6752749223fe7e0b904d242494b24f08.ppt
- Количество слайдов: 22
 Mitochondrial complex III deficiency associated with a homozygous mutation in UQCRQ Ortal Barel Laboratory of Dr. Ohad Birk Ben Gurion University Beer-Sheva, Israel
Mitochondrial complex III deficiency associated with a homozygous mutation in UQCRQ Ortal Barel Laboratory of Dr. Ohad Birk Ben Gurion University Beer-Sheva, Israel
 The Israeli Bedouin § Isolated inbred population § High rate of consanguineous marriage § Negev region of southern Israel § High frequency of autosomal recessive diseases § Effective in mapping genes of rare Recessive diseases
The Israeli Bedouin § Isolated inbred population § High rate of consanguineous marriage § Negev region of southern Israel § High frequency of autosomal recessive diseases § Effective in mapping genes of rare Recessive diseases
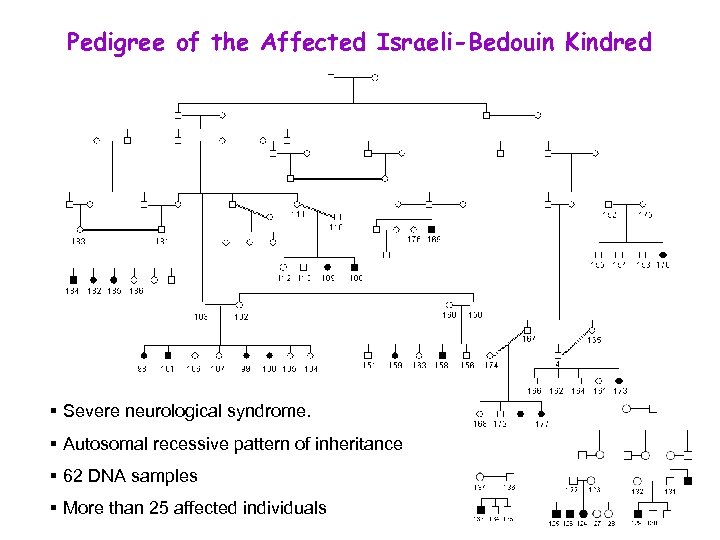 Pedigree of the Affected Israeli-Bedouin Kindred § Severe neurological syndrome. B § Autosomal recessive pattern of inheritance § 62 DNA samples § More than 25 affected individuals
Pedigree of the Affected Israeli-Bedouin Kindred § Severe neurological syndrome. B § Autosomal recessive pattern of inheritance § 62 DNA samples § More than 25 affected individuals
 The hallmarks of the phenotype: § Markedly uniform phenotype § Early onset in infancy § Severe psychomotor retardation § Extrapyramidal signs (age 2 -3 years): Dystonic postures, athetoid movements and ataxia § Severe defects in verbal receptive communication § Near total absence of expressive communication skills (verbal) § Hypotonia (inability to walk unsupported)
