bb1c5db3ddb07121950540a035f41cb8.ppt
- Количество слайдов: 77
Microbial Genetics and biotechnology
Define Terms o o o o Genetics Genome / Genomics Chromosomes Gene Genotype Phenotype Recombination
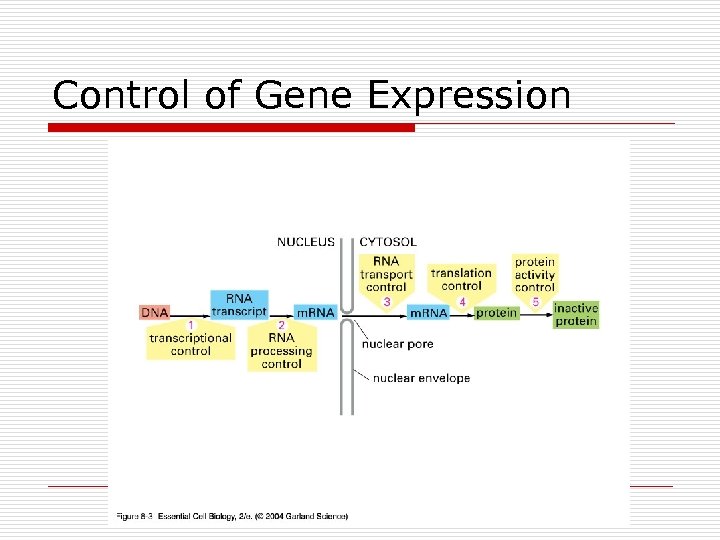
Control of Gene Expression
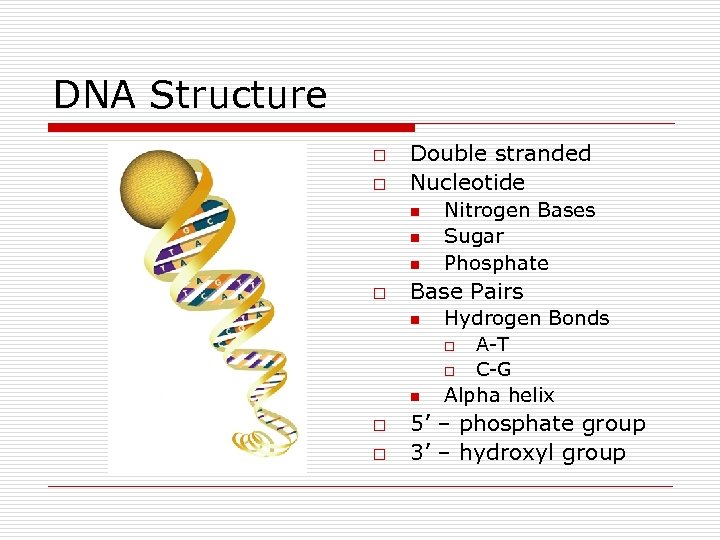
DNA Structure o o Double stranded Nucleotide n n n o Nitrogen Bases Sugar Phosphate Base Pairs n Hydrogen Bonds o o n o o A-T C-G Alpha helix 5’ – phosphate group 3’ – hydroxyl group
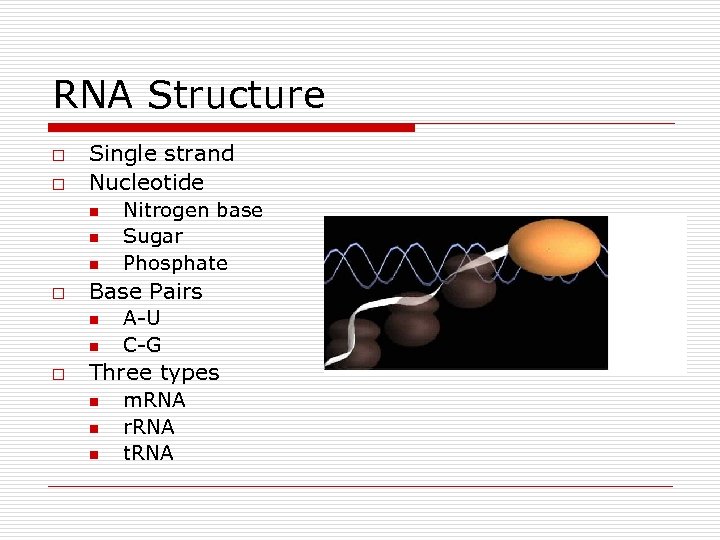
RNA Structure o o Single strand Nucleotide n n n o Base Pairs n n o Nitrogen base Sugar Phosphate A-U C-G Three types n n n m. RNA r. RNA t. RNA
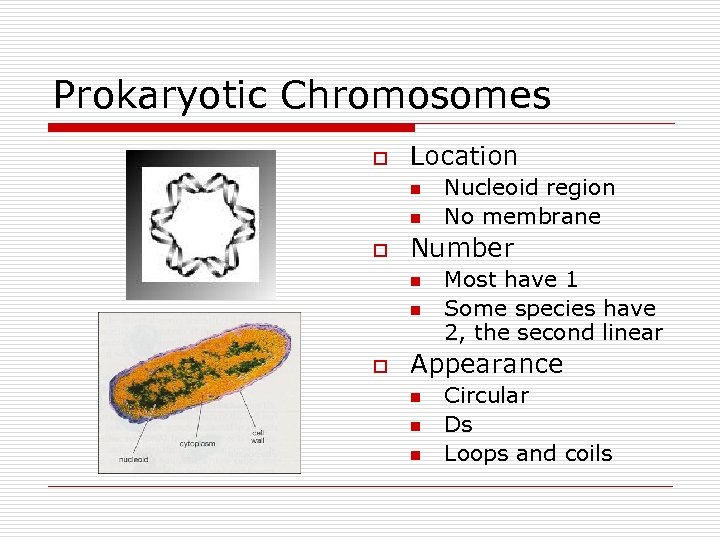
Prokaryotic Chromosomes o Location n n o Number n n o Nucleoid region No membrane Most have 1 Some species have 2, the second linear Appearance n n n Circular Ds Loops and coils
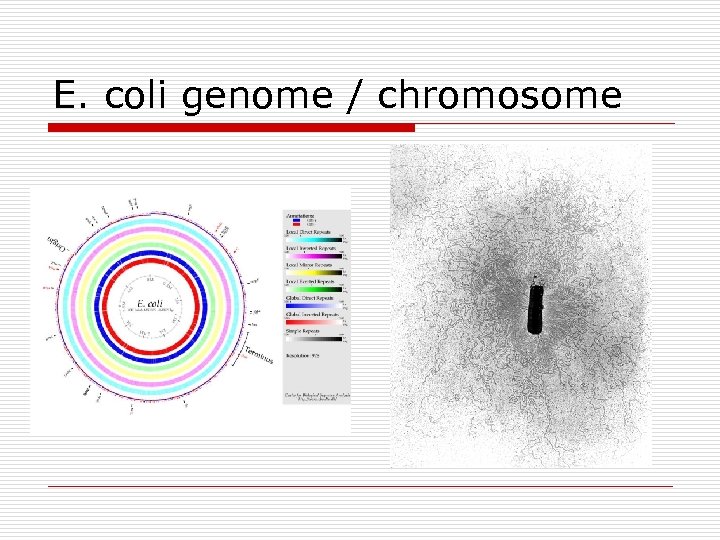
E. coli genome / chromosome
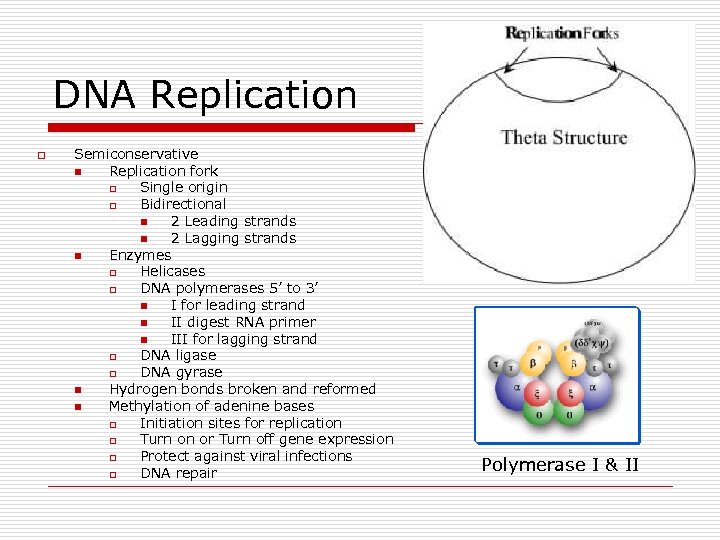
