e88bf501d5cc0b57ebd95a3c10371fac.ppt
- Количество слайдов: 89
 Mining the functional genomics data III Data integration: Gene Ontology, PPI, URLMAP Jaak Vilo vilo@egeen. ee Havana, Cuba, 21. 11. 2003
Mining the functional genomics data III Data integration: Gene Ontology, PPI, URLMAP Jaak Vilo vilo@egeen. ee Havana, Cuba, 21. 11. 2003
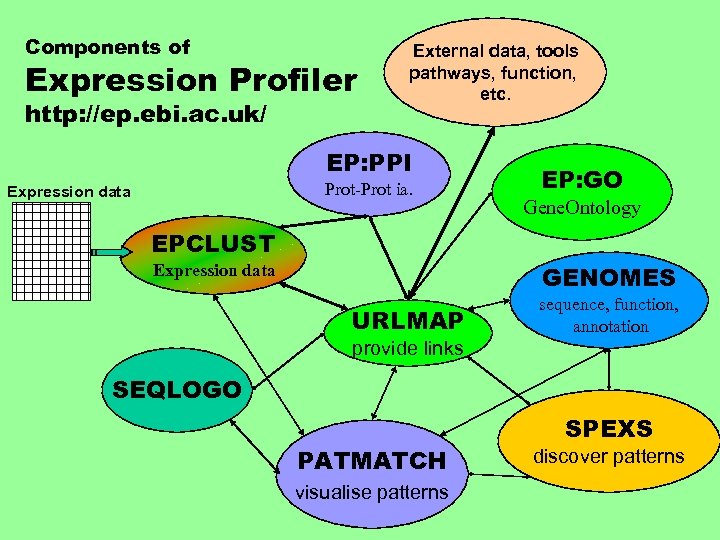 Components of Expression Profiler http: //ep. ebi. ac. uk/ External data, tools pathways, function, etc. EP: PPI Prot-Prot ia. Expression data EP: GO Gene. Ontology EPCLUST Expression data GENOMES URLMAP sequence, function, annotation provide links SEQLOGO PATMATCH visualise patterns SPEXS discover patterns
Components of Expression Profiler http: //ep. ebi. ac. uk/ External data, tools pathways, function, etc. EP: PPI Prot-Prot ia. Expression data EP: GO Gene. Ontology EPCLUST Expression data GENOMES URLMAP sequence, function, annotation provide links SEQLOGO PATMATCH visualise patterns SPEXS discover patterns
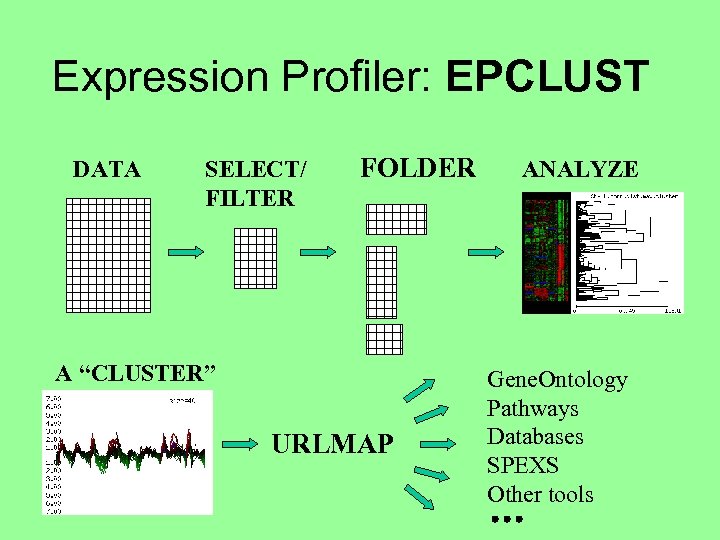 Expression Profiler: EPCLUST DATA SELECT/ FILTER FOLDER A “CLUSTER” URLMAP ANALYZE Gene. Ontology Pathways Databases SPEXS Other tools
Expression Profiler: EPCLUST DATA SELECT/ FILTER FOLDER A “CLUSTER” URLMAP ANALYZE Gene. Ontology Pathways Databases SPEXS Other tools
 URLMAP • Given a cluster of genes - many web based tools and databases to consult/follow up. How to link to them? • How to manage many links, many tools? • Answer: Centralize that linking
URLMAP • Given a cluster of genes - many web based tools and databases to consult/follow up. How to link to them? • How to manage many links, many tools? • Answer: Centralize that linking
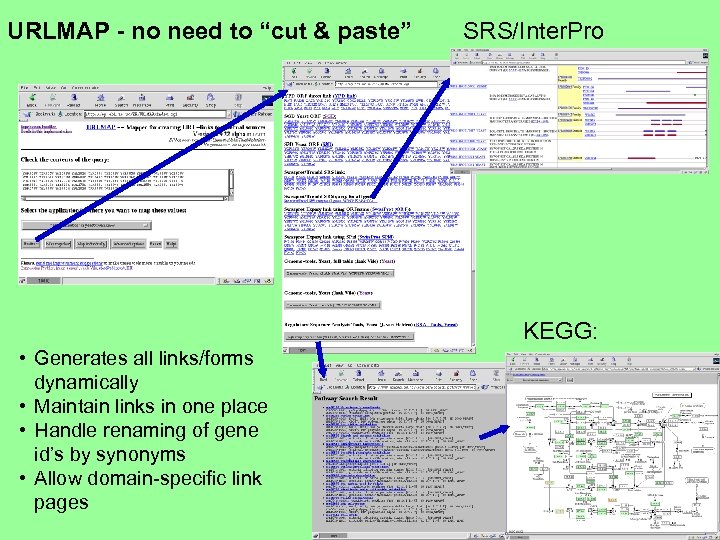 URLMAP - no need to “cut & paste” SRS/Inter. Pro KEGG: • Generates all links/forms dynamically • Maintain links in one place • Handle renaming of gene id’s by synonyms • Allow domain-specific link pages
URLMAP - no need to “cut & paste” SRS/Inter. Pro KEGG: • Generates all links/forms dynamically • Maintain links in one place • Handle renaming of gene id’s by synonyms • Allow domain-specific link pages
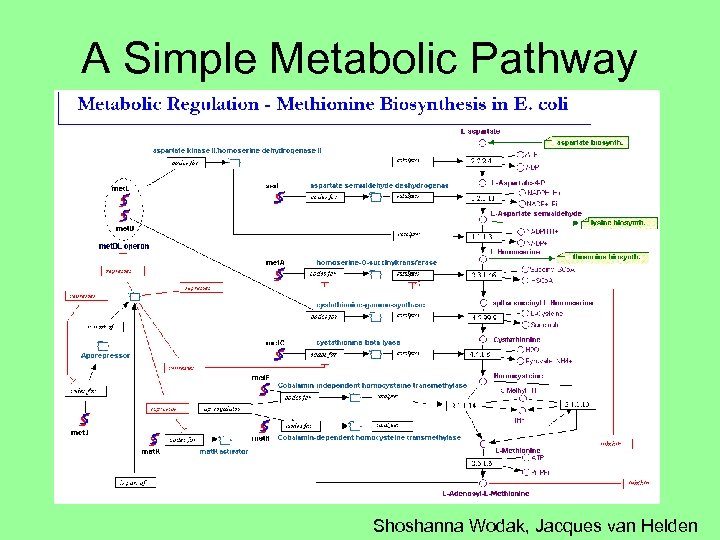 A Simple Metabolic Pathway Shoshanna Wodak, Jacques van Helden
A Simple Metabolic Pathway Shoshanna Wodak, Jacques van Helden
 Links for each item type • Yeast S. cerevisiae gene ID-s (ORFname, SP id, SGD ID, …) • Pattern collections, e. g. substrings to profile generation by SEQLOGO • Keyword searches from web based search engines
Links for each item type • Yeast S. cerevisiae gene ID-s (ORFname, SP id, SGD ID, …) • Pattern collections, e. g. substrings to profile generation by SEQLOGO • Keyword searches from web based search engines
 Management of links • Hierarchies of link collections • One can point to any (sub)hierarchy directly • LINK = • • URL, title, form parameters modifications/code DB lookups for synonyms
Management of links • Hierarchies of link collections • One can point to any (sub)hierarchy directly • LINK = • • URL, title, form parameters modifications/code DB lookups for synonyms
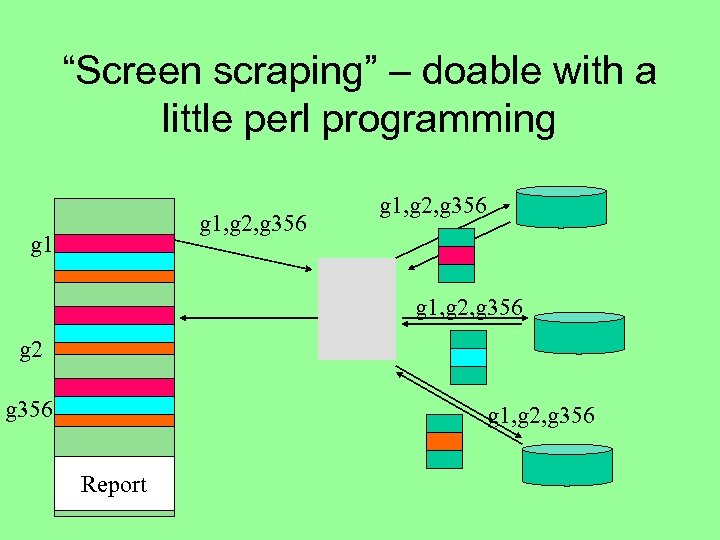 “Screen scraping” – doable with a little perl programming g 1, g 2, g 356 g 2 g 356 g 1, g 2, g 356 Report
“Screen scraping” – doable with a little perl programming g 1, g 2, g 356 g 2 g 356 g 1, g 2, g 356 Report
 Gene Ontology. TM www. geneontology. org • GO is a systematic effort for data annotation • Three independent ontologies • Molecular Function • Biological Process • Cellular component • How to integrate that into analysis tools?
Gene Ontology. TM www. geneontology. org • GO is a systematic effort for data annotation • Three independent ontologies • Molecular Function • Biological Process • Cellular component • How to integrate that into analysis tools?