The hallmarks of the phenotype: § Markedly uniform phenotype § Early onset in infancy § Severe psychomotor retardation § Extrapyramidal signs (age 2 -3 years): Dystonic postures, athetoid movements and ataxia § Severe defects in verbal receptive communication § Near total absence of expressive communication skills (verbal) § Hypotonia (inability to walk unsupported)
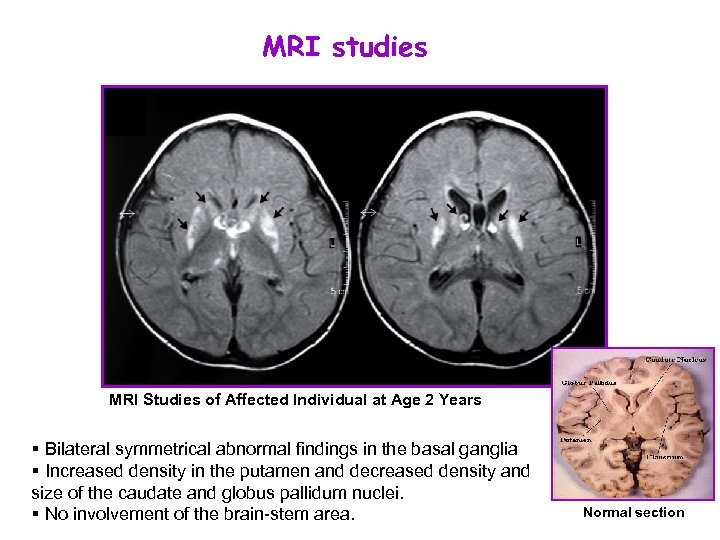 MRI studies MRI Studies of Affected Individual at Age 2 Years § Bilateral symmetrical abnormal findings in the basal ganglia § Increased density in the putamen and decreased density and size of the caudate and globus pallidum nuclei. § No involvement of the brain-stem area. Normal section
MRI studies MRI Studies of Affected Individual at Age 2 Years § Bilateral symmetrical abnormal findings in the basal ganglia § Increased density in the putamen and decreased density and size of the caudate and globus pallidum nuclei. § No involvement of the brain-stem area. Normal section
 Muscle biopsy § Muscle biopsies were done in three of the affected individuals (ages 3, 15 and 2 years). - Preserved architecture of the skeletal muscle - Nonspecific histological findings § Metabolic work up: - All laboratories values were within the normal range except: Elevated serum lactate and pyruvate Elevated Cerbrospinal fluid (CSF) lactate
Muscle biopsy § Muscle biopsies were done in three of the affected individuals (ages 3, 15 and 2 years). - Preserved architecture of the skeletal muscle - Nonspecific histological findings § Metabolic work up: - All laboratories values were within the normal range except: Elevated serum lactate and pyruvate Elevated Cerbrospinal fluid (CSF) lactate