DNA Replication o Semiconservative n Replication fork o Single origin o Bidirectional n 2 Leading strands n 2 Lagging strands n Enzymes o Helicases o DNA polymerases 5’ to 3’ n I for leading strand n II digest RNA primer n III for lagging strand o DNA ligase o DNA gyrase n Hydrogen bonds broken and reformed n Methylation of adenine bases o Initiation sites for replication o Turn on or Turn off gene expression o Protect against viral infections o DNA repair Polymerase I & II
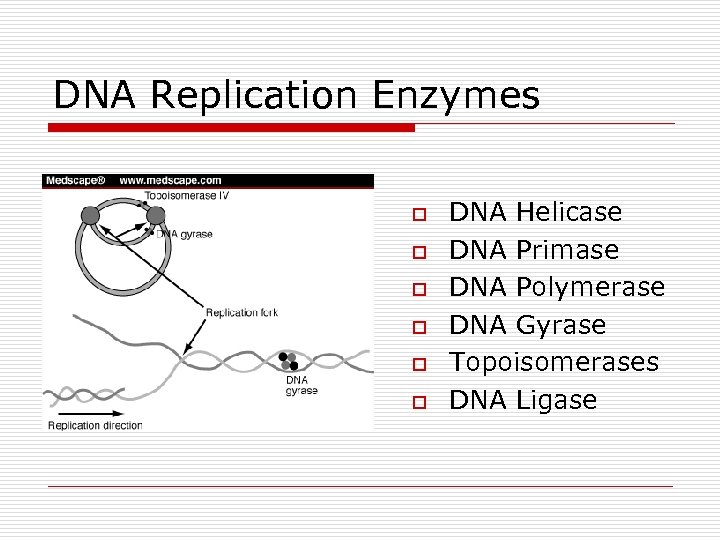
DNA Replication Enzymes o o o DNA Helicase DNA Primase DNA Polymerase DNA Gyrase Topoisomerases DNA Ligase
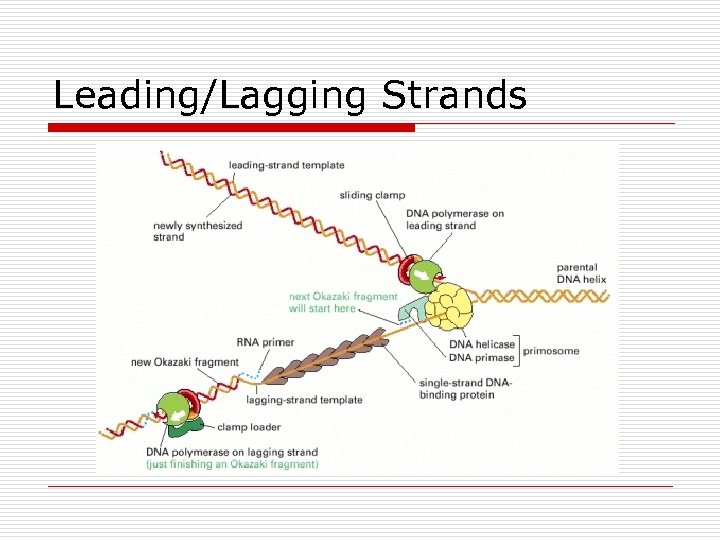
Leading/Lagging Strands
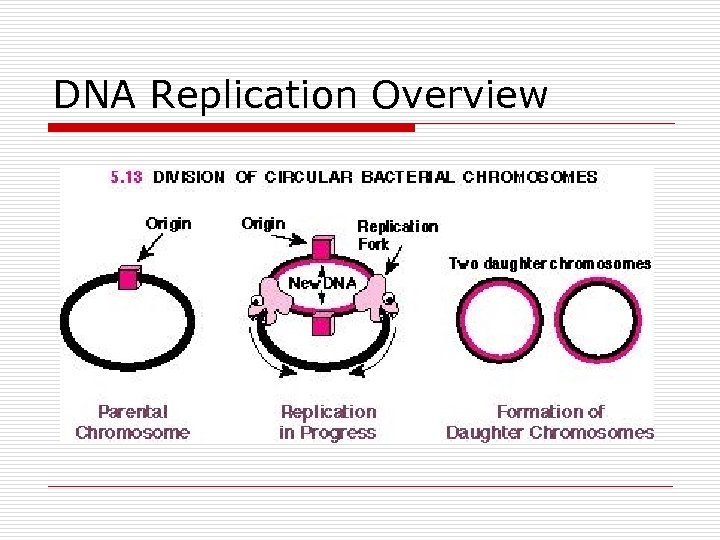
DNA Replication Overview
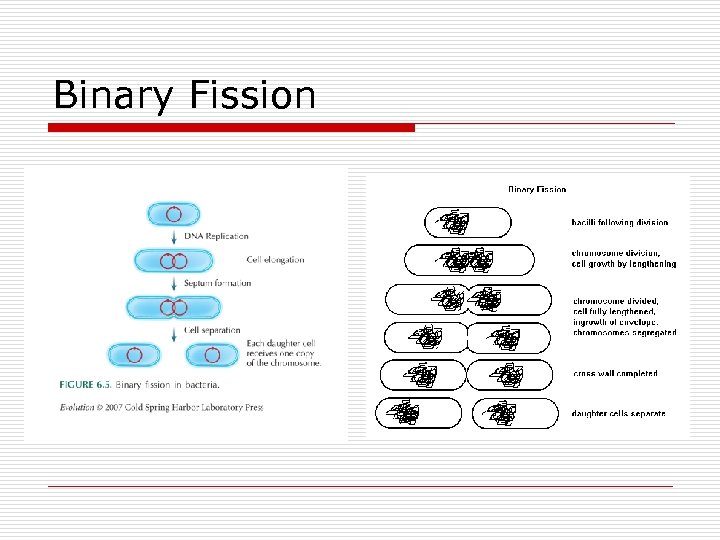
Binary Fission
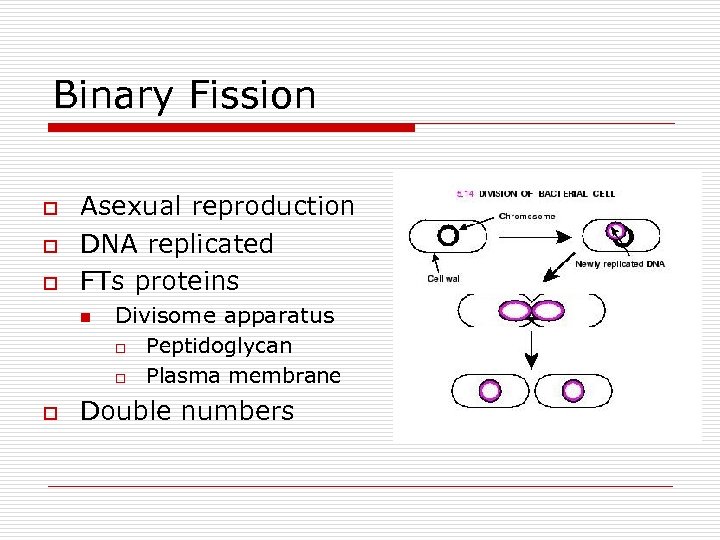
Binary Fission o o o Asexual reproduction DNA replicated FTs proteins n o Divisome apparatus o Peptidoglycan o Plasma membrane Double numbers
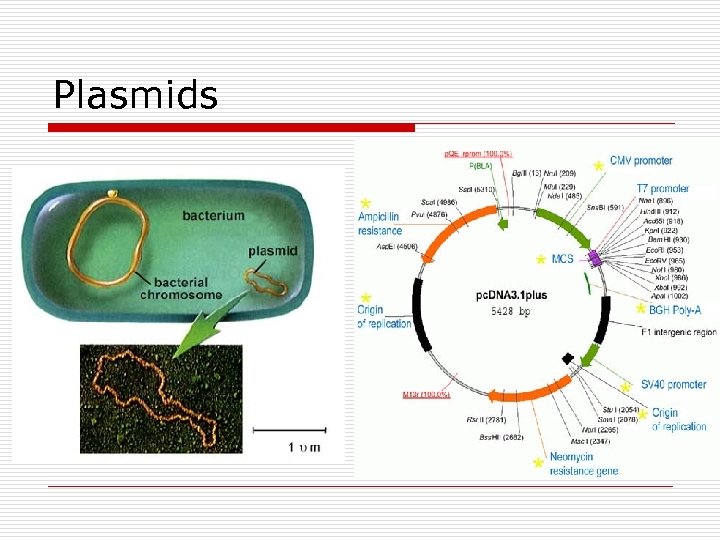