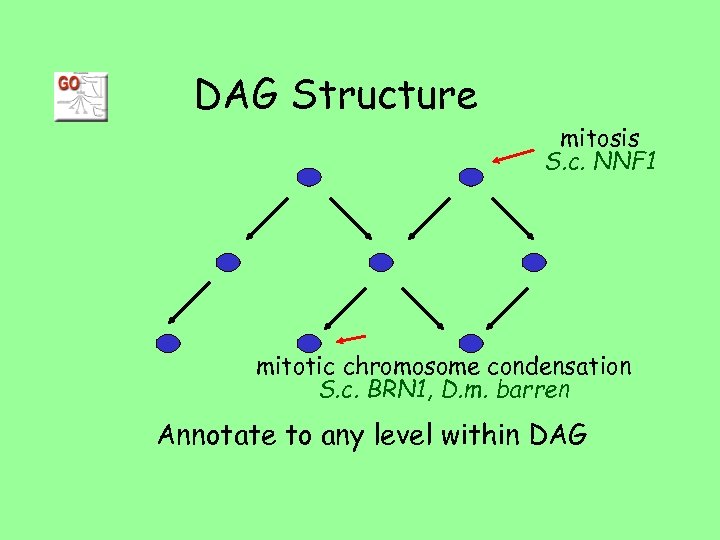 DAG Structure mitosis S. c. NNF 1 mitotic chromosome condensation S. c. BRN 1, D. m. barren Annotate to any level within DAG
DAG Structure mitosis S. c. NNF 1 mitotic chromosome condensation S. c. BRN 1, D. m. barren Annotate to any level within DAG
 GO Annotation: Data • Database object: gene or gene product • GO term ID • Reference • publication or computational method • Evidence supporting annotation
GO Annotation: Data • Database object: gene or gene product • GO term ID • Reference • publication or computational method • Evidence supporting annotation
 GO Evidence Codes IDA - Inferred from Direct Assay IMP - Inferred from Mutant Phenotype TAS - Traceable Author Statement NAS - Non-traceable Author Statement IGI - Inferred from Genetic Interaction IC - Inferred by Curator IPI - Inferred from Physical Interaction ISS - Inferred from Sequence or structural Similarity IEP - Inferred from Expression Pattern IEA - Inferred from Electronic Annotation ND - Not Determined
GO Evidence Codes IDA - Inferred from Direct Assay IMP - Inferred from Mutant Phenotype TAS - Traceable Author Statement NAS - Non-traceable Author Statement IGI - Inferred from Genetic Interaction IC - Inferred by Curator IPI - Inferred from Physical Interaction ISS - Inferred from Sequence or structural Similarity IEP - Inferred from Expression Pattern IEA - Inferred from Electronic Annotation ND - Not Determined
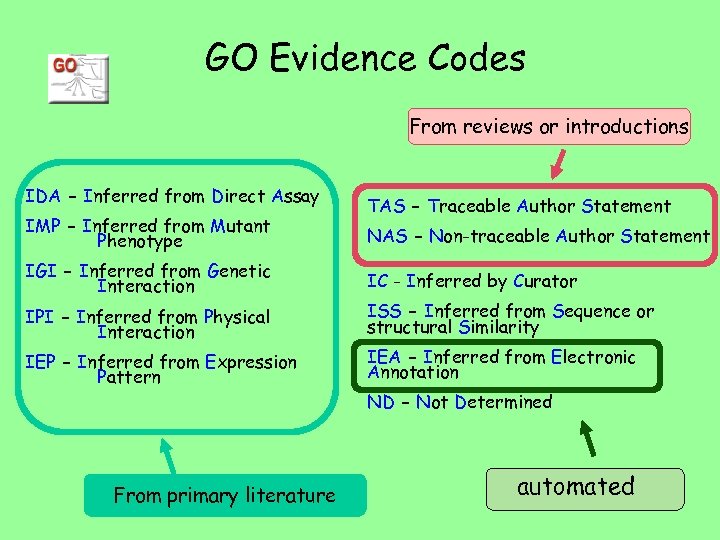 GO Evidence Codes From reviews or introductions IDA - Inferred from Direct Assay IMP - Inferred from Mutant Phenotype TAS - Traceable Author Statement NAS - Non-traceable Author Statement IGI - Inferred from Genetic Interaction IC - Inferred by Curator IPI - Inferred from Physical Interaction ISS - Inferred from Sequence or structural Similarity IEP - Inferred from Expression Pattern IEA - Inferred from Electronic Annotation ND - Not Determined From primary literature automated
GO Evidence Codes From reviews or introductions IDA - Inferred from Direct Assay IMP - Inferred from Mutant Phenotype TAS - Traceable Author Statement NAS - Non-traceable Author Statement IGI - Inferred from Genetic Interaction IC - Inferred by Curator IPI - Inferred from Physical Interaction ISS - Inferred from Sequence or structural Similarity IEP - Inferred from Expression Pattern IEA - Inferred from Electronic Annotation ND - Not Determined From primary literature automated
 Example (Go. Miner)
Example (Go. Miner)
 EP: GO tool for Gene. Ontology • Browse • Search by keywords; EC, term. etc. . • Get associated genes • Submit associated genes to URLMAP • Annotate gene clusters using GO terms
EP: GO tool for Gene. Ontology • Browse • Search by keywords; EC, term. etc. . • Get associated genes • Submit associated genes to URLMAP • Annotate gene clusters using GO terms
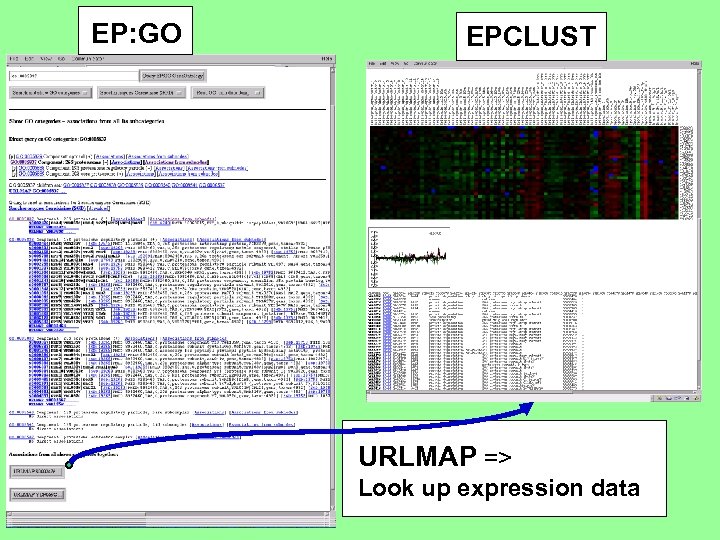 EP: GO EPCLUST URLMAP => Look up expression data
EP: GO EPCLUST URLMAP => Look up expression data
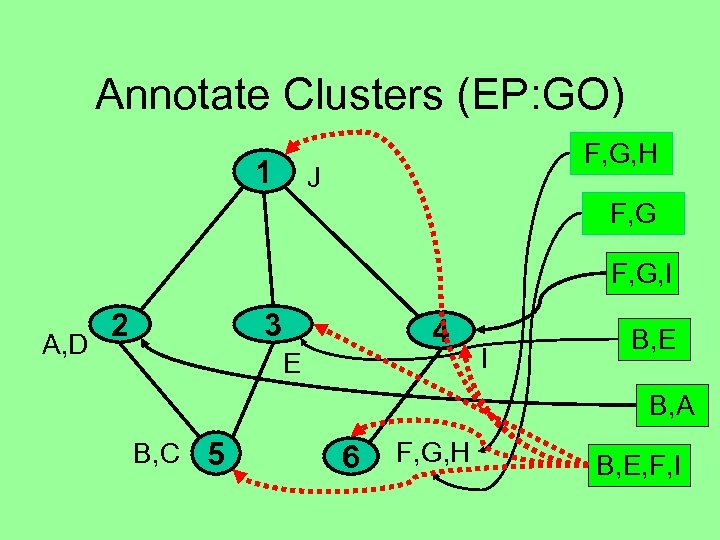 Annotate Clusters (EP: GO) 1 F, G, H J F, G, I A, D 2 3 4 E I B, E B, A B, C 5 6 F, G, H B, E, F, I
Annotate Clusters (EP: GO) 1 F, G, H J F, G, I A, D 2 3 4 E I B, E B, A B, C 5 6 F, G, H B, E, F, I
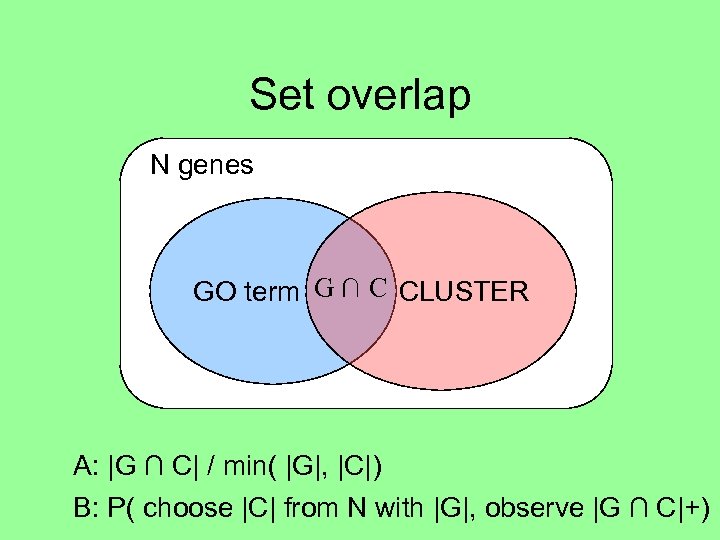 Set overlap N genes GO term G ∩ C CLUSTER A: |G ∩ C| / min( |G|, |C|) B: P( choose |C| from N with |G|, observe |G ∩ C|+)
Set overlap N genes GO term G ∩ C CLUSTER A: |G ∩ C| / min( |G|, |C|) B: P( choose |C| from N with |G|, observe |G ∩ C|+)
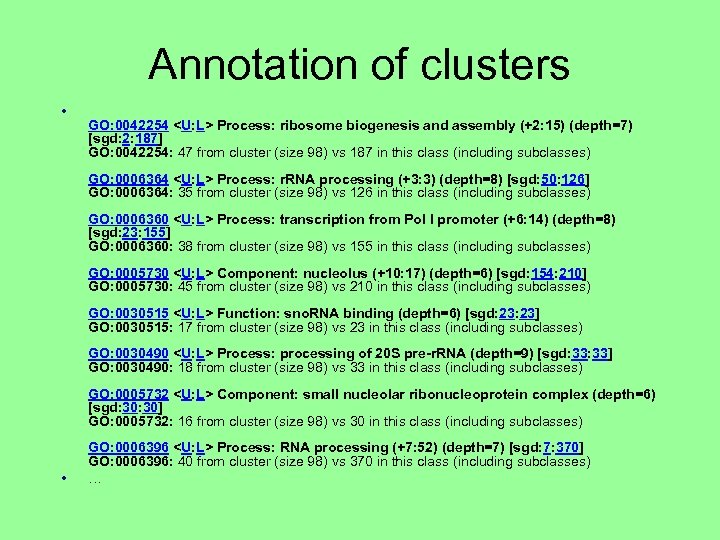 Annotation of clusters • GO: 0042254
Annotation of clusters • GO: 0042254
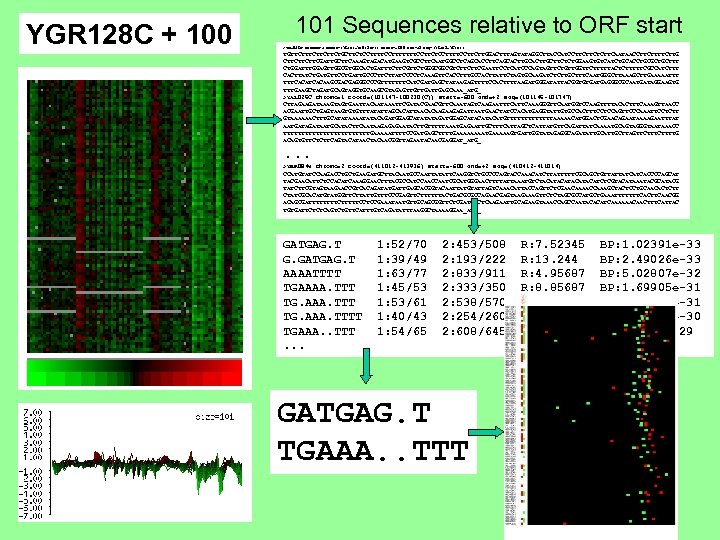 YGR 128 C + 100 101 Sequences relative to ORF start >YAL 036 C chromo=1 coord=(76154 -75048(C)) start=-600 end=+2 seq=(76152 -76754) TGTTCTTCTTCTGCTTCTCCTTTTTTTCCTTCTCCTTTTCCTTCTTGGACTTTAGTATAGGCTTACCATCCTTCTTCAATAACCTTCTTG CTTCTTCTTCGATTGCTTCAAAGTAGACATGAAGTCGCCTTCAATGGCCTCAGCACCTTCAGCACTTGCTTCTCTGGAAGTGTCATCTGCACCTGCGCTGCTTT CTGGATTTGGAGTTGGCGTGGCACTGATTTCTTCGTTCTGGGCGGCGTCTTCTTCGAATTCCTCATCCCAGTAGTTCTGTTGGTTCTTTTTACTCTTTTTCGCCATCTTT CACTTATCTGATGTTCCTGATTGCCCTTCTTATCCCCTCAAAGTTCACCTTTGCCACTTATTCTAGTGCAAGATCTCTTGCTTTCAATGGGCTTAAAGCTTGAAAAATTT TTTCACAAGCGACGAGGGCCCGTTTTTTTCATCGATGAGCTATAAGAGTTTTCCACTTTTAAGATGGGATATTACGGTGTGATGAGGGCGCAATGATAGGAAGTG TTTGAAGCTAGATGCAGTAGGTGCAAGCGTAGAGTTGTTGAGCAAA_ATG_ >YAL 025 C chromo=1 coord=(101147 -100230(C)) start=-600 end=+2 seq=(101145 -101747) CTTAGAAGATAAAGTAGTGAATTACAATAAATTCGATACGAACGTTCAAATAGTCAAGAATTTCAAAGGGTTCAATGGTCCAAGTTTTACACTTTCAAAGTTAACC ACGAATTGCTGAGTAAGTGTGTTTATATTAGCACATTAACACAAGAAGAGATTAATGAACTATCCACATGAGGTATTGTGCCACTTTCCTCCAGTTCCCAAATTCCTCTT GTAAAAAACTTTGCATATAAAATATACAGATGGAGCATATATAGATGGAGCATACATGTTTTTTTAAAAACATGGACTCGAACAGAATAAAAGAATTTAT AATGATAATGCATACTTCAATAAGAGAGAATACTTGTTTTTAAATGAGAATTGCTTTCATTAGCTCATTATGTTCAGATTATCAAAATGCAGTAGGGTAATAAACC TTTTTTTTTTTTGAAAAATTTTCCGATGAGCTTTTGAAAATGAAAAAGTGATTGGTATAGAGGCAGATATTGCTTAGTTCTTTTG ACAGTGTTCTCTTCAGTACATAACTACAACGGTTAGAATACAACGAGGAT_ATG_ . . . >YBR 084 W chromo=2 coord=(411012 -413936) start=-600 end=+2 seq=(410412 -411014) CCATGTATCCAAGACCTGCTGAAGATGCTTACAATGCCAATTATATTCAAGGTCTGCCCCAGTACCAAACATCTTATTTTTCGCAGCTGTTATTATCATCACCCCAGCAT TACGAACATTCTCCACATCAAAGGAACTTTACGCCATCCAATCGCATGGGAACTTTTATTAAATGTCTACATACATCTCGTACATAAATACGCATACG TATCTTCGTAGTAAGAACCGTCACAGATATGATTGAGCACGGTACAATTATGTATTAGTCAAACATTACCAGTTCTCGAACAAAACCAAAGCTACTCCTGCAACACTCTT CTATCGCACATGTATGGTTCTTATTGTTTCCCGAGTTCTTTTTTACTGACGCGCCAGAACGAGTAAGAAAGTTCTCTAGCGCCATGCTGAAATTTTTTTCACTTCAACGG ACAGCGATTTTCTTTTTCCTCCGAAATAATGTTGCAGCGGTTCTCGATGCCTCAAGAATTGCAGAAGTAAACCAGCCAATACACATCAAAAAACAACTTTCATTAC TGTGATTCTCTCAGTCTGTTCATTTGTCAGATATTTAAGGCTAAAAGGAA_ATG_ GATGAG. T G. GATGAG. T AAAATTTT TGAAAA. TTT TG. AAA. TTTT TGAAA. . TTT. . . 1: 52/70 1: 39/49 1: 63/77 1: 45/53 1: 53/61 1: 40/43 1: 54/65 2: 453/508 2: 193/222 2: 833/911 2: 333/350 2: 538/570 2: 254/260 2: 608/645 GATGAG. T TGAAA. . TTT R: 7. 52345 R: 13. 244 R: 4. 95687 R: 8. 85687 R: 6. 45662 R: 10. 3214 R: 5. 82106 BP: 1. 02391 e-33 BP: 2. 49026 e-33 BP: 5. 02807 e-32 BP: 1. 69905 e-31 BP: 3. 24836 e-31 BP: 3. 84624 e-30 BP: 1. 0887 e-29