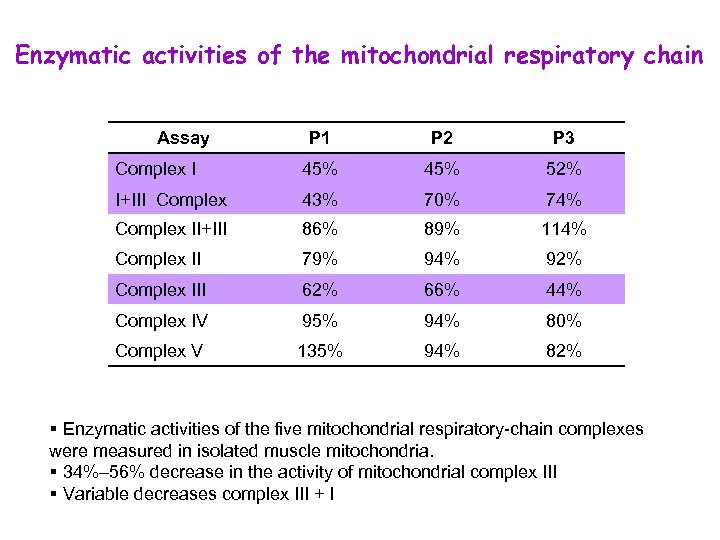 Enzymatic activities of the mitochondrial respiratory chain Assay P 1 P 2 P 3 Complex I 45% 52% I+III Complex 43% 70% 74% Complex II+III 86% 89% 114% Complex II 79% 94% 92% Complex III 62% 66% 44% Complex IV 95% 94% 80% Complex V 135% 94% 82% § Enzymatic activities of the five mitochondrial respiratory-chain complexes were measured in isolated muscle mitochondria. § 34%– 56% decrease in the activity of mitochondrial complex III § Variable decreases complex III + I
Enzymatic activities of the mitochondrial respiratory chain Assay P 1 P 2 P 3 Complex I 45% 52% I+III Complex 43% 70% 74% Complex II+III 86% 89% 114% Complex II 79% 94% 92% Complex III 62% 66% 44% Complex IV 95% 94% 80% Complex V 135% 94% 82% § Enzymatic activities of the five mitochondrial respiratory-chain complexes were measured in isolated muscle mitochondria. § 34%– 56% decrease in the activity of mitochondrial complex III § Variable decreases complex III + I
 Mitochondrial disease Summary of the phenotype and the clinical features: § Autosomal recessive § Elevated serum lactate and pyruvate § Decreased activity of mitochondrial complex III & I § Neurological abnormalities § Involvement of tissues characteristic of mitochondrial diseases (high energy consumption)
Mitochondrial disease Summary of the phenotype and the clinical features: § Autosomal recessive § Elevated serum lactate and pyruvate § Decreased activity of mitochondrial complex III & I § Neurological abnormalities § Involvement of tissues characteristic of mitochondrial diseases (high energy consumption)