Plasmids
Plasmids o o 2% of genetic information (5 -100 genes) ds, circular extra chromosomal DNA Independent replication Cellular Traits n n n F-Fertility R-Resistance : inactivate AB, toxins, heavy metals Dissimilation: catabolism of unusual substances Bacteriocins Virulence : enzymes, toxins, attachment
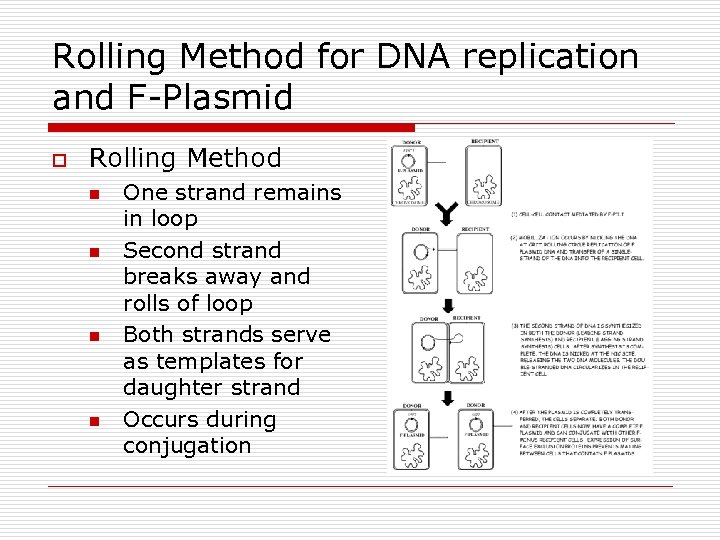
Rolling Method for DNA replication and F-Plasmid o Rolling Method n n One strand remains in loop Second strand breaks away and rolls of loop Both strands serve as templates for daughter strand Occurs during conjugation
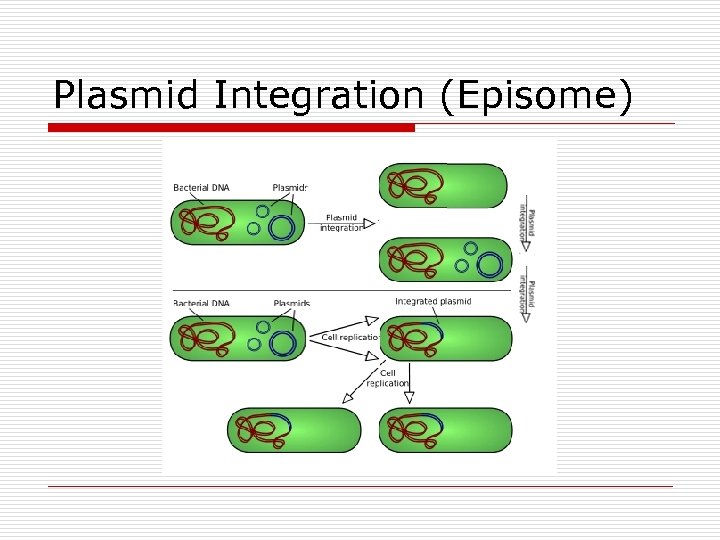
Plasmid Integration (Episome)
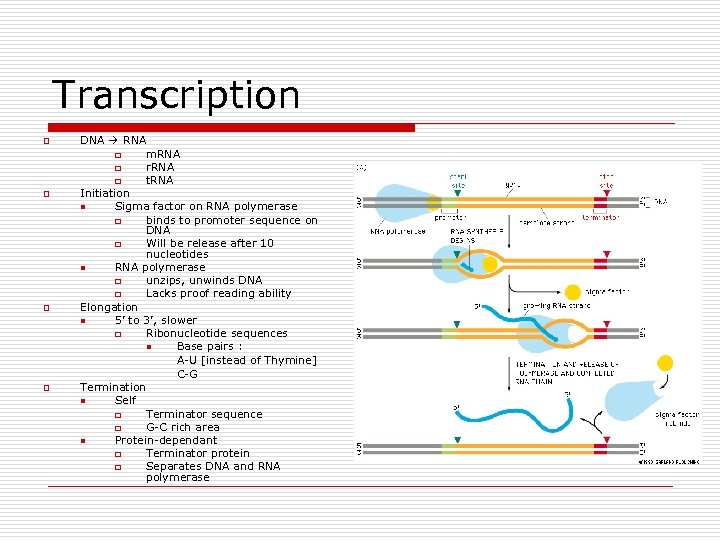
Transcription o o DNA RNA o m. RNA o r. RNA o t. RNA Initiation n Sigma factor on RNA polymerase o binds to promoter sequence on DNA o Will be release after 10 nucleotides n RNA polymerase o unzips, unwinds DNA o Lacks proof reading ability Elongation n 5’ to 3’, slower o Ribonucleotide sequences n Base pairs : A-U [instead of Thymine] C-G Termination n Self o Terminator sequence o G-C rich area n Protein-dependant o Terminator protein o Separates DNA and RNA polymerase
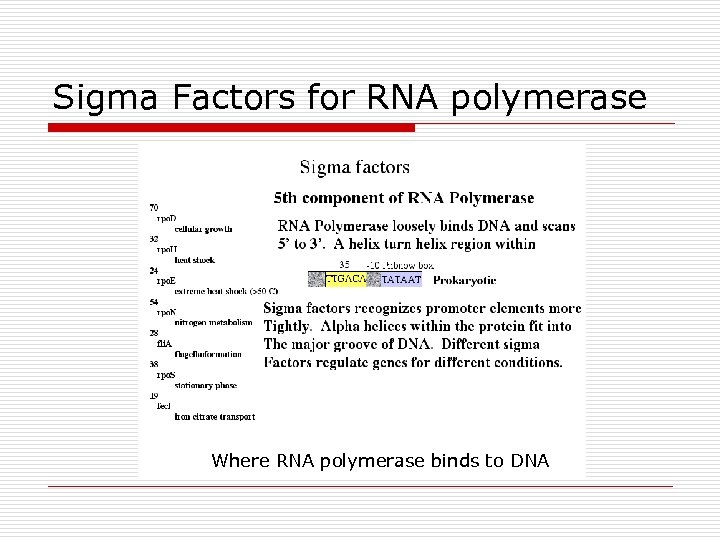
Sigma Factors for RNA polymerase Where RNA polymerase binds to DNA
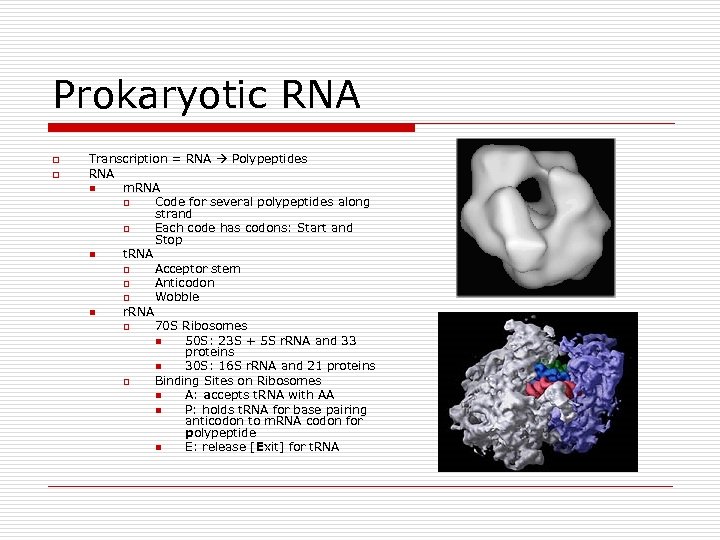
Prokaryotic RNA o o Transcription = RNA Polypeptides RNA n m. RNA o Code for several polypeptides along strand o Each code has codons: Start and Stop n t. RNA o Acceptor stem o Anticodon o Wobble n r. RNA o 70 S Ribosomes n 50 S: 23 S + 5 S r. RNA and 33 proteins n 30 S: 16 S r. RNA and 21 proteins o Binding Sites on Ribosomes n A: accepts t. RNA with AA n P: holds t. RNA for base pairing anticodon to m. RNA codon for polypeptide n E: release [Exit] for t. RNA
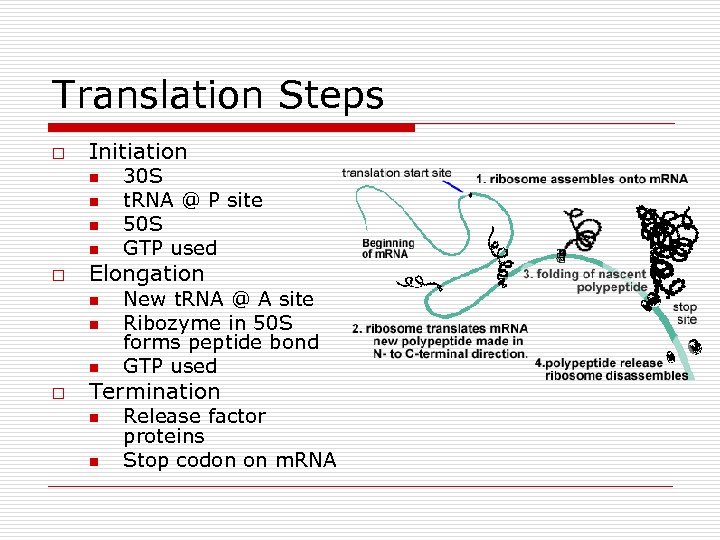
Translation Steps o Initiation n n o Elongation n o 30 S t. RNA @ P site 50 S GTP used New t. RNA @ A site Ribozyme in 50 S forms peptide bond GTP used Termination n n Release factor proteins Stop codon on m. RNA