YGR 128 C + 100 101 Sequences relative to ORF start >YAL 036 C chromo=1 coord=(76154 -75048(C)) start=-600 end=+2 seq=(76152 -76754) TGTTCTTCTTCTGCTTCTCCTTTTTTTCCTTCTCCTTTTCCTTCTTGGACTTTAGTATAGGCTTACCATCCTTCTTCAATAACCTTCTTG CTTCTTCTTCGATTGCTTCAAAGTAGACATGAAGTCGCCTTCAATGGCCTCAGCACCTTCAGCACTTGCTTCTCTGGAAGTGTCATCTGCACCTGCGCTGCTTT CTGGATTTGGAGTTGGCGTGGCACTGATTTCTTCGTTCTGGGCGGCGTCTTCTTCGAATTCCTCATCCCAGTAGTTCTGTTGGTTCTTTTTACTCTTTTTCGCCATCTTT CACTTATCTGATGTTCCTGATTGCCCTTCTTATCCCCTCAAAGTTCACCTTTGCCACTTATTCTAGTGCAAGATCTCTTGCTTTCAATGGGCTTAAAGCTTGAAAAATTT TTTCACAAGCGACGAGGGCCCGTTTTTTTCATCGATGAGCTATAAGAGTTTTCCACTTTTAAGATGGGATATTACGGTGTGATGAGGGCGCAATGATAGGAAGTG TTTGAAGCTAGATGCAGTAGGTGCAAGCGTAGAGTTGTTGAGCAAA_ATG_ >YAL 025 C chromo=1 coord=(101147 -100230(C)) start=-600 end=+2 seq=(101145 -101747) CTTAGAAGATAAAGTAGTGAATTACAATAAATTCGATACGAACGTTCAAATAGTCAAGAATTTCAAAGGGTTCAATGGTCCAAGTTTTACACTTTCAAAGTTAACC ACGAATTGCTGAGTAAGTGTGTTTATATTAGCACATTAACACAAGAAGAGATTAATGAACTATCCACATGAGGTATTGTGCCACTTTCCTCCAGTTCCCAAATTCCTCTT GTAAAAAACTTTGCATATAAAATATACAGATGGAGCATATATAGATGGAGCATACATGTTTTTTTAAAAACATGGACTCGAACAGAATAAAAGAATTTAT AATGATAATGCATACTTCAATAAGAGAGAATACTTGTTTTTAAATGAGAATTGCTTTCATTAGCTCATTATGTTCAGATTATCAAAATGCAGTAGGGTAATAAACC TTTTTTTTTTTTGAAAAATTTTCCGATGAGCTTTTGAAAATGAAAAAGTGATTGGTATAGAGGCAGATATTGCTTAGTTCTTTTG ACAGTGTTCTCTTCAGTACATAACTACAACGGTTAGAATACAACGAGGAT_ATG_ . . . >YBR 084 W chromo=2 coord=(411012 -413936) start=-600 end=+2 seq=(410412 -411014) CCATGTATCCAAGACCTGCTGAAGATGCTTACAATGCCAATTATATTCAAGGTCTGCCCCAGTACCAAACATCTTATTTTTCGCAGCTGTTATTATCATCACCCCAGCAT TACGAACATTCTCCACATCAAAGGAACTTTACGCCATCCAATCGCATGGGAACTTTTATTAAATGTCTACATACATCTCGTACATAAATACGCATACG TATCTTCGTAGTAAGAACCGTCACAGATATGATTGAGCACGGTACAATTATGTATTAGTCAAACATTACCAGTTCTCGAACAAAACCAAAGCTACTCCTGCAACACTCTT CTATCGCACATGTATGGTTCTTATTGTTTCCCGAGTTCTTTTTTACTGACGCGCCAGAACGAGTAAGAAAGTTCTCTAGCGCCATGCTGAAATTTTTTTCACTTCAACGG ACAGCGATTTTCTTTTTCCTCCGAAATAATGTTGCAGCGGTTCTCGATGCCTCAAGAATTGCAGAAGTAAACCAGCCAATACACATCAAAAAACAACTTTCATTAC TGTGATTCTCTCAGTCTGTTCATTTGTCAGATATTTAAGGCTAAAAGGAA_ATG_ GATGAG. T G. GATGAG. T AAAATTTT TGAAAA. TTT TG. AAA. TTTT TGAAA. . TTT. . . 1: 52/70 1: 39/49 1: 63/77 1: 45/53 1: 53/61 1: 40/43 1: 54/65 2: 453/508 2: 193/222 2: 833/911 2: 333/350 2: 538/570 2: 254/260 2: 608/645 GATGAG. T TGAAA. . TTT R: 7. 52345 R: 13. 244 R: 4. 95687 R: 8. 85687 R: 6. 45662 R: 10. 3214 R: 5. 82106 BP: 1. 02391 e-33 BP: 2. 49026 e-33 BP: 5. 02807 e-32 BP: 1. 69905 e-31 BP: 3. 24836 e-31 BP: 3. 84624 e-30 BP: 1. 0887 e-29
 EP: PPI Protein-protein interaction • There are high-throughput technologies for identifying hypothetical protein-protein interactions • Which ones of these are more likely to be true? • Can these predictions help predicting gene function?
EP: PPI Protein-protein interaction • There are high-throughput technologies for identifying hypothetical protein-protein interactions • Which ones of these are more likely to be true? • Can these predictions help predicting gene function?

 PPI pairs
PPI pairs
 We have expression data
We have expression data
 Cluster
Cluster
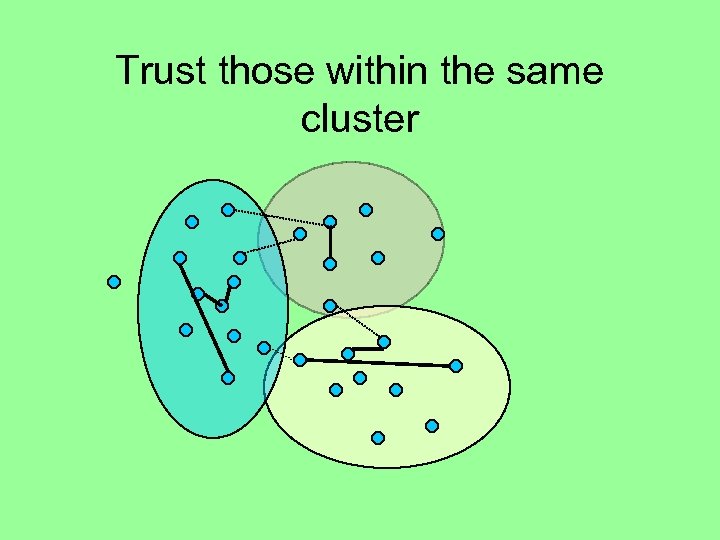 Trust those within the same cluster
Trust those within the same cluster
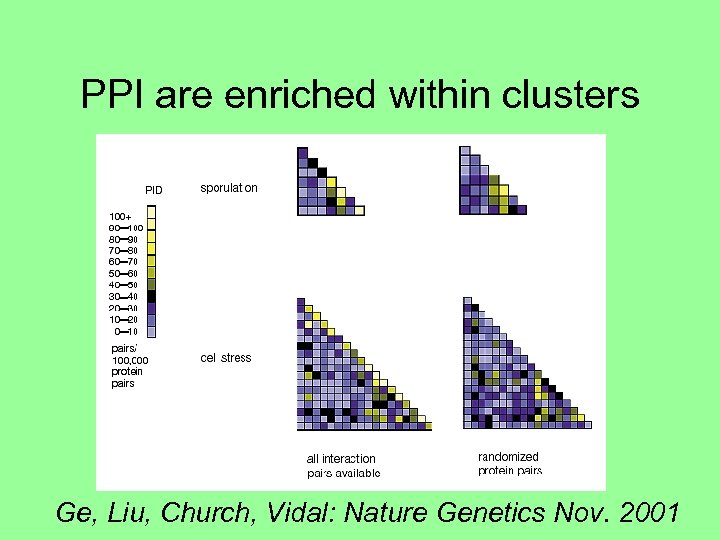 PPI are enriched within clusters Ge, Liu, Church, Vidal: Nature Genetics Nov. 2001
PPI are enriched within clusters Ge, Liu, Church, Vidal: Nature Genetics Nov. 2001
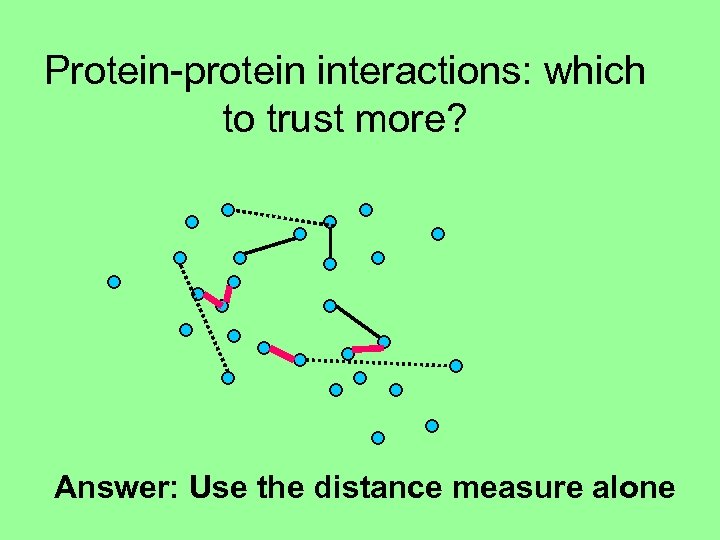 Protein-protein interactions: which to trust more? Answer: Use the distance measure alone
Protein-protein interactions: which to trust more? Answer: Use the distance measure alone
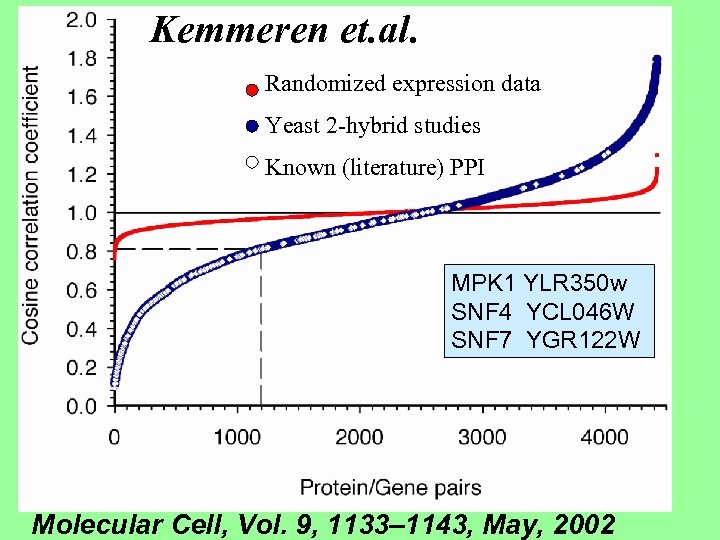 Kemmeren et. al. Randomized expression data Yeast 2 -hybrid studies Known (literature) PPI MPK 1 YLR 350 w SNF 4 YCL 046 W SNF 7 YGR 122 W Molecular Cell, Vol. 9, 1133– 1143, May, 2002
Kemmeren et. al. Randomized expression data Yeast 2 -hybrid studies Known (literature) PPI MPK 1 YLR 350 w SNF 4 YCL 046 W SNF 7 YGR 122 W Molecular Cell, Vol. 9, 1133– 1143, May, 2002
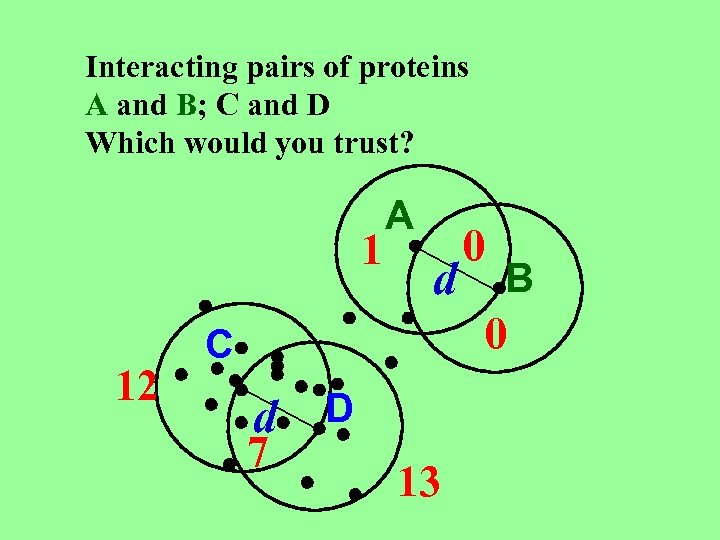 Interacting pairs of proteins A and B; C and D Which would you trust? 1 12 A d 0 B 0 C d 7 D 13
Interacting pairs of proteins A and B; C and D Which would you trust? 1 12 A d 0 B 0 C d 7 D 13
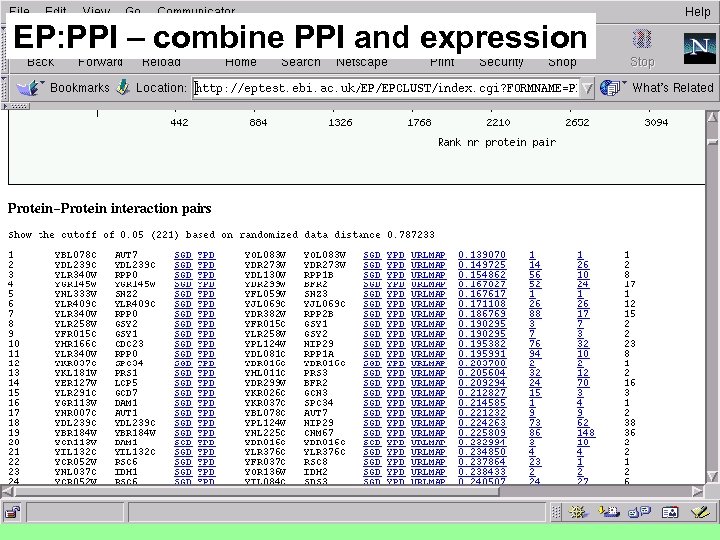 EP: PPI – combine PPI and expression
EP: PPI – combine PPI and expression
 Results • Confidence in 973 out of 5342 putative two-hybrid interactions from S. cerevisiae is increased. • Besides verification, integration of expression and interaction data is employed to provide functional annotation for over 300 previously uncharacterized genes. • The robustness of these approaches is demonstrated by experiments that test the in silico predictions made. • This study shows how integration improves the utility of different types of functional genomic data and how well this contributes to functional annotation.