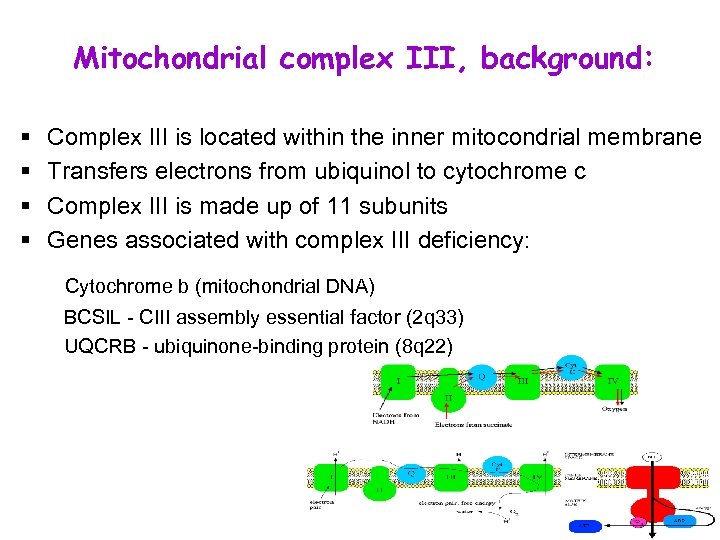 Mitochondrial complex III, background: § § Complex III is located within the inner mitocondrial membrane Transfers electrons from ubiquinol to cytochrome c Complex III is made up of 11 subunits Genes associated with complex III deficiency: Cytochrome b (mitochondrial DNA) BCSIL - CIII assembly essential factor (2 q 33) UQCRB - ubiquinone-binding protein (8 q 22)
Mitochondrial complex III, background: § § Complex III is located within the inner mitocondrial membrane Transfers electrons from ubiquinol to cytochrome c Complex III is made up of 11 subunits Genes associated with complex III deficiency: Cytochrome b (mitochondrial DNA) BCSIL - CIII assembly essential factor (2 q 33) UQCRB - ubiquinone-binding protein (8 q 22)
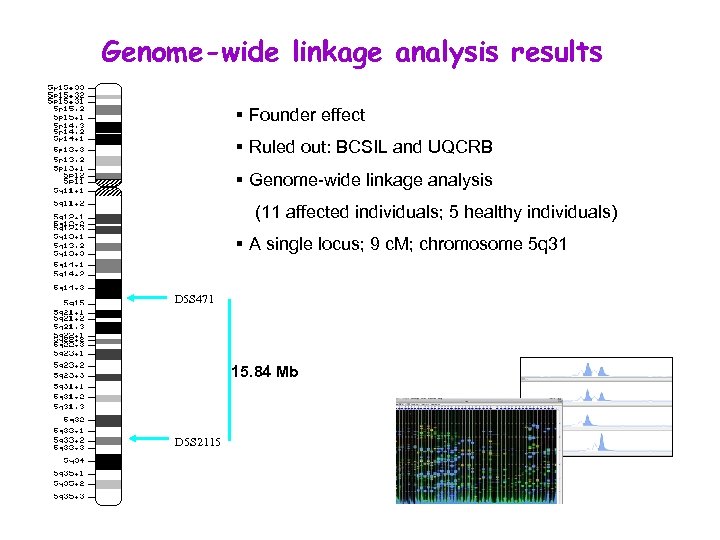 Genome-wide linkage analysis results § Founder effect § Ruled out: BCSIL and UQCRB § Genome-wide linkage analysis (11 affected individuals; 5 healthy individuals) § A single locus; 9 c. M; chromosome 5 q 31 D 5 S 471 15. 84 Mb D 5 S 2115
Genome-wide linkage analysis results § Founder effect § Ruled out: BCSIL and UQCRB § Genome-wide linkage analysis (11 affected individuals; 5 healthy individuals) § A single locus; 9 c. M; chromosome 5 q 31 D 5 S 471 15. 84 Mb D 5 S 2115
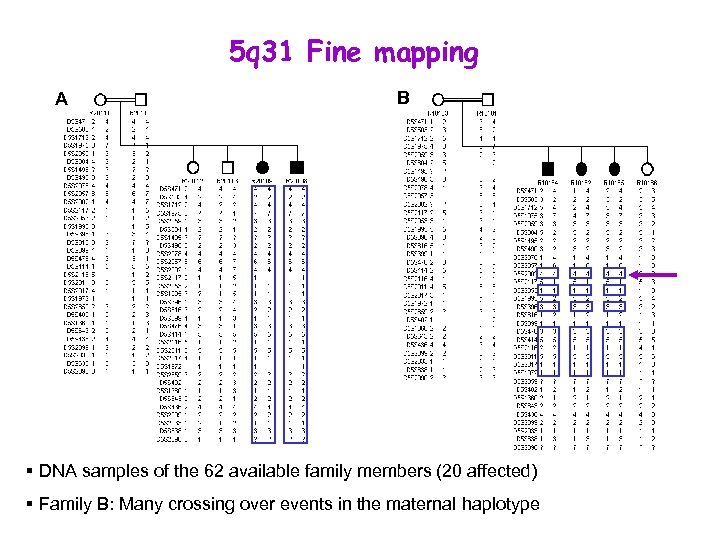 5 q 31 Fine mapping A B § DNA samples of the 62 available family members (20 affected) § Family B: Many crossing over events in the maternal haplotype
5 q 31 Fine mapping A B § DNA samples of the 62 available family members (20 affected) § Family B: Many crossing over events in the maternal haplotype