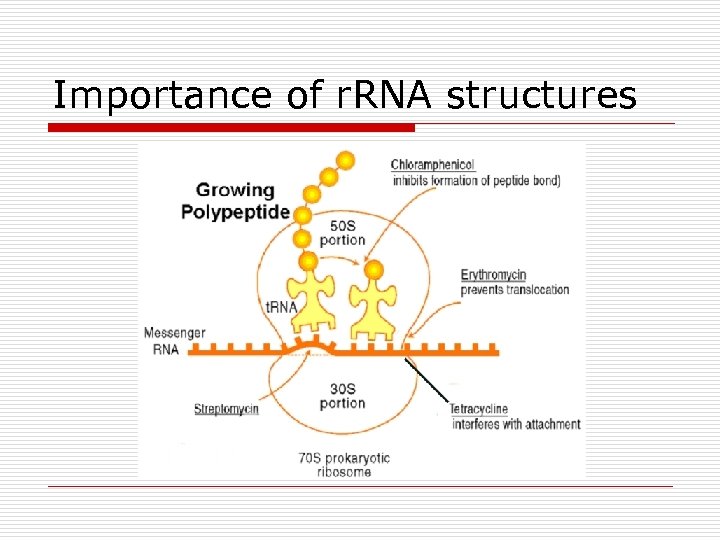
Importance of r. RNA structures
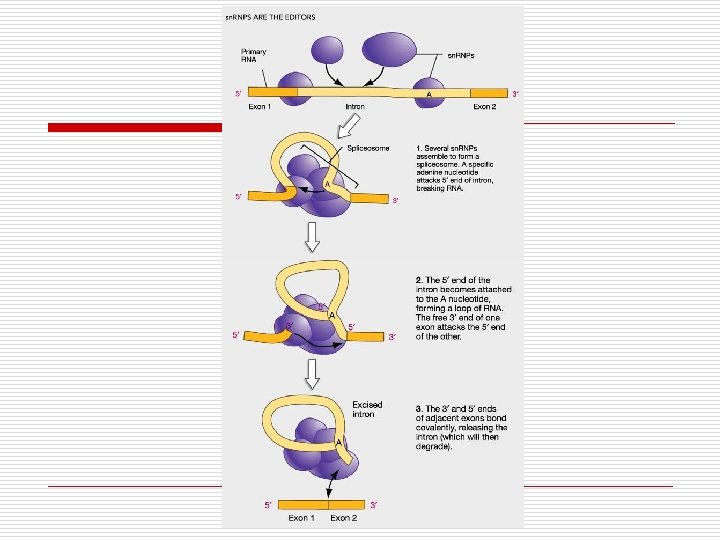
Regulation of Gene Expression o Constitutive n n o Not regulated Always “on” at fixed rate o Transcription o Translation 60 -80% Polypeptides need in large amounts Regulated n n n Only when needed Control synthesis of genes for enzymes o Induction o Repression Control enzyme activity: feedback o Noncompetitive inhibition o Competitive inhibition
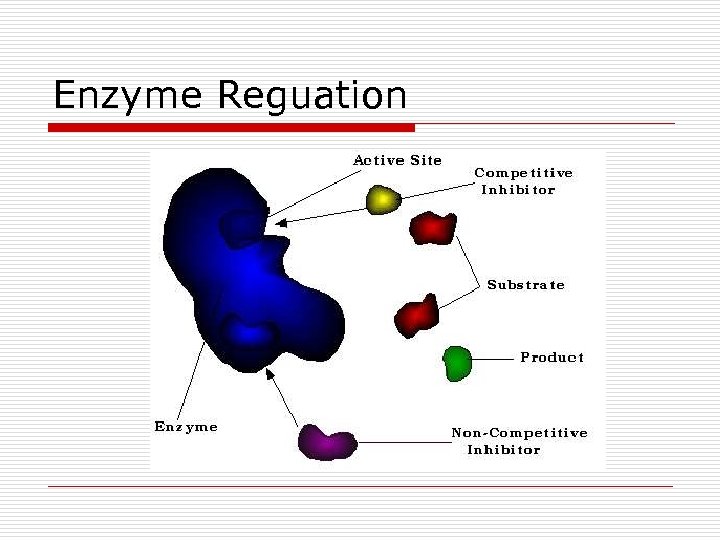
Enzyme Reguation
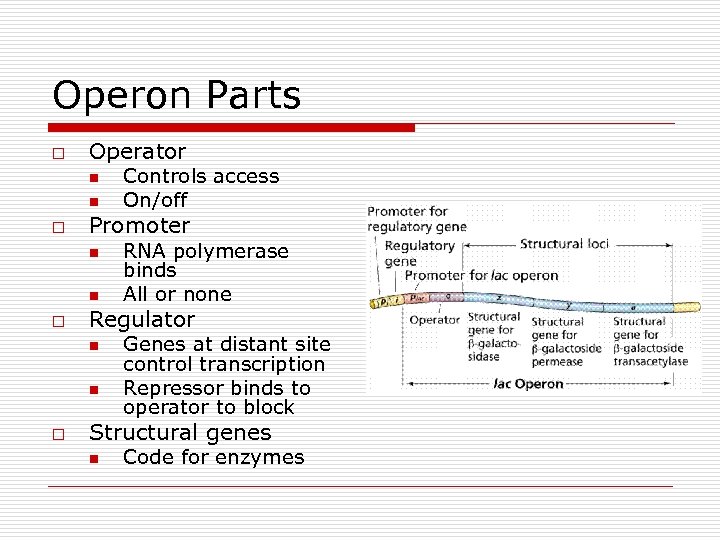
Operon Parts o Operator n n o Promoter n n o RNA polymerase binds All or none Regulator n n o Controls access On/off Genes at distant site control transcription Repressor binds to operator to block Structural genes n Code for enzymes
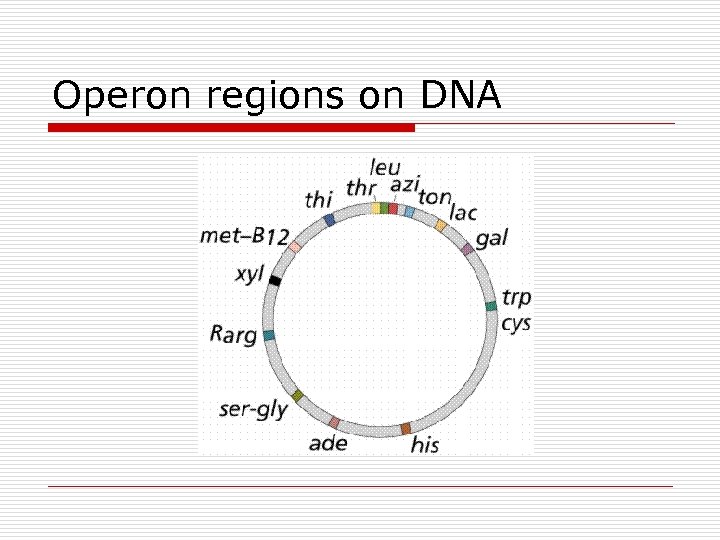
Operon regions on DNA
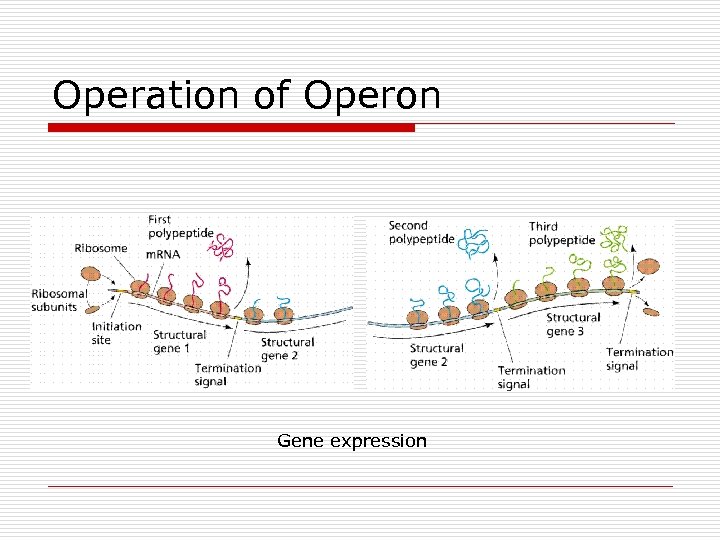
Operation of Operon Gene expression

Operator Always “on” unless switched off by repressor