Results • Confidence in 973 out of 5342 putative two-hybrid interactions from S. cerevisiae is increased. • Besides verification, integration of expression and interaction data is employed to provide functional annotation for over 300 previously uncharacterized genes. • The robustness of these approaches is demonstrated by experiments that test the in silico predictions made. • This study shows how integration improves the utility of different types of functional genomic data and how well this contributes to functional annotation.
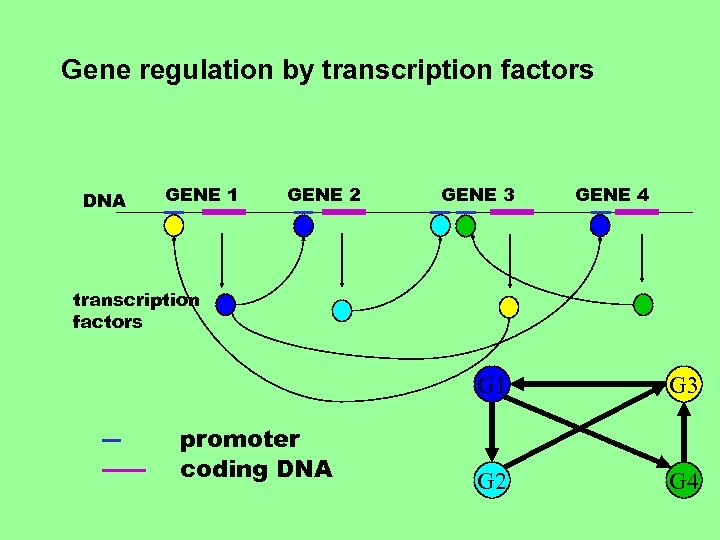 Gene regulation by transcription factors DNA GENE 1 GENE 2 GENE 3 GENE 4 transcription factors G 1 promoter coding DNA G 3 G 2 G 4
Gene regulation by transcription factors DNA GENE 1 GENE 2 GENE 3 GENE 4 transcription factors G 1 promoter coding DNA G 3 G 2 G 4
 Networks • Graphical models • Directed labelled graph • Nodes • Arcs/Edges • Labels genes relationships types of relationships
Networks • Graphical models • Directed labelled graph • Nodes • Arcs/Edges • Labels genes relationships types of relationships
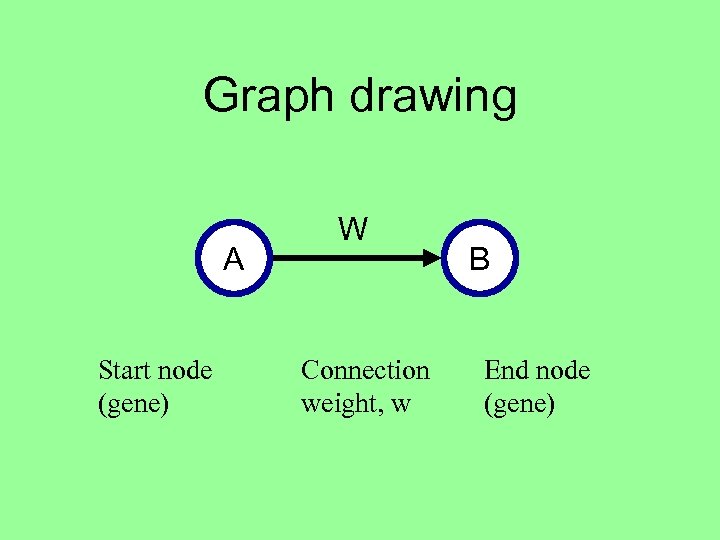 Graph drawing A Start node (gene) W Connection weight, w B End node (gene)
Graph drawing A Start node (gene) W Connection weight, w B End node (gene)
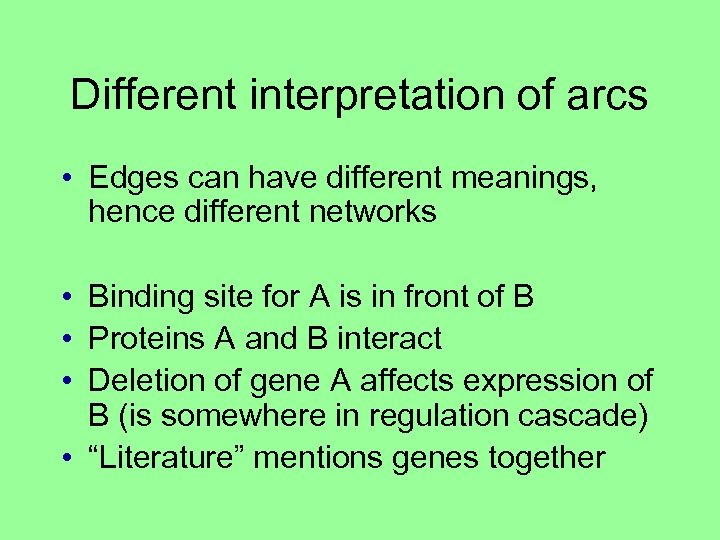 Different interpretation of arcs • Edges can have different meanings, hence different networks • Binding site for A is in front of B • Proteins A and B interact • Deletion of gene A affects expression of B (is somewhere in regulation cascade) • “Literature” mentions genes together
Different interpretation of arcs • Edges can have different meanings, hence different networks • Binding site for A is in front of B • Proteins A and B interact • Deletion of gene A affects expression of B (is somewhere in regulation cascade) • “Literature” mentions genes together
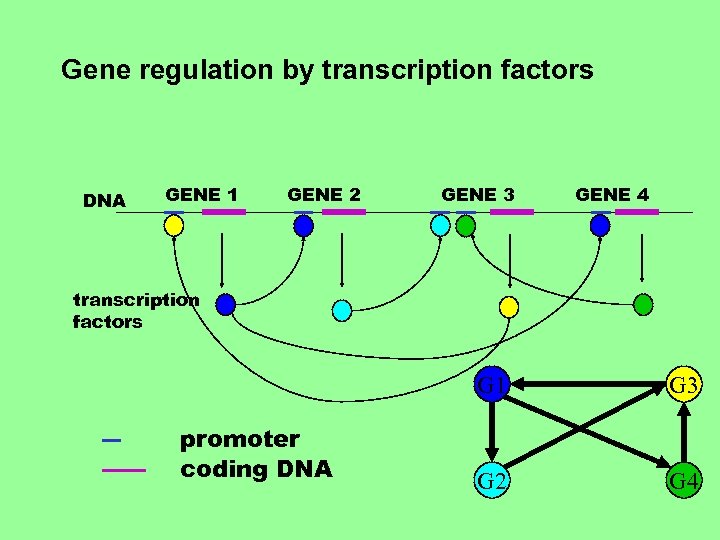 Gene regulation by transcription factors DNA GENE 1 GENE 2 GENE 3 GENE 4 transcription factors G 1 promoter coding DNA G 3 G 2 G 4
Gene regulation by transcription factors DNA GENE 1 GENE 2 GENE 3 GENE 4 transcription factors G 1 promoter coding DNA G 3 G 2 G 4
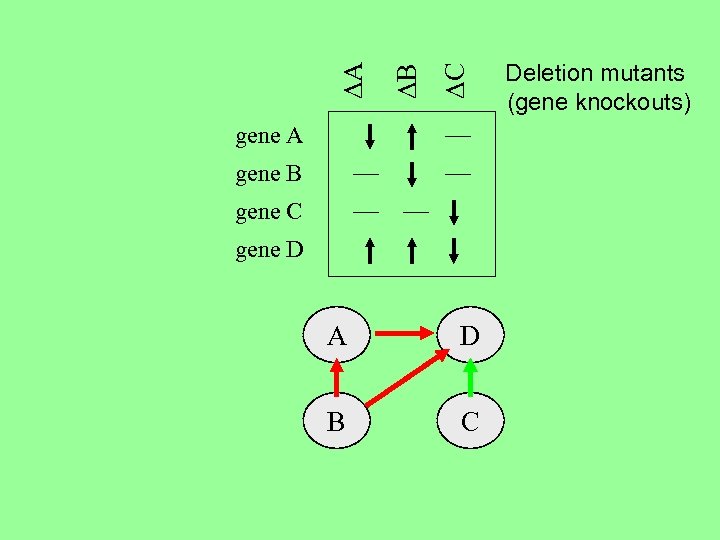 DC DB DA gene B gene C gene D A D B C Deletion mutants (gene knockouts)
DC DB DA gene B gene C gene D A D B C Deletion mutants (gene knockouts)
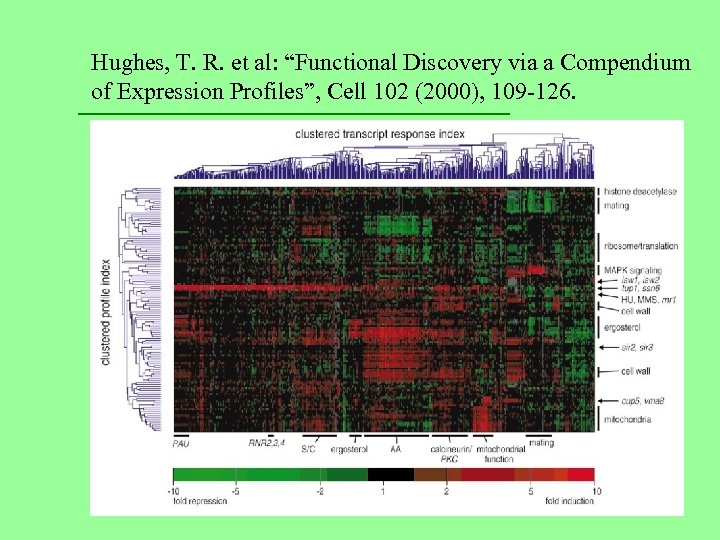 Hughes, T. R. et al: “Functional Discovery via a Compendium of Expression Profiles”, Cell 102 (2000), 109 -126.
Hughes, T. R. et al: “Functional Discovery via a Compendium of Expression Profiles”, Cell 102 (2000), 109 -126.