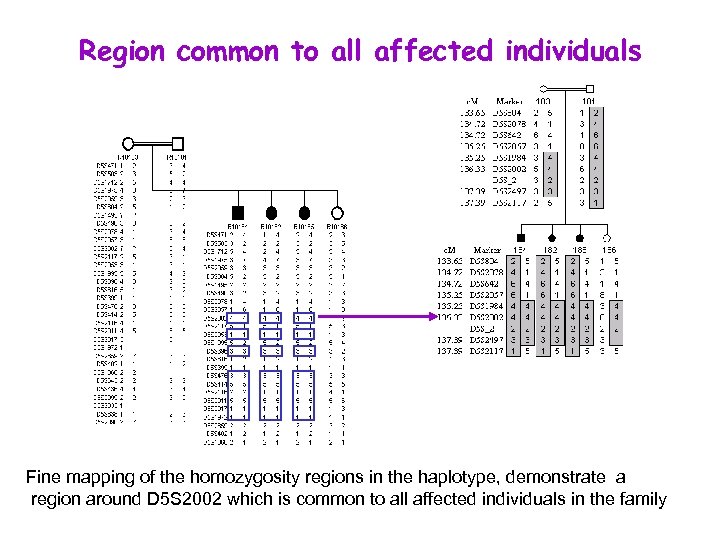 Region common to all affected individuals Fine mapping of the homozygosity regions in the haplotype, demonstrate a region around D 5 S 2002 which is common to all affected individuals in the family
Region common to all affected individuals Fine mapping of the homozygosity regions in the haplotype, demonstrate a region around D 5 S 2002 which is common to all affected individuals in the family
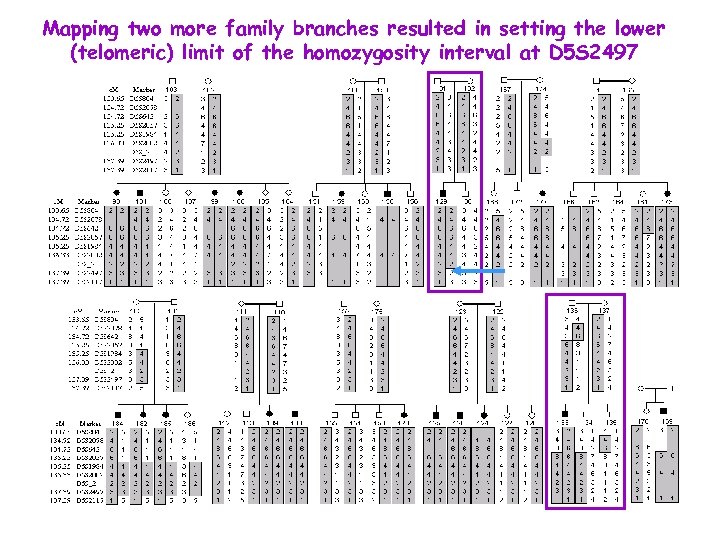 Mapping two more family branches resulted in setting the lower (telomeric) limit of the homozygosity interval at D 5 S 2497
Mapping two more family branches resulted in setting the lower (telomeric) limit of the homozygosity interval at D 5 S 2497
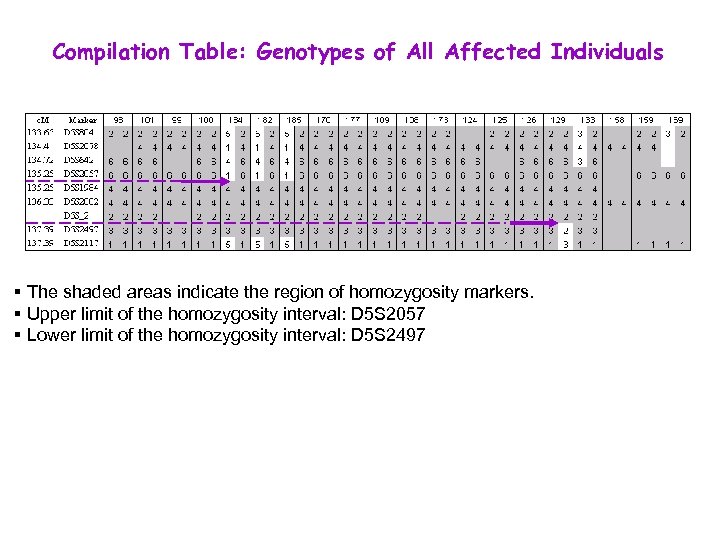 Compilation Table: Genotypes of All Affected Individuals § The shaded areas indicate the region of homozygosity markers. § Upper limit of the homozygosity interval: D 5 S 2057 § Lower limit of the homozygosity interval: D 5 S 2497
Compilation Table: Genotypes of All Affected Individuals § The shaded areas indicate the region of homozygosity markers. § Upper limit of the homozygosity interval: D 5 S 2057 § Lower limit of the homozygosity interval: D 5 S 2497
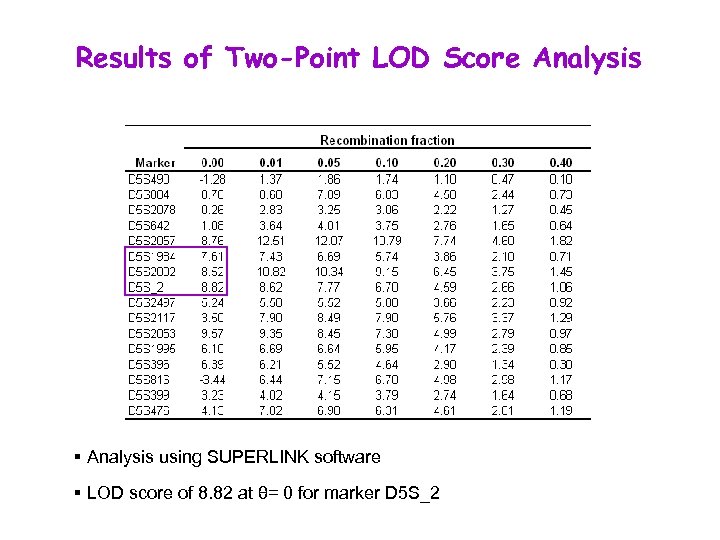 Results of Two-Point LOD Score Analysis § Analysis using SUPERLINK software § LOD score of 8. 82 at θ= 0 for marker D 5 S_2
Results of Two-Point LOD Score Analysis § Analysis using SUPERLINK software § LOD score of 8. 82 at θ= 0 for marker D 5 S_2
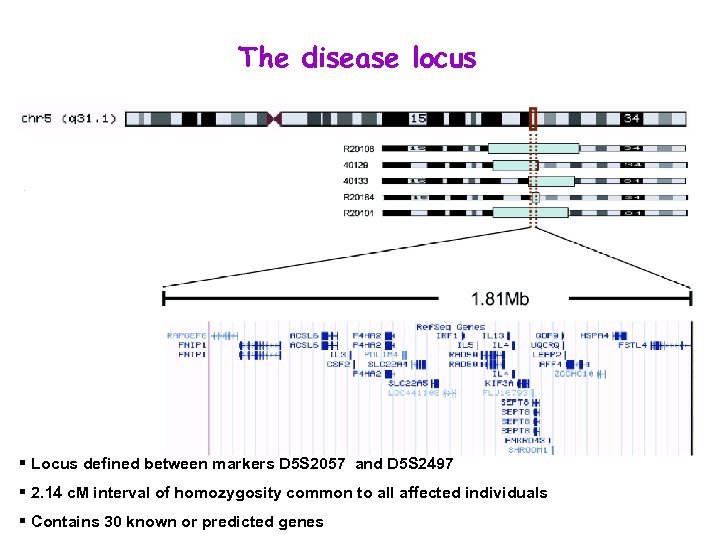 The disease locus § Locus defined between markers D 5 S 2057 and D 5 S 2497 § 2. 14 c. M interval of homozygosity common to all affected individuals § Contains 30 known or predicted genes