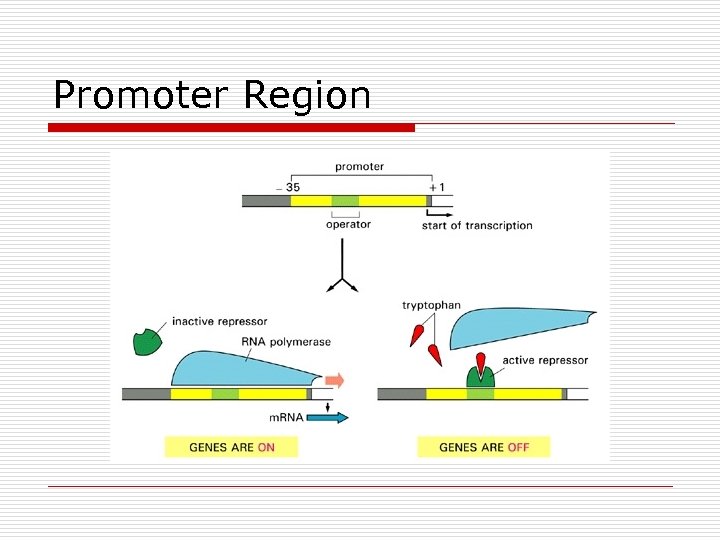
Promoter Region
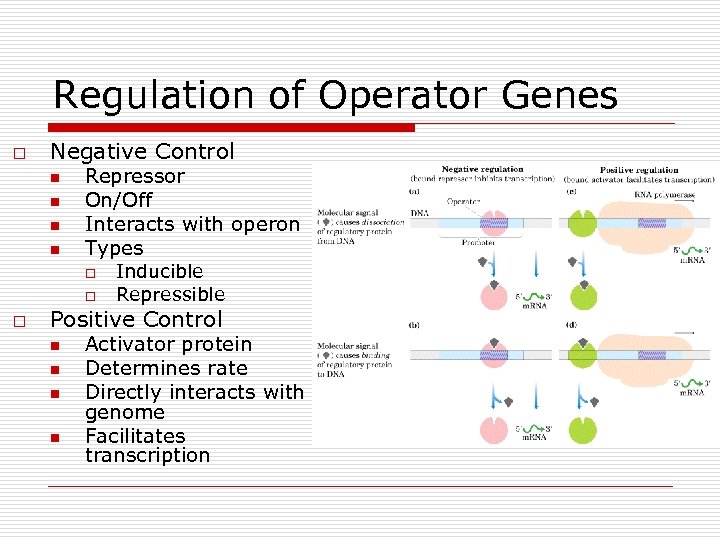
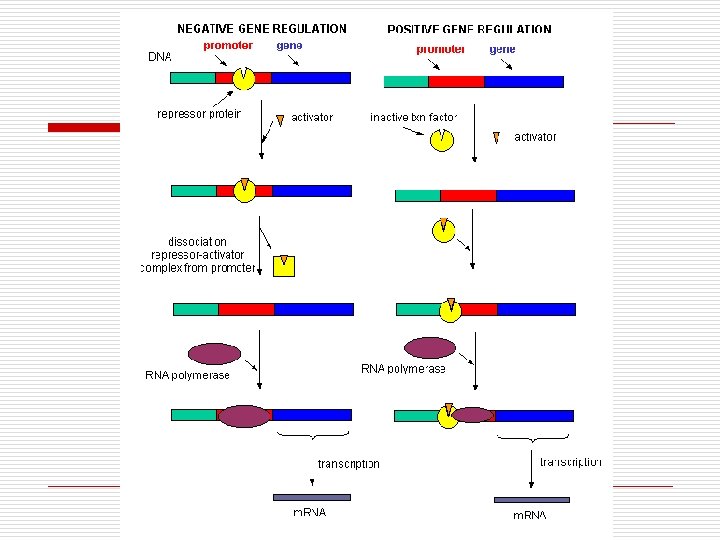
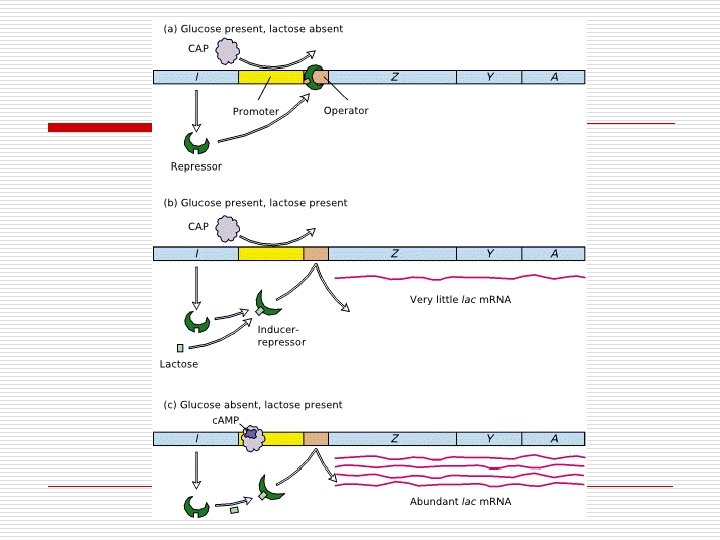
Regulation of Operator Genes o Negative Control n n Repressor On/Off Interacts with operon Types o o o Inducible Repressible Positive Control n n Activator protein Determines rate Directly interacts with genome Facilitates transcription
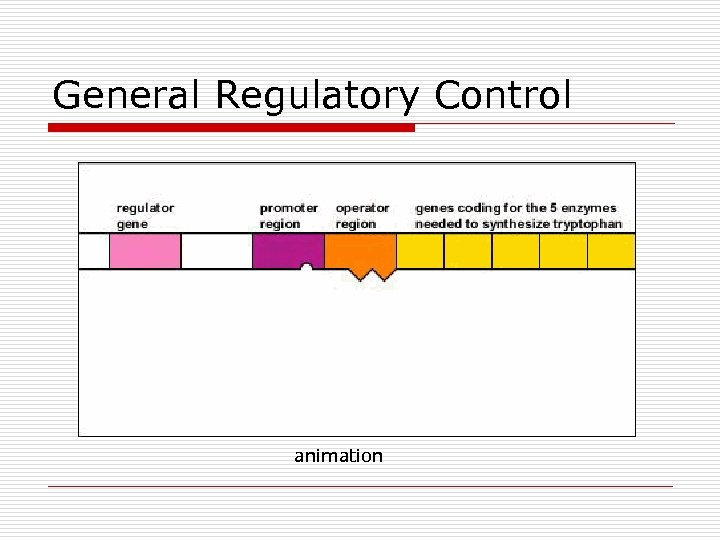
General Regulatory Control animation
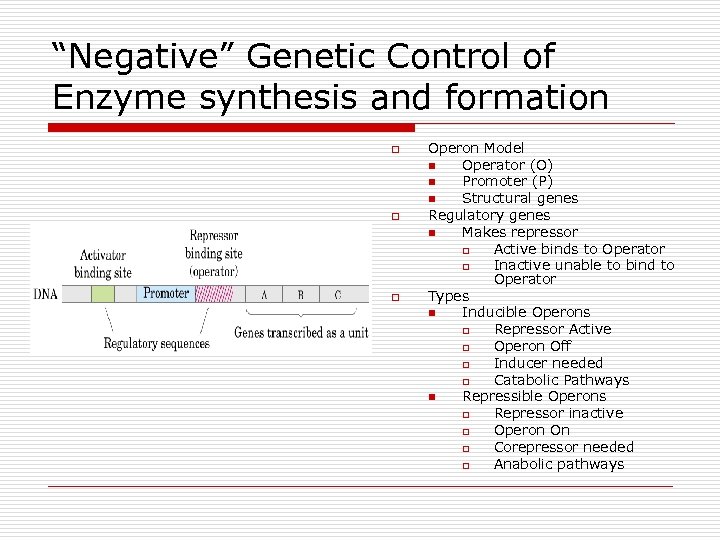
“Negative” Genetic Control of Enzyme synthesis and formation o o o Operon Model n Operator (O) n Promoter (P) n Structural genes Regulatory genes n Makes repressor o Active binds to Operator o Inactive unable to bind to Operator Types n Inducible Operons o Repressor Active o Operon Off o Inducer needed o Catabolic Pathways n Repressible Operons o Repressor inactive o Operon On o Corepressor needed o Anabolic pathways
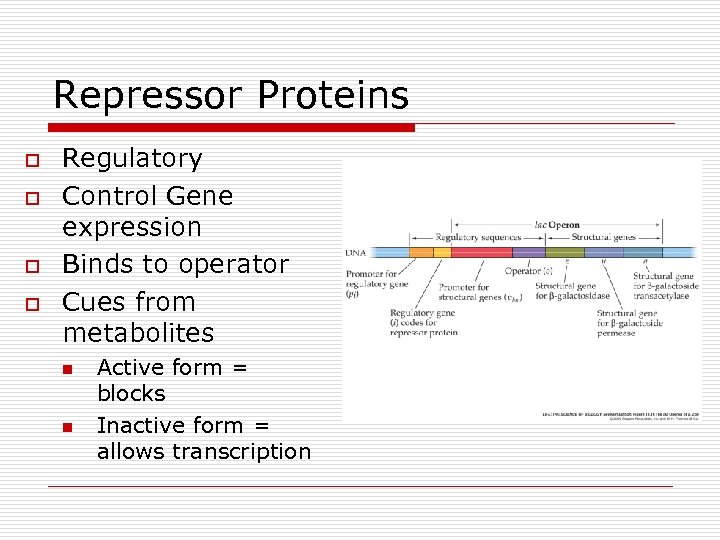