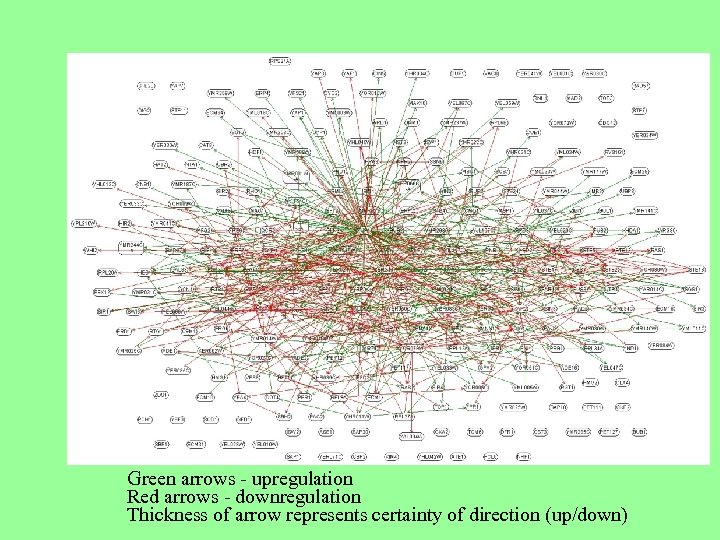 Green arrows - upregulation Red arrows - downregulation Thickness of arrow represents certainty of direction (up/down)
Green arrows - upregulation Red arrows - downregulation Thickness of arrow represents certainty of direction (up/down)
 A complete graph
A complete graph
 Features/distributions that do not depend on discretisation thresholds • Visual inspection, biological interpretation • General statistics and features of the graphs • Indegree/Outdegree • Complexity of the networks • What is the modularity? • How many components? • Deletion of hot-spots, does it break the net?
Features/distributions that do not depend on discretisation thresholds • Visual inspection, biological interpretation • General statistics and features of the graphs • Indegree/Outdegree • Complexity of the networks • What is the modularity? • How many components? • Deletion of hot-spots, does it break the net?
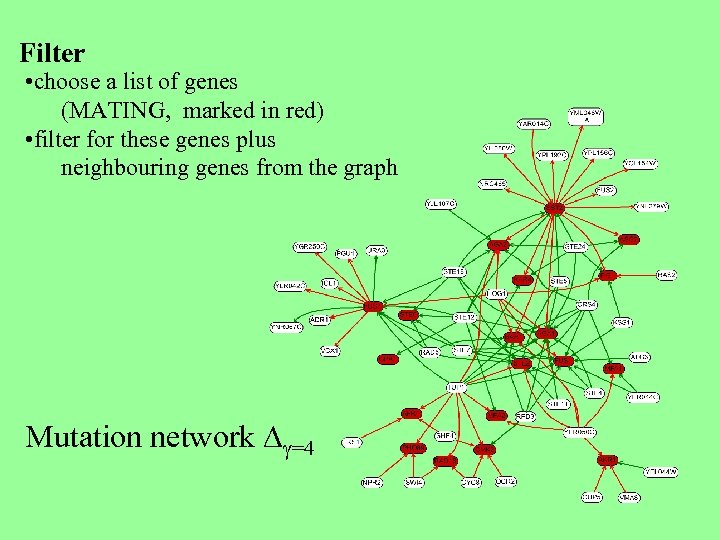 Filter • choose a list of genes (MATING, marked in red) • filter for these genes plus neighbouring genes from the graph Mutation network D =4
Filter • choose a list of genes (MATING, marked in red) • filter for these genes plus neighbouring genes from the graph Mutation network D =4
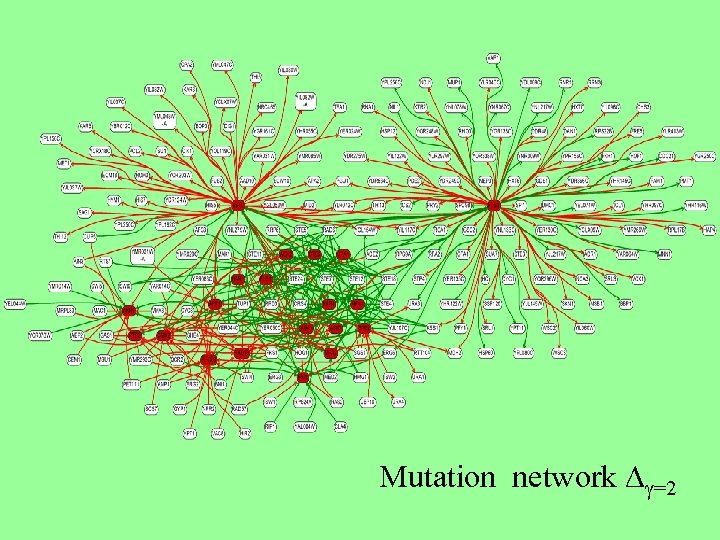 Mutation network D =2
Mutation network D =2
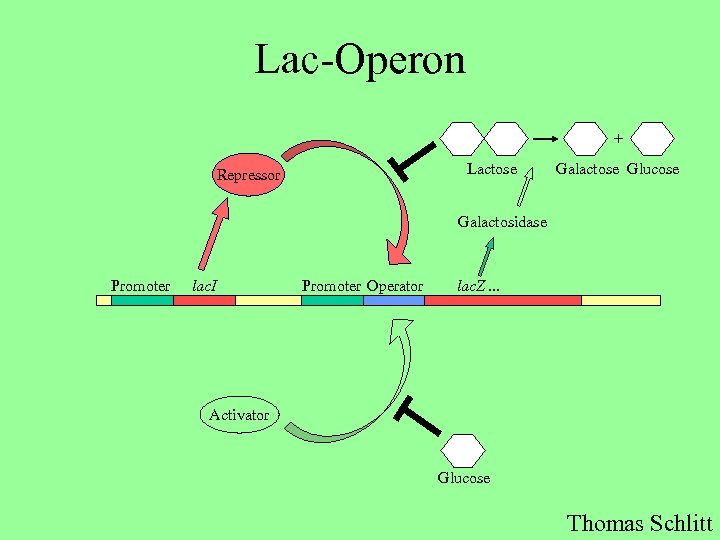 Lac-Operon + Lactose Repressor Galactose Glucose Galactosidase Promoter lac. I Promoter Operator lac. Z. . . Activator Glucose Thomas Schlitt
Lac-Operon + Lactose Repressor Galactose Glucose Galactosidase Promoter lac. I Promoter Operator lac. Z. . . Activator Glucose Thomas Schlitt
 Gene regulatory networks • What formalisms to use to describe them? • When does model correspond to biological reality? • How to simulate models on computer • Is it possible to verify models by experiments? • How to restore networks from raw data without knowing the structure or parameters?
Gene regulatory networks • What formalisms to use to describe them? • When does model correspond to biological reality? • How to simulate models on computer • Is it possible to verify models by experiments? • How to restore networks from raw data without knowing the structure or parameters?
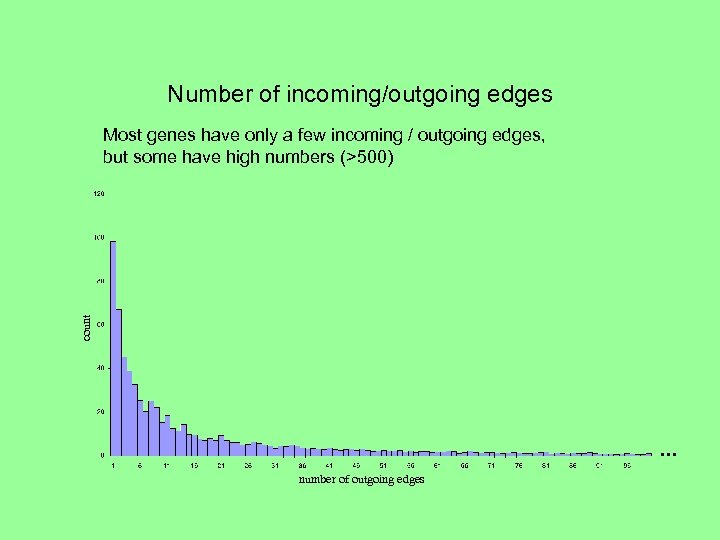 Number of incoming/outgoing edges count Most genes have only a few incoming / outgoing edges, but some have high numbers (>500) . . . number of outgoing edges
Number of incoming/outgoing edges count Most genes have only a few incoming / outgoing edges, but some have high numbers (>500) . . . number of outgoing edges
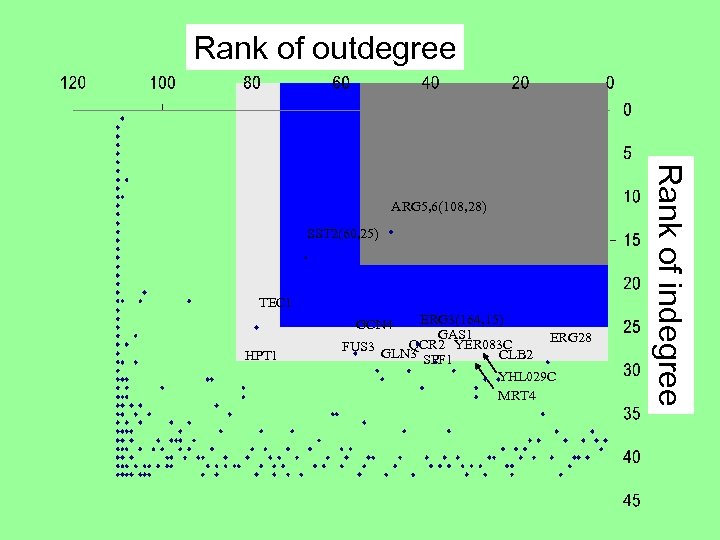 Rank of outdegree SST 2(60, 25) TEC 1 ERG 3(164, 15) GAS 1 QCR 2 YER 083 C FUS 3 GLN 3 CLB 2 SPF 1 GCN 4 HPT 1 ERG 28 YHL 029 C MRT 4 Rank of indegree ARG 5, 6(108, 28)
Rank of outdegree SST 2(60, 25) TEC 1 ERG 3(164, 15) GAS 1 QCR 2 YER 083 C FUS 3 GLN 3 CLB 2 SPF 1 GCN 4 HPT 1 ERG 28 YHL 029 C MRT 4 Rank of indegree ARG 5, 6(108, 28)
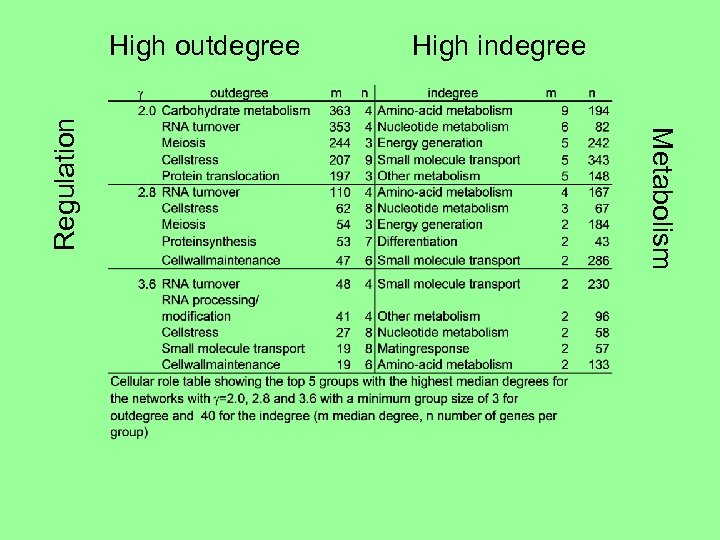 High indegree Metabolism Regulation High outdegree
High indegree Metabolism Regulation High outdegree
 Network modularity • Is there one “big” dominant connected component and possibly a number of small components, or several components of comparable sizes? • Can the network be broken down in several components of comparable size by removing nodes of high degree (i. e. , nodes with many incoming or outgoing edges)?
Network modularity • Is there one “big” dominant connected component and possibly a number of small components, or several components of comparable sizes? • Can the network be broken down in several components of comparable size by removing nodes of high degree (i. e. , nodes with many incoming or outgoing edges)?