The disease locus § Locus defined between markers D 5 S 2057 and D 5 S 2497 § 2. 14 c. M interval of homozygosity common to all affected individuals § Contains 30 known or predicted genes
 UQCRQ Official Full Name: Ubiquinol-cytochrome c reductase, complex III subunit VII, 9. 5 k. Da (QCR 8; QP-C ) Summary: § Encodes a ubiquinone-binding protein of low molecular mass. § The protein is a small core-associated protein and a subunit of mitochondrial complex III. § The bc 1 complex (complex III ) contains 11 subunits: - 3 respiratory subunits (cytochrome b, cytochrome c 1 and Rieske) - 2 core proteins (UQCRC 1 and UQCRC 2) - 6 low-molecular weight proteins (UQCRH, UQCRB, UQCRQ, UQCR 10, UQCR 11 and a cleavage product of Rieske)
UQCRQ Official Full Name: Ubiquinol-cytochrome c reductase, complex III subunit VII, 9. 5 k. Da (QCR 8; QP-C ) Summary: § Encodes a ubiquinone-binding protein of low molecular mass. § The protein is a small core-associated protein and a subunit of mitochondrial complex III. § The bc 1 complex (complex III ) contains 11 subunits: - 3 respiratory subunits (cytochrome b, cytochrome c 1 and Rieske) - 2 core proteins (UQCRC 1 and UQCRC 2) - 6 low-molecular weight proteins (UQCRH, UQCRB, UQCRQ, UQCR 10, UQCR 11 and a cleavage product of Rieske)
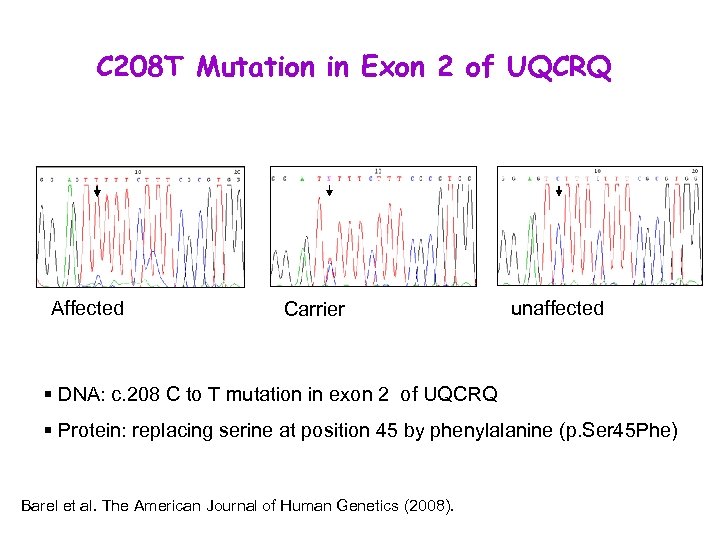 C 208 T Mutation in Exon 2 of UQCRQ Affected Carrier unaffected § DNA: c. 208 C to T mutation in exon 2 of UQCRQ § Protein: replacing serine at position 45 by phenylalanine (p. Ser 45 Phe) Barel et al. The American Journal of Human Genetics (2008).