Repressor Proteins o o Regulatory Control Gene expression Binds to operator Cues from metabolites n n Active form = blocks Inactive form = allows transcription
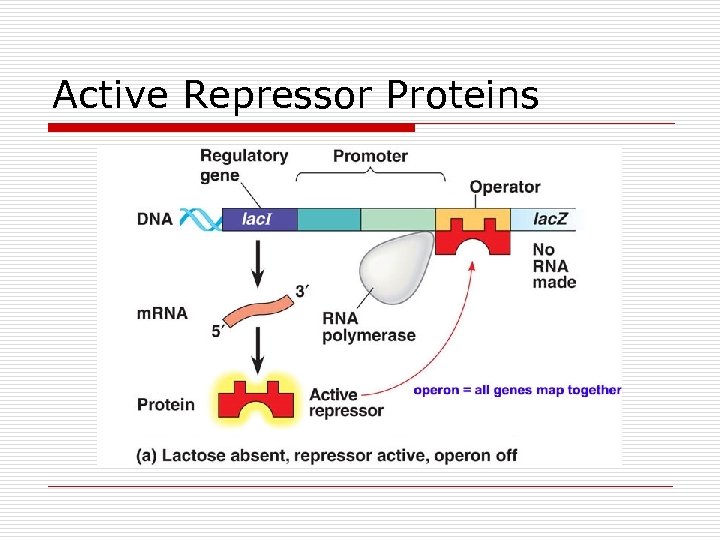
Active Repressor Proteins
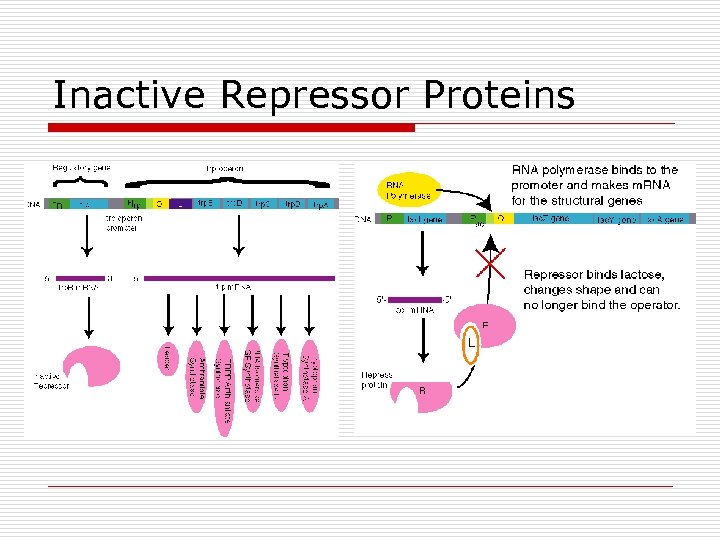
Inactive Repressor Proteins
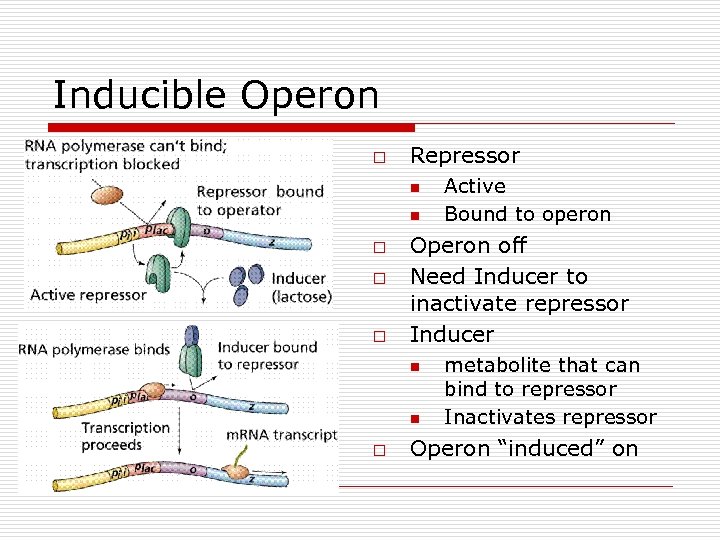
Inducible Operon o Repressor n n o o o Operon off Need Inducer to inactivate repressor Inducer n n o Active Bound to operon metabolite that can bind to repressor Inactivates repressor Operon “induced” on
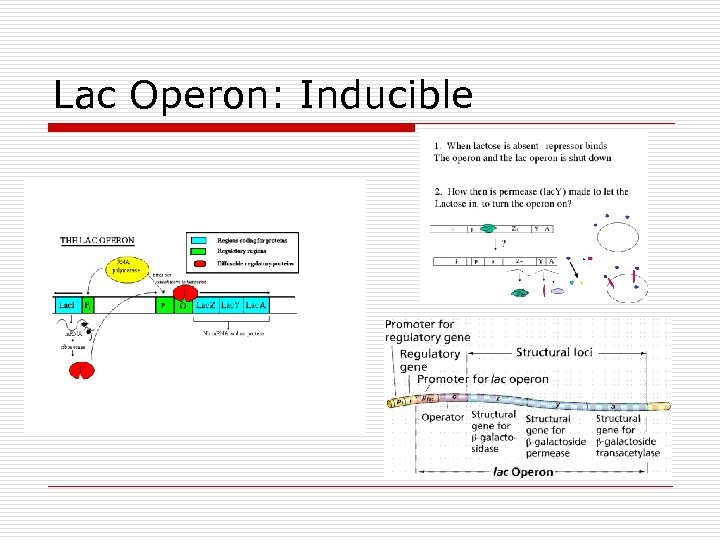
Lac Operon: Inducible
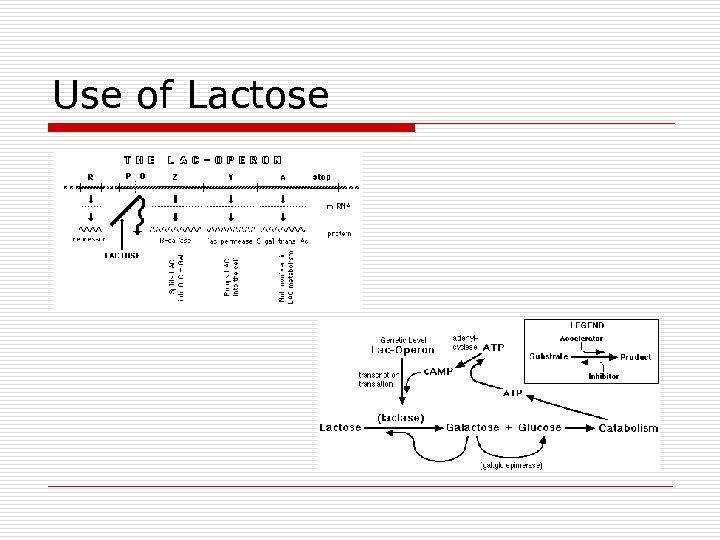
Use of Lactose
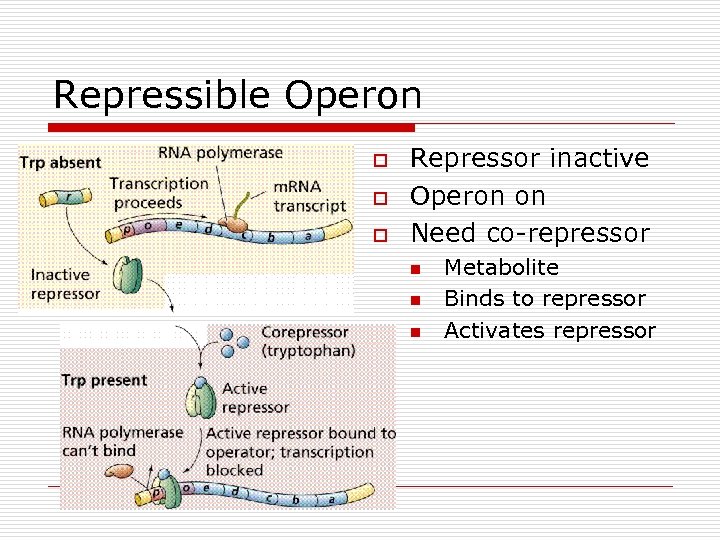
Repressible Operon o o o Repressor inactive Operon on Need co-repressor n n n Metabolite Binds to repressor Activates repressor
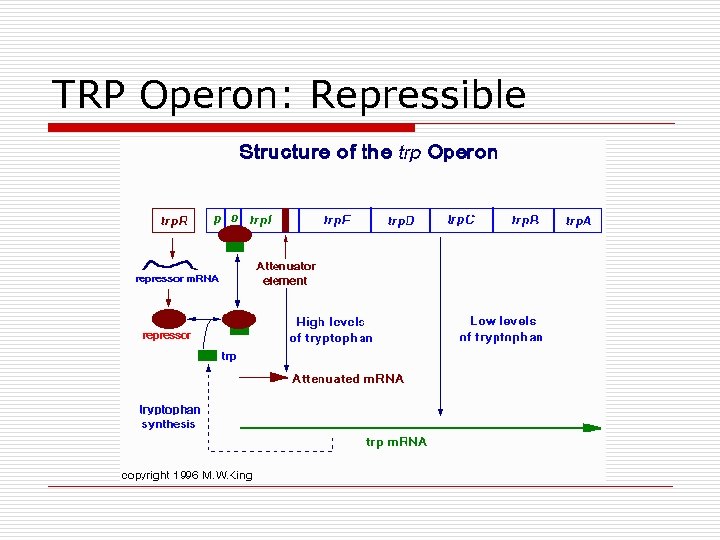
TRP Operon: Repressible
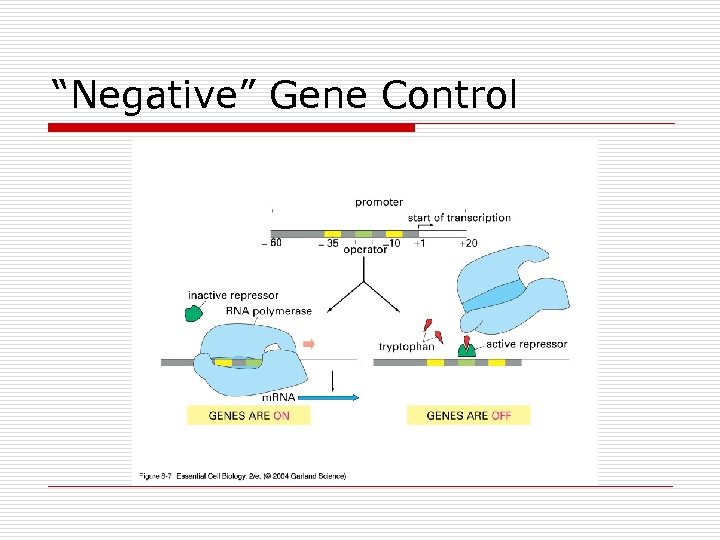
“Negative” Gene Control
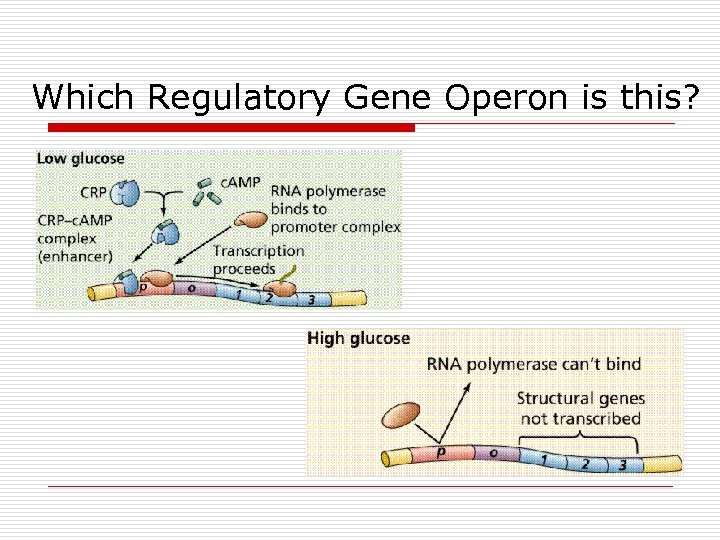
Which Regulatory Gene Operon is this?