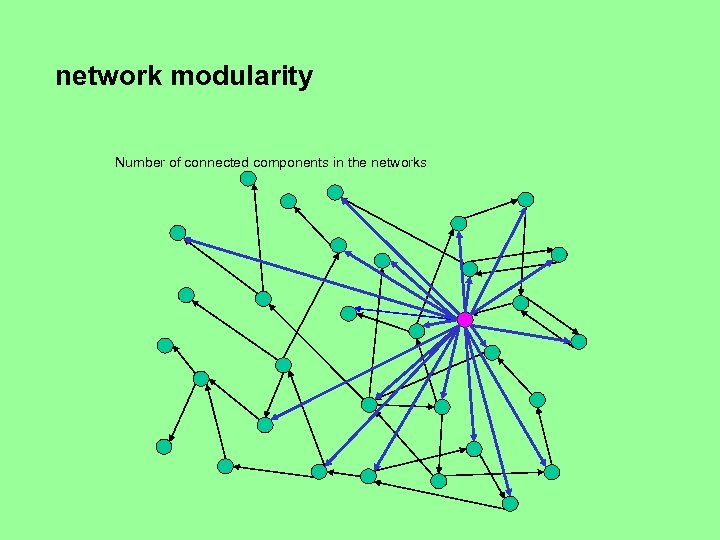 network modularity Number of connected components in the networks
network modularity Number of connected components in the networks
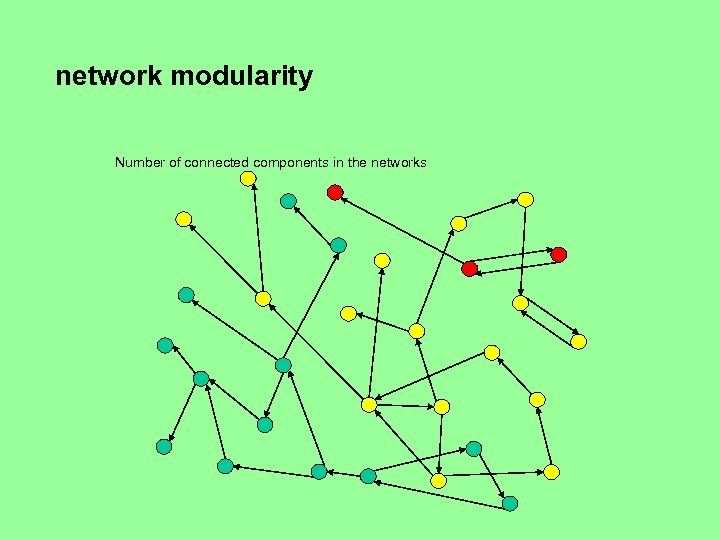 network modularity Number of connected components in the networks
network modularity Number of connected components in the networks
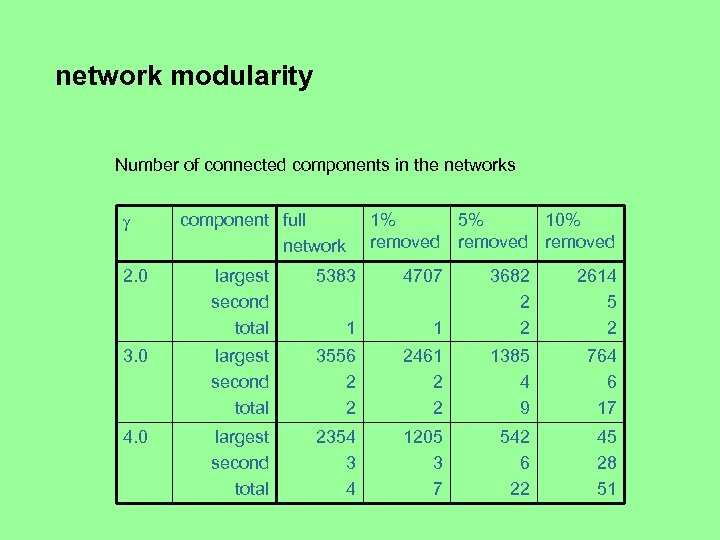 network modularity Number of connected components in the networks 2. 0 component full network 1% 5% 10% removed largest second total 5383 4707 1 3682 2 2 2614 5 2 1 3. 0 largest second total 3556 2 2 2461 2 2 1385 4 9 764 6 17 4. 0 largest second total 2354 3 4 1205 3 7 542 6 22 45 28 51
network modularity Number of connected components in the networks 2. 0 component full network 1% 5% 10% removed largest second total 5383 4707 1 3682 2 2 2614 5 2 1 3. 0 largest second total 3556 2 2 2461 2 2 1385 4 9 764 6 17 4. 0 largest second total 2354 3 4 1205 3 7 542 6 22 45 28 51
 Modularity other opinions • Wagner, Genome Research 2002 – there exist many independent modules • Featherstone and Broadie, Bioessays 2002 - there is only one giant module • All depends on the definition of the ‘module’
Modularity other opinions • Wagner, Genome Research 2002 – there exist many independent modules • Featherstone and Broadie, Bioessays 2002 - there is only one giant module • All depends on the definition of the ‘module’
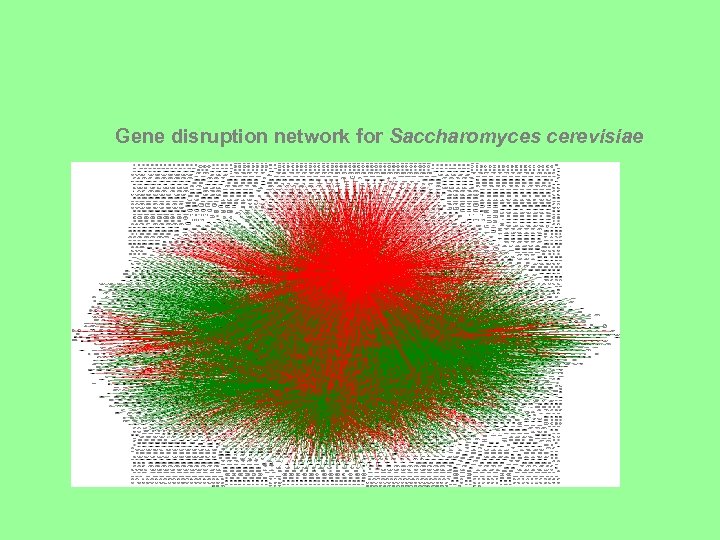 Gene disruption network for Saccharomyces cerevisiae
Gene disruption network for Saccharomyces cerevisiae
 a closer look
a closer look
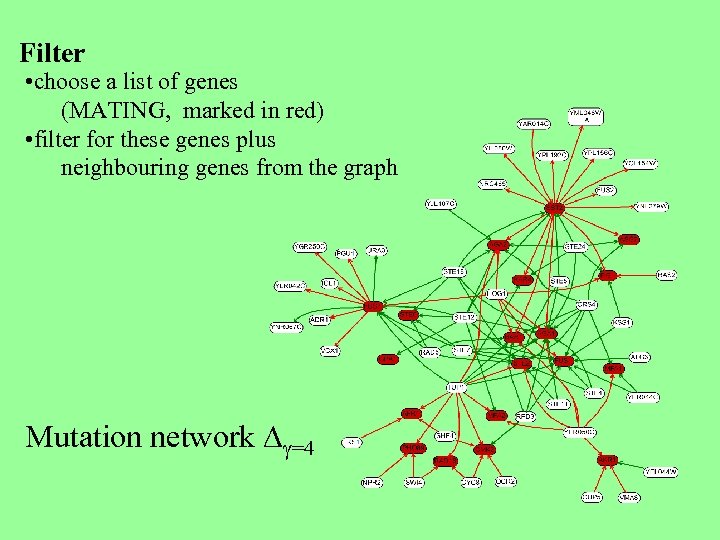 Filter • choose a list of genes (MATING, marked in red) • filter for these genes plus neighbouring genes from the graph Mutation network D =4
Filter • choose a list of genes (MATING, marked in red) • filter for these genes plus neighbouring genes from the graph Mutation network D =4
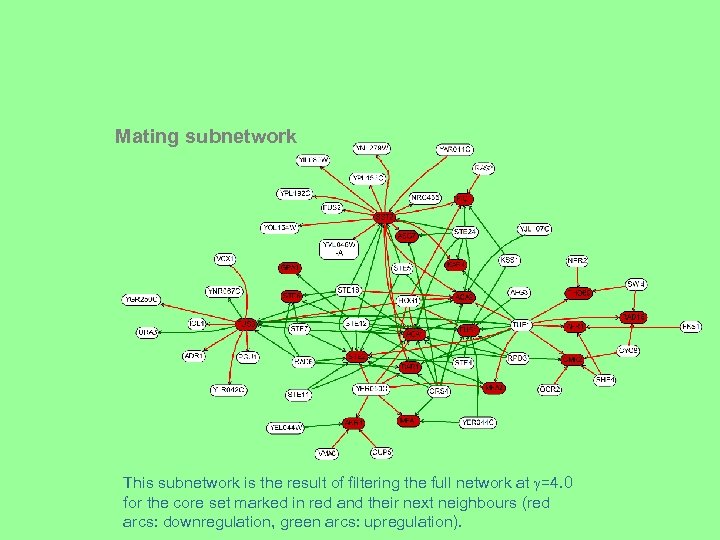 Mating subnetwork This subnetwork is the result of filtering the full network at =4. 0 for the core set marked in red and their next neighbours (red arcs: downregulation, green arcs: upregulation).
Mating subnetwork This subnetwork is the result of filtering the full network at =4. 0 for the core set marked in red and their next neighbours (red arcs: downregulation, green arcs: upregulation).
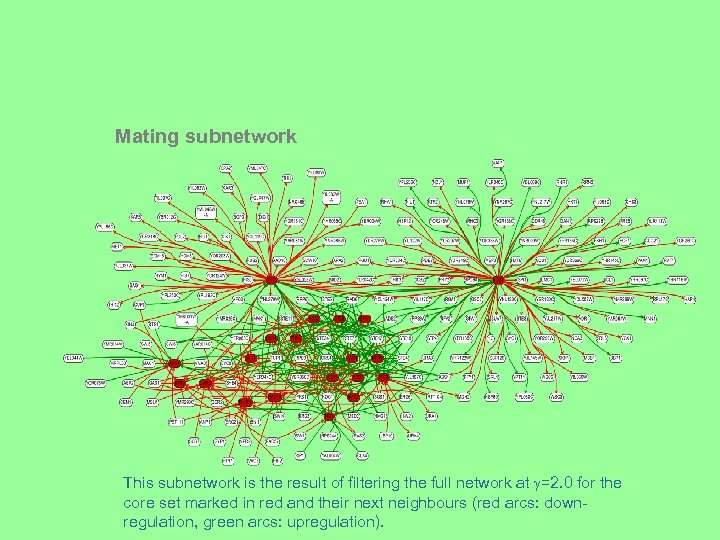 Mating subnetwork This subnetwork is the result of filtering the full network at =2. 0 for the core set marked in red and their next neighbours (red arcs: downregulation, green arcs: upregulation).
Mating subnetwork This subnetwork is the result of filtering the full network at =2. 0 for the core set marked in red and their next neighbours (red arcs: downregulation, green arcs: upregulation).