C 208 T Mutation in Exon 2 of UQCRQ Affected Carrier unaffected § DNA: c. 208 C to T mutation in exon 2 of UQCRQ § Protein: replacing serine at position 45 by phenylalanine (p. Ser 45 Phe) Barel et al. The American Journal of Human Genetics (2008).
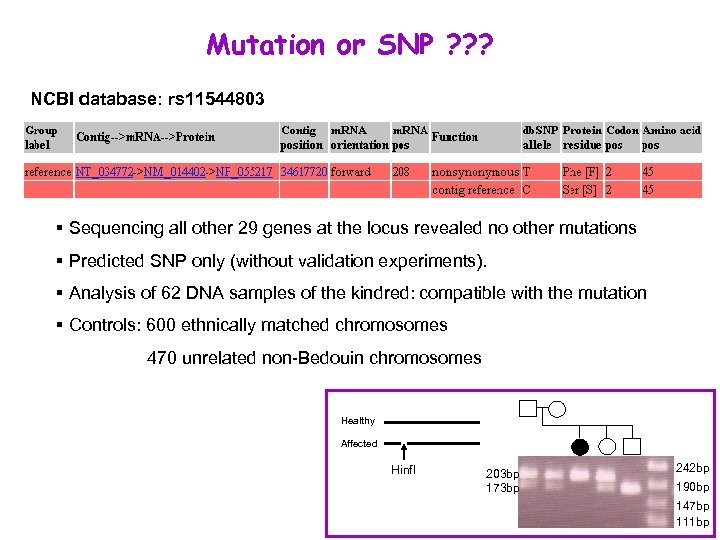 Mutation or SNP ? ? ? NCBI database: rs 11544803 § Sequencing all other 29 genes at the locus revealed no other mutations § Predicted SNP only (without validation experiments). § Analysis of 62 DNA samples of the kindred: compatible with the mutation § Controls: 600 ethnically matched chromosomes 470 unrelated non-Bedouin chromosomes Healthy Affected Hinf. I 203 bp 173 bp 242 bp 190 bp 147 bp 111 bp
Mutation or SNP ? ? ? NCBI database: rs 11544803 § Sequencing all other 29 genes at the locus revealed no other mutations § Predicted SNP only (without validation experiments). § Analysis of 62 DNA samples of the kindred: compatible with the mutation § Controls: 600 ethnically matched chromosomes 470 unrelated non-Bedouin chromosomes Healthy Affected Hinf. I 203 bp 173 bp 242 bp 190 bp 147 bp 111 bp
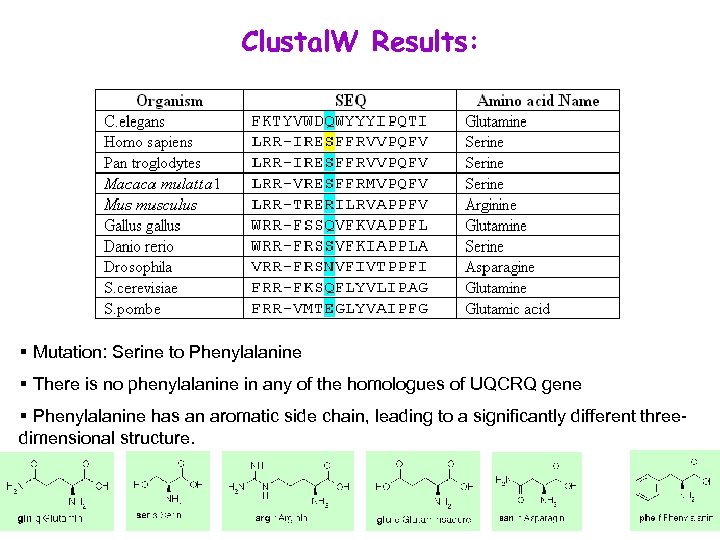 Clustal. W Results: § Mutation: Serine to Phenylalanine § There is no phenylalanine in any of the homologues of UQCRQ gene § Phenylalanine has an aromatic side chain, leading to a significantly different threedimensional structure.
