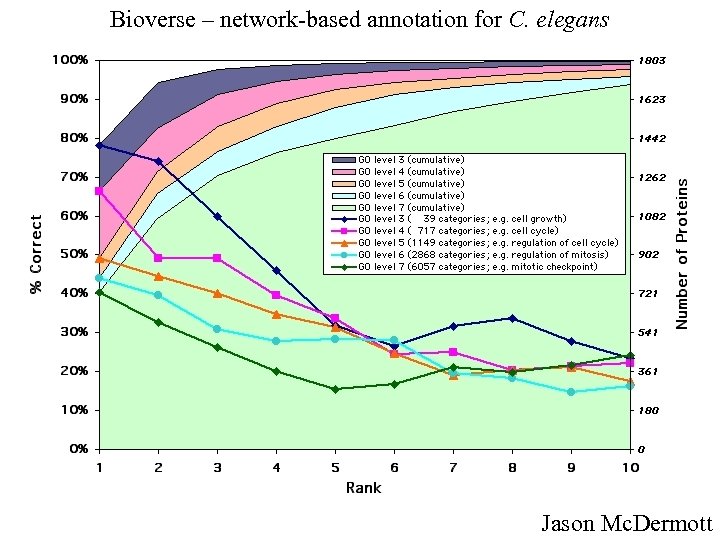 Bioverse – network-based annotation for C. elegans Jason Mc. Dermott
Bioverse – network-based annotation for C. elegans Jason Mc. Dermott
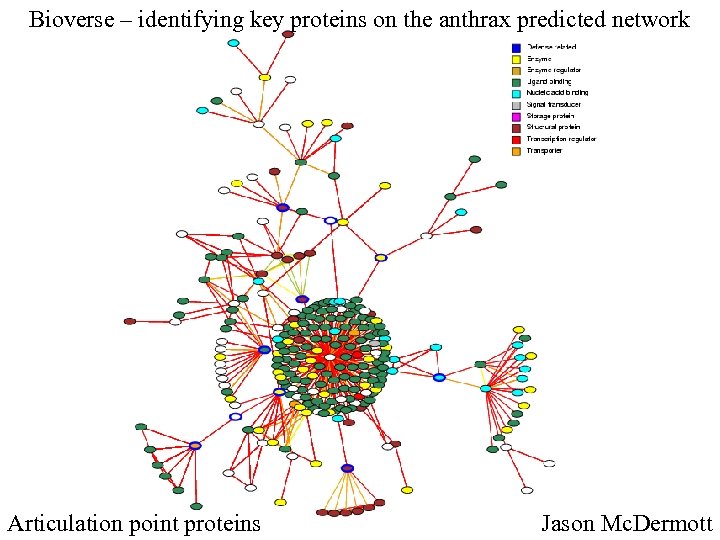 Bioverse – identifying key proteins on the anthrax predicted network Articulation point proteins Jason Mc. Dermott
Bioverse – identifying key proteins on the anthrax predicted network Articulation point proteins Jason Mc. Dermott
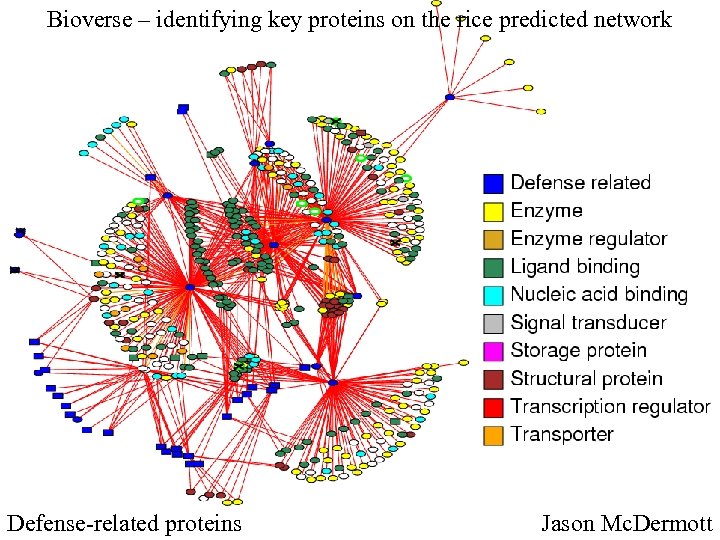 Bioverse – identifying key proteins on the rice predicted network Defense-related proteins Jason Mc. Dermott
Bioverse – identifying key proteins on the rice predicted network Defense-related proteins Jason Mc. Dermott
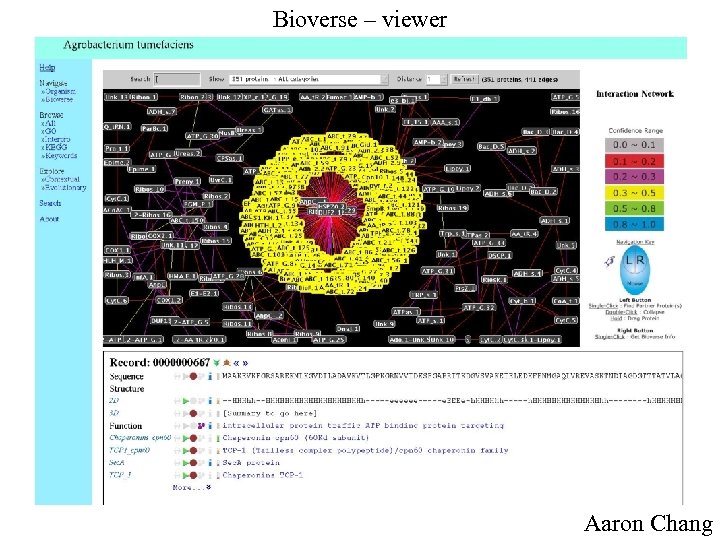 Bioverse – viewer Aaron Chang
Bioverse – viewer Aaron Chang
 Take home message Prediction of protein structure, function, and networks may be used to model whole genomes to understand organismal function and evolution
Take home message Prediction of protein structure, function, and networks may be used to model whole genomes to understand organismal function and evolution
 Acknowledgements Aaron Chang Ashley Lam Ekachai Jenwitheesuk Gong Cheng Jason Mc. Dermott Kai Wang Ling-Hong Hung Lynne Townsend Marissa La. Madrid Mike Inouye Stewart Moughon Shing-Chung Ngan Yi-Ling Cheng Zach Frazier National Institutes of Health National Science Foundation Searle Scholars Program (Kinship Foundation) UW Advanced Technology Initative in Infectious Diseases
Acknowledgements Aaron Chang Ashley Lam Ekachai Jenwitheesuk Gong Cheng Jason Mc. Dermott Kai Wang Ling-Hong Hung Lynne Townsend Marissa La. Madrid Mike Inouye Stewart Moughon Shing-Chung Ngan Yi-Ling Cheng Zach Frazier National Institutes of Health National Science Foundation Searle Scholars Program (Kinship Foundation) UW Advanced Technology Initative in Infectious Diseases