 Conclusion • more information than randomised networks • no optimal • powerlaw distribution of arcs • no obvious modules • local networks make sense
Conclusion • more information than randomised networks • no optimal • powerlaw distribution of arcs • no obvious modules • local networks make sense
 Lac-Operon + Lactose Repressor Galactose Glucose Galactosidase Promoter lac. I Promoter Operator lac. Z. . . Activator Glucose Thomas Schlitt
Lac-Operon + Lactose Repressor Galactose Glucose Galactosidase Promoter lac. I Promoter Operator lac. Z. . . Activator Glucose Thomas Schlitt
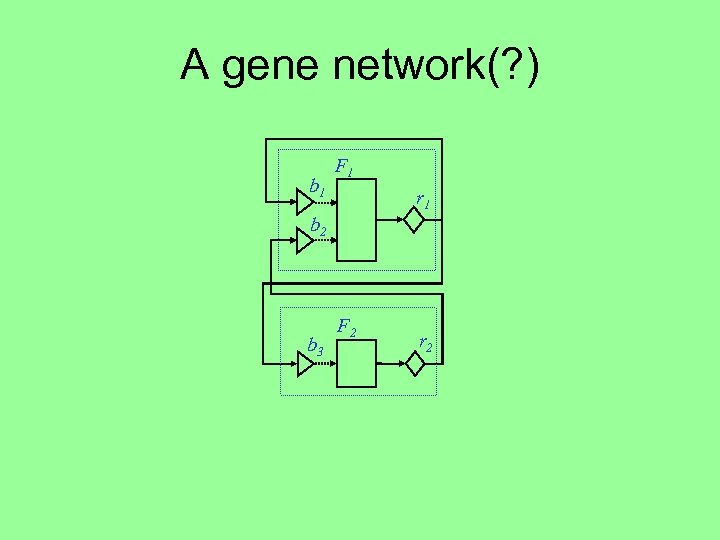 A gene network(? ) b 1 F 1 r 1 b 2 b 3 F 2 r 2
A gene network(? ) b 1 F 1 r 1 b 2 b 3 F 2 r 2
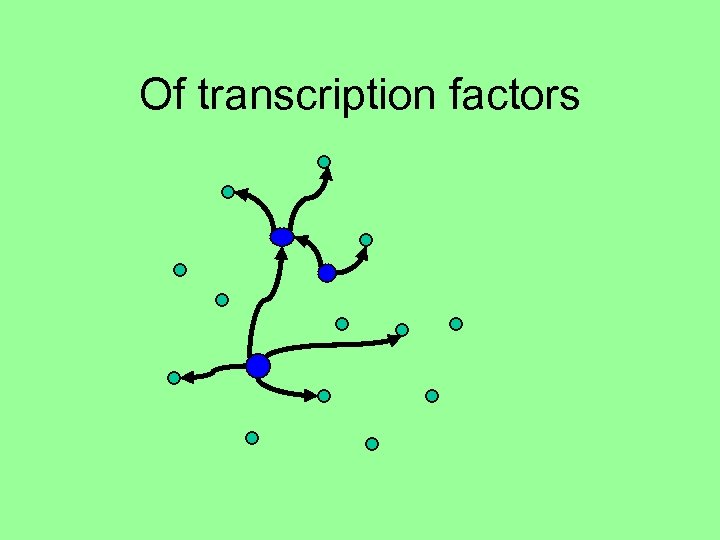 Of transcription factors
Of transcription factors
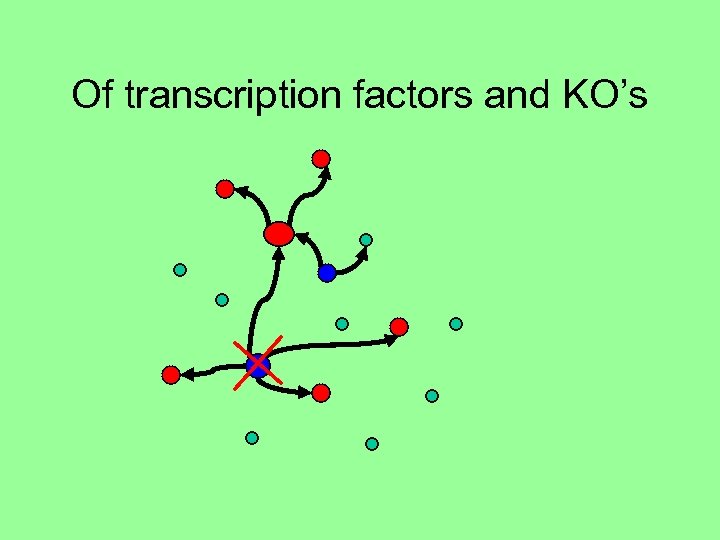 Of transcription factors and KO’s
Of transcription factors and KO’s
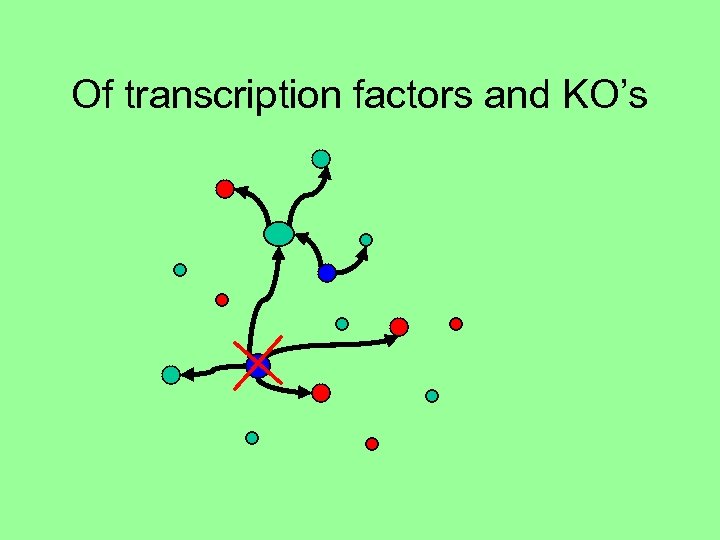 Of transcription factors and KO’s
Of transcription factors and KO’s
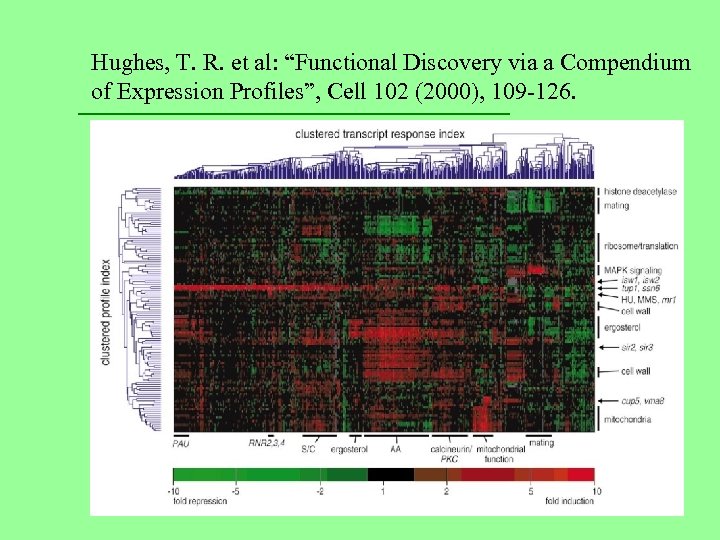 Hughes, T. R. et al: “Functional Discovery via a Compendium of Expression Profiles”, Cell 102 (2000), 109 -126.
Hughes, T. R. et al: “Functional Discovery via a Compendium of Expression Profiles”, Cell 102 (2000), 109 -126.
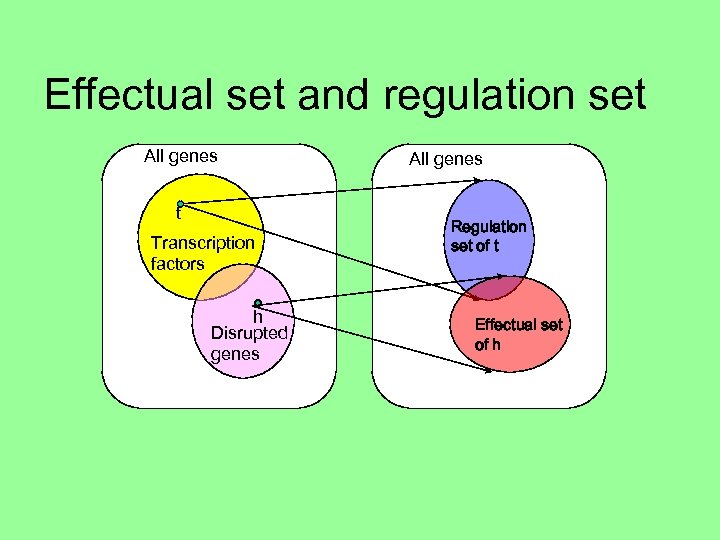 Effectual set and regulation set All genes t Transcription factors h Disrupted genes All genes Regulation set of t Effectual set of h
Effectual set and regulation set All genes t Transcription factors h Disrupted genes All genes Regulation set of t Effectual set of h
 Effectual set and regulation set All genes Transcription factors g Disrupted genes Regulation set of t Effectual set of h
Effectual set and regulation set All genes Transcription factors g Disrupted genes Regulation set of t Effectual set of h
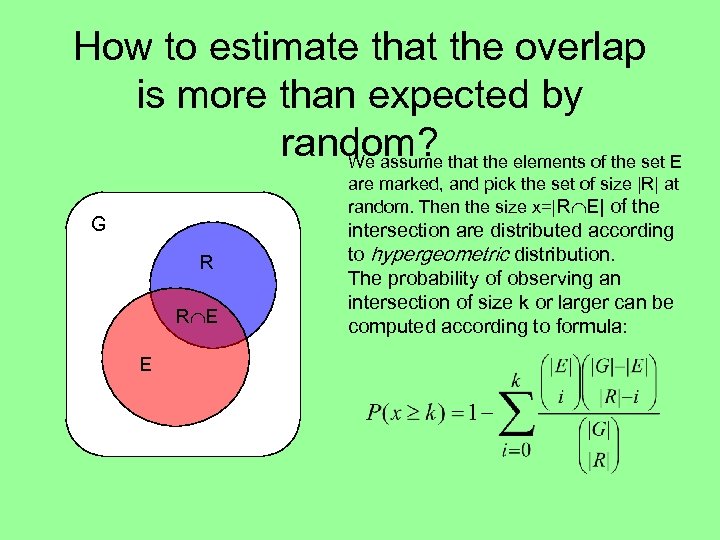 How to estimate that the overlap is more than expected by random? that the elements of the set E We assume are marked, and pick the set of size |R| at random. Then the size x=|R E| of the G R R E E intersection are distributed according to hypergeometric distribution. The probability of observing an intersection of size k or larger can be computed according to formula:
How to estimate that the overlap is more than expected by random? that the elements of the set E We assume are marked, and pick the set of size |R| at random. Then the size x=|R E| of the G R R E E intersection are distributed according to hypergeometric distribution. The probability of observing an intersection of size k or larger can be computed according to formula:
 Data • Disrupted genes – 263 disrupted genes excluding drug treatments and haploid states (Hughes et al) • Transcription factor binding sites – 356 binding sites, from these 37 experimentally proved (Pilpel et al, 2001)
Data • Disrupted genes – 263 disrupted genes excluding drug treatments and haploid states (Hughes et al) • Transcription factor binding sites – 356 binding sites, from these 37 experimentally proved (Pilpel et al, 2001)
 Disrupted TF • Only 5 transcription factors from our set (of known binding sites) were disrupted on the experiments – mbp 1, yaf 1, swi 5, gcn 4 • For three of them – mbp 1, yap 1, gcn 4 –the regulation and effectual sets were highly correlating • yaf 1 is activated with oleate, while in oleate free environment Yaf 1 (alias OAF 1) disruption does not have significant effect • swi 5 affects only haploid state, while we use only diploid
Disrupted TF • Only 5 transcription factors from our set (of known binding sites) were disrupted on the experiments – mbp 1, yaf 1, swi 5, gcn 4 • For three of them – mbp 1, yap 1, gcn 4 –the regulation and effectual sets were highly correlating • yaf 1 is activated with oleate, while in oleate free environment Yaf 1 (alias OAF 1) disruption does not have significant effect • swi 5 affects only haploid state, while we use only diploid
 Effectual sets correlating with other TF binding sites • From 37 of the experimentally proven binding sites, 20 correlate with one or more effectual sets • If the disrupted gene correlate with a regulation set of a different gene, the correlation should be explained
Effectual sets correlating with other TF binding sites • From 37 of the experimentally proven binding sites, 20 correlate with one or more effectual sets • If the disrupted gene correlate with a regulation set of a different gene, the correlation should be explained
 Possible explanations why disruption of gene A may correlate with regulation set of a different gene (TF) T: • T belongs to the disruption set of A (cascade)
Possible explanations why disruption of gene A may correlate with regulation set of a different gene (TF) T: • T belongs to the disruption set of A (cascade)
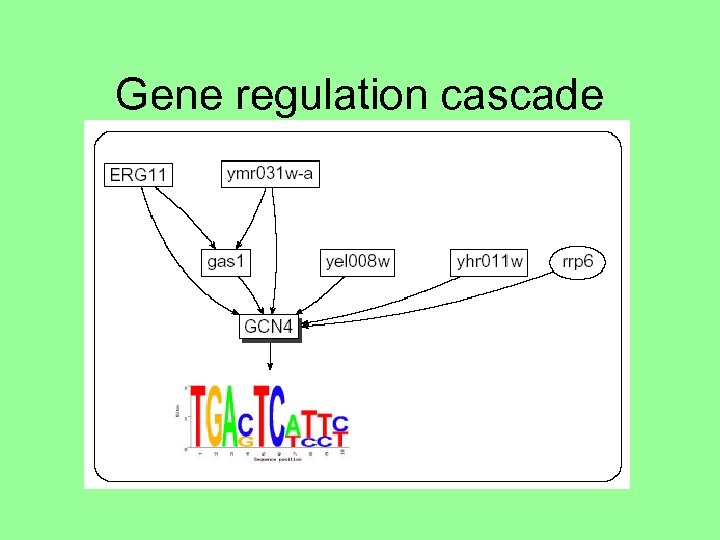 Gene regulation cascade
Gene regulation cascade
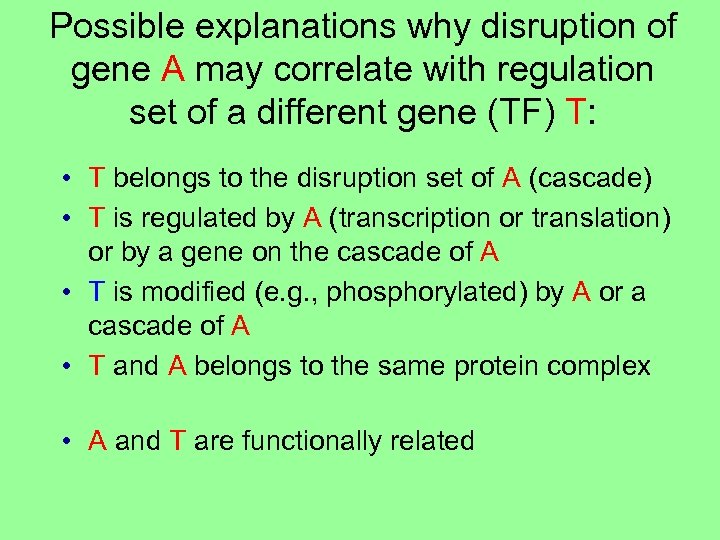 Possible explanations why disruption of gene A may correlate with regulation set of a different gene (TF) T: • T belongs to the disruption set of A (cascade) • T is regulated by A (transcription or translation) or by a gene on the cascade of A • T is modified (e. g. , phosphorylated) by A or a cascade of A • T and A belongs to the same protein complex • A and T are functionally related
Possible explanations why disruption of gene A may correlate with regulation set of a different gene (TF) T: • T belongs to the disruption set of A (cascade) • T is regulated by A (transcription or translation) or by a gene on the cascade of A • T is modified (e. g. , phosphorylated) by A or a cascade of A • T and A belongs to the same protein complex • A and T are functionally related
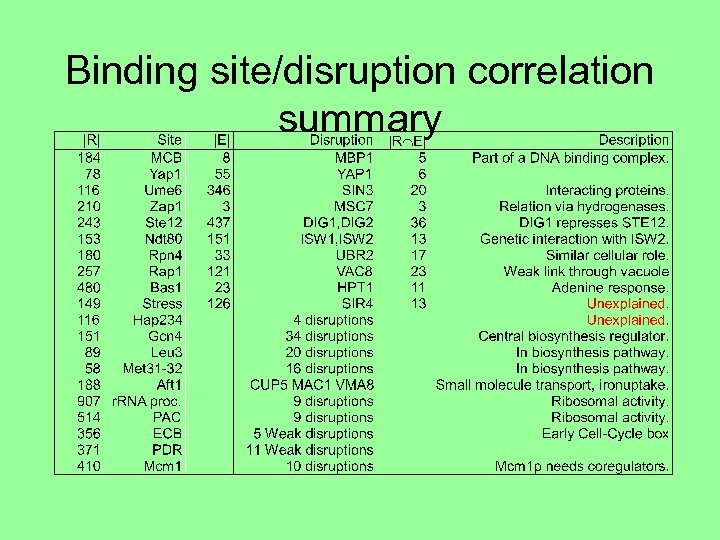 Binding site/disruption correlation summary
Binding site/disruption correlation summary