Clustal. W Results: § Mutation: Serine to Phenylalanine § There is no phenylalanine in any of the homologues of UQCRQ gene § Phenylalanine has an aromatic side chain, leading to a significantly different threedimensional structure.
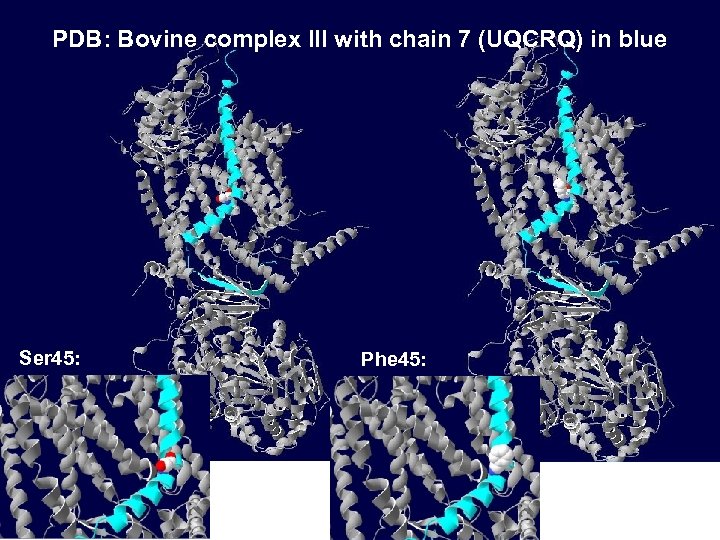 PDB: Bovine complex III with chain 7 (UQCRQ) in blue Ser 45: Phe 45:
PDB: Bovine complex III with chain 7 (UQCRQ) in blue Ser 45: Phe 45:
 Acknowledgements: Dr. Ohad Birk’s group Dr. Rivka Ofir Dr. Sara Sivan The Lab members Dr. Zamir Shorer Pediatric Neurology, Soroka Medical Center, Beer-Sheva, Israel Dr. Ann Saada (Reisch) Metabolic Disease Unit, Hadassah Medical Center, Jerusalem Dr. Stavit Shalev Genetics Institute, Ha. Emek Medical Center, Afula Asi Cohen, Dr. Noam Zilberberg Lab, Department of Life Sciences, BGU The Kahn Family Foundation The Ori’s Foundation - In Memory of Ori Levi Foundation that supports research for the diagnosis, treatment and cure of mitochondrial disorder (www. orifund. org)
Acknowledgements: Dr. Ohad Birk’s group Dr. Rivka Ofir Dr. Sara Sivan The Lab members Dr. Zamir Shorer Pediatric Neurology, Soroka Medical Center, Beer-Sheva, Israel Dr. Ann Saada (Reisch) Metabolic Disease Unit, Hadassah Medical Center, Jerusalem Dr. Stavit Shalev Genetics Institute, Ha. Emek Medical Center, Afula Asi Cohen, Dr. Noam Zilberberg Lab, Department of Life Sciences, BGU The Kahn Family Foundation The Ori’s Foundation - In Memory of Ori Levi Foundation that supports research for the diagnosis, treatment and cure of mitochondrial disorder (www. orifund. org)