 Conclusion • Most of the binding site/disruption set correlations can be explained via • Regulation cascades • Protein complexes (K. Palin et al, to appear in ECCB 2002, special issue of Bioinformatics)
Conclusion • Most of the binding site/disruption set correlations can be explained via • Regulation cascades • Protein complexes (K. Palin et al, to appear in ECCB 2002, special issue of Bioinformatics)
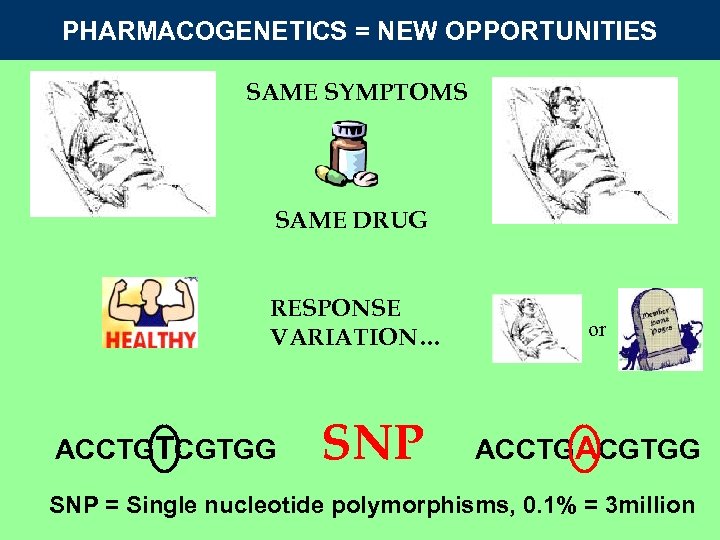 PHARMACOGENETICS = NEW OPPORTUNITIES SAME SYMPTOMS SAME DRUG RESPONSE VARIATION… ACCTGTCGTGG SNP or ACCTGACGTGG SNP = Single nucleotide polymorphisms, 0. 1% = 3 million
PHARMACOGENETICS = NEW OPPORTUNITIES SAME SYMPTOMS SAME DRUG RESPONSE VARIATION… ACCTGTCGTGG SNP or ACCTGACGTGG SNP = Single nucleotide polymorphisms, 0. 1% = 3 million
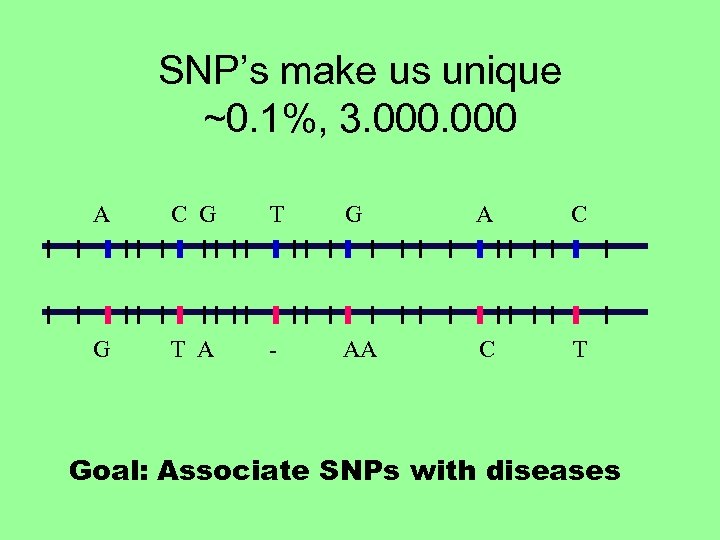 SNP’s make us unique ~0. 1%, 3. 000 A C G T G A C G T A - AA C T Goal: Associate SNPs with diseases
SNP’s make us unique ~0. 1%, 3. 000 A C G T G A C G T A - AA C T Goal: Associate SNPs with diseases
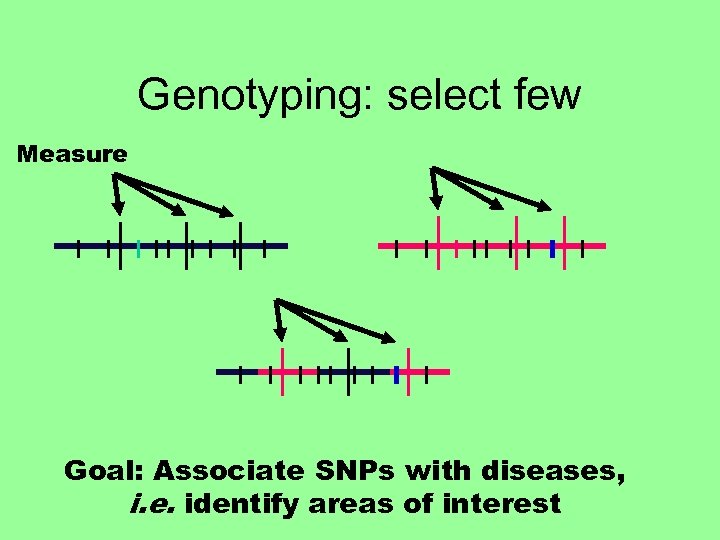 Genotyping: select few Measure Goal: Associate SNPs with diseases, i. e. identify areas of interest
Genotyping: select few Measure Goal: Associate SNPs with diseases, i. e. identify areas of interest
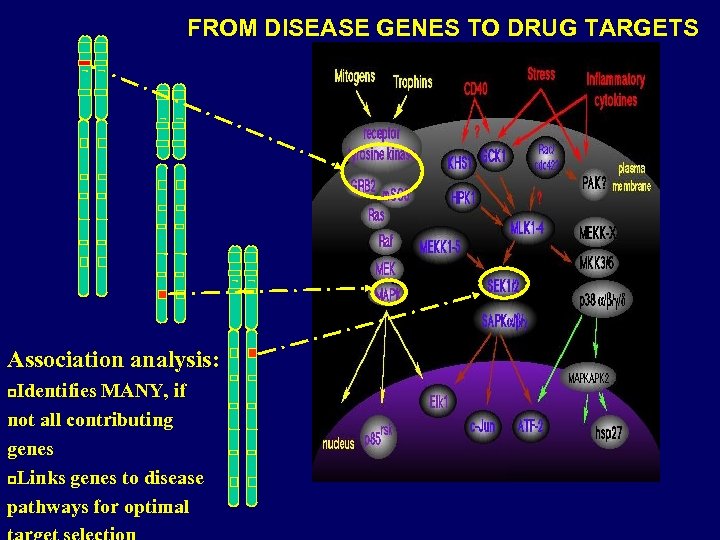 FROM DISEASE GENES TO DRUG TARGETS Association analysis: Identifies MANY, if not all contributing genes p. Links genes to disease pathways for optimal p
FROM DISEASE GENES TO DRUG TARGETS Association analysis: Identifies MANY, if not all contributing genes p. Links genes to disease pathways for optimal p
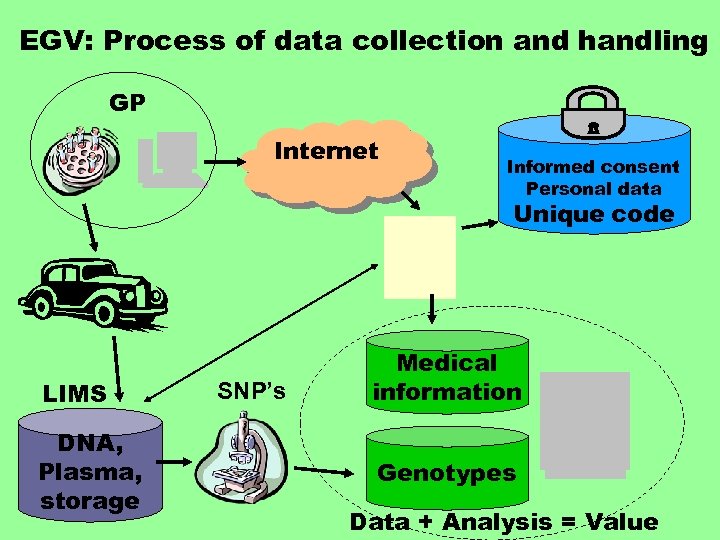 EGV: Process of data collection and handling GP Internet Informed consent Personal data Unique code LIMS DNA, Plasma, storage SNP’s Medical information Genotypes Data + Analysis = Value
EGV: Process of data collection and handling GP Internet Informed consent Personal data Unique code LIMS DNA, Plasma, storage SNP’s Medical information Genotypes Data + Analysis = Value
 Bioinformatics: Where does IT stand? • Data modelling, storage, access • Inference from data • Hypotheses generation and testing • Allow novel types of questions to be asked by providing analysis methods that are able to cope with all the information that is available today
Bioinformatics: Where does IT stand? • Data modelling, storage, access • Inference from data • Hypotheses generation and testing • Allow novel types of questions to be asked by providing analysis methods that are able to cope with all the information that is available today
 Compute Infrastructure
Compute Infrastructure
 Bioinformatics: Challenges • • • Knowledge representation, data semantics Data size and its speed of growth New/emerging data collection technologies Integration of different data types Discovery of useful knowledge Modeling living systems as a whole • Improved health care products • Medical informatics – bringing the knowledge to doctor’s bench
Bioinformatics: Challenges • • • Knowledge representation, data semantics Data size and its speed of growth New/emerging data collection technologies Integration of different data types Discovery of useful knowledge Modeling living systems as a whole • Improved health care products • Medical informatics – bringing the knowledge to doctor’s bench
 References for this talk http: //www. egeen. ee/u/vilo/Publications/ Jaak Vilo, Misha Kapushesky, Patrick Kemmeren, Ugis Sarkans, Alvis Brazma. Expression Profiler. In Parmigiani, G. , Garrett, E. S. , Irizarry, R. and Zeger, S. L. (eds), The Analysis of Gene Expression Data: Methods and Software, Springer Verlag, New York, NY. Patrick Kemmeren, Nynke L. van Berkum, Jaak Vilo, Theo Bijma, Rogier Donders, Alvis Brazma, and Frank C. P. Holstege Protein Interaction Verification and Functional Annotation by Integrated Analysis of Genome-Scale Data Molecular Cell 2002, May 24; 9(5) pp. 1133 -1143 Johan Rung, Thomas Schlitt, Alvis Brazma, Karlis Freivalds, Jaak Vilo Building and analysing genome-wide gene disruption networks Bioinformatics 2002 Oct; 18 Suppl 2: S 202 -210 European Conference on Computational Biology (ECCB 2002) Kimmo Palin, Esko Ukkonen, Alvis Brazma, Jaak Vilo Correlating gene promoters and expression in gene disruption experiments Bioinformatics 2002 Oct; 18 Suppl 2: S 172 -180; European Conference on Computational Biology (ECCB, 2002)
References for this talk http: //www. egeen. ee/u/vilo/Publications/ Jaak Vilo, Misha Kapushesky, Patrick Kemmeren, Ugis Sarkans, Alvis Brazma. Expression Profiler. In Parmigiani, G. , Garrett, E. S. , Irizarry, R. and Zeger, S. L. (eds), The Analysis of Gene Expression Data: Methods and Software, Springer Verlag, New York, NY. Patrick Kemmeren, Nynke L. van Berkum, Jaak Vilo, Theo Bijma, Rogier Donders, Alvis Brazma, and Frank C. P. Holstege Protein Interaction Verification and Functional Annotation by Integrated Analysis of Genome-Scale Data Molecular Cell 2002, May 24; 9(5) pp. 1133 -1143 Johan Rung, Thomas Schlitt, Alvis Brazma, Karlis Freivalds, Jaak Vilo Building and analysing genome-wide gene disruption networks Bioinformatics 2002 Oct; 18 Suppl 2: S 202 -210 European Conference on Computational Biology (ECCB 2002) Kimmo Palin, Esko Ukkonen, Alvis Brazma, Jaak Vilo Correlating gene promoters and expression in gene disruption experiments Bioinformatics 2002 Oct; 18 Suppl 2: S 172 -180; European Conference on Computational Biology (ECCB, 2002)
 Acknowledgements Alvis Brazma Patrick Kemmeren, EBI, UMC Utrecht Frank Holstege, UMC Utrecht Thomas Schlitt, Johan Rung EBI Kimmo Palin, Esko Ukkonen, U. Helsinki + the rest of the EBI microarray team
Acknowledgements Alvis Brazma Patrick Kemmeren, EBI, UMC Utrecht Frank Holstege, UMC Utrecht Thomas Schlitt, Johan Rung EBI Kimmo Palin, Esko Ukkonen, U. Helsinki + the rest of the EBI microarray team