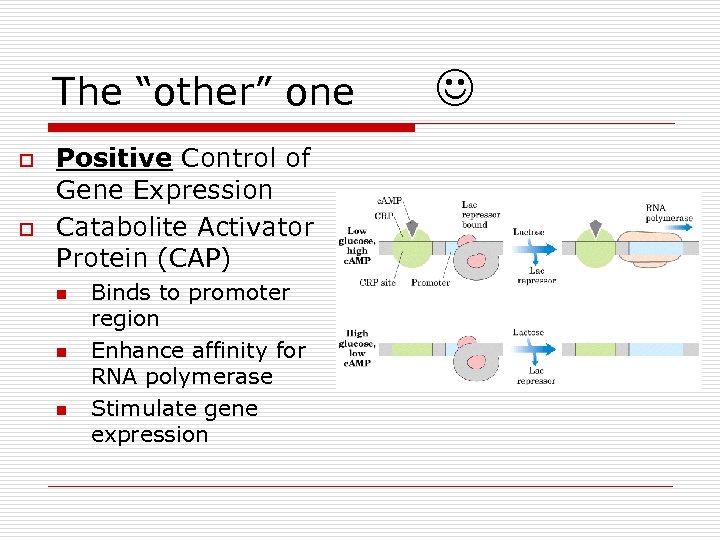
The “other” one o o Positive Control of Gene Expression Catabolite Activator Protein (CAP) n n n Binds to promoter region Enhance affinity for RNA polymerase Stimulate gene expression
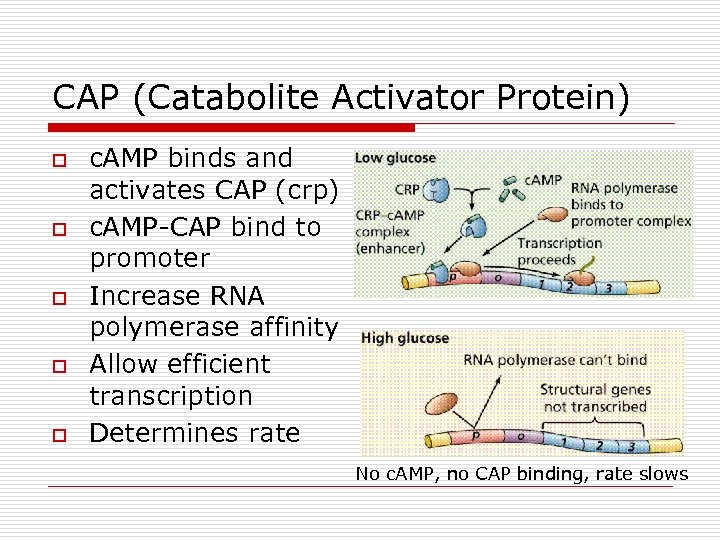
CAP (Catabolite Activator Protein) o o o c. AMP binds and activates CAP (crp) c. AMP-CAP bind to promoter Increase RNA polymerase affinity Allow efficient transcription Determines rate No c. AMP, no CAP binding, rate slows
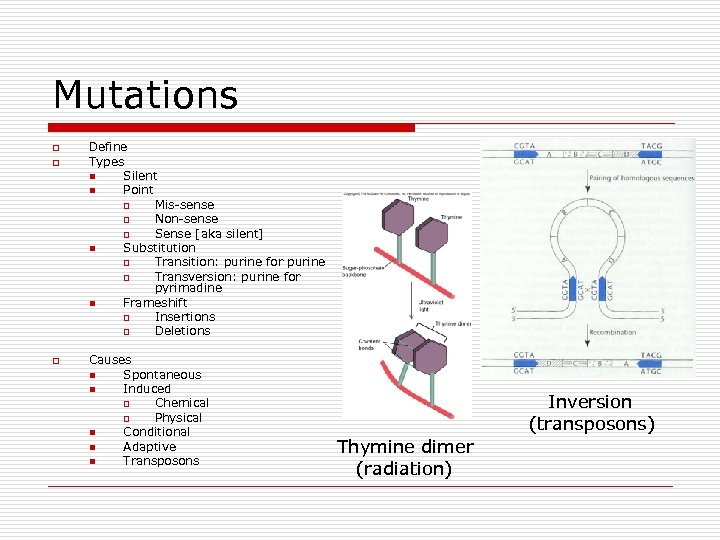
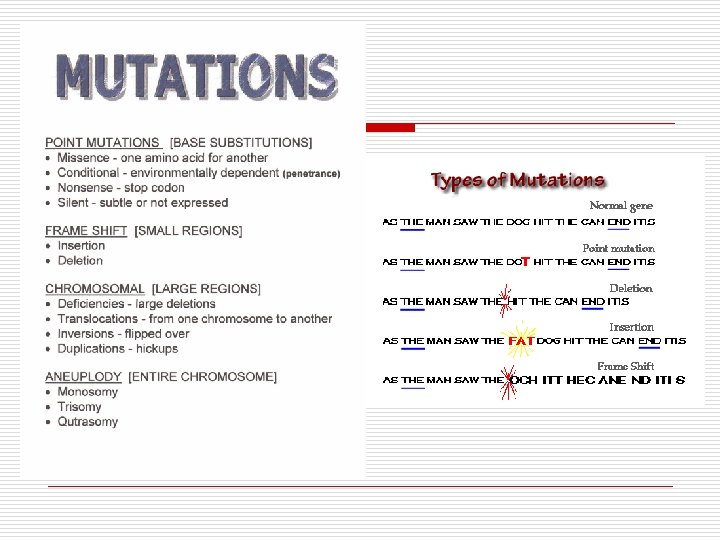
Mutations o o o Define Types n Silent n Point o Mis-sense o Non-sense o Sense [aka silent] n Substitution o Transition: purine for purine o Transversion: purine for pyrimadine n Frameshift o Insertions o Deletions Causes n Spontaneous n Induced o Chemical o Physical n Conditional n Adaptive n Transposons Inversion (transposons) Thymine dimer (radiation)
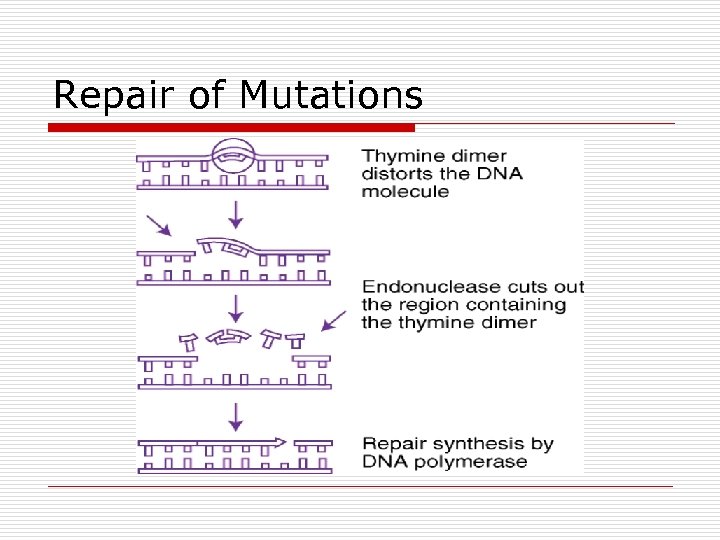
Repair of Mutations
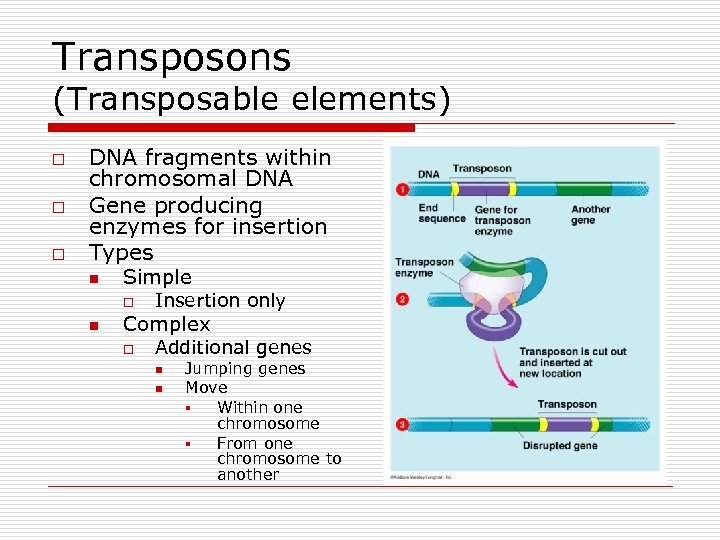
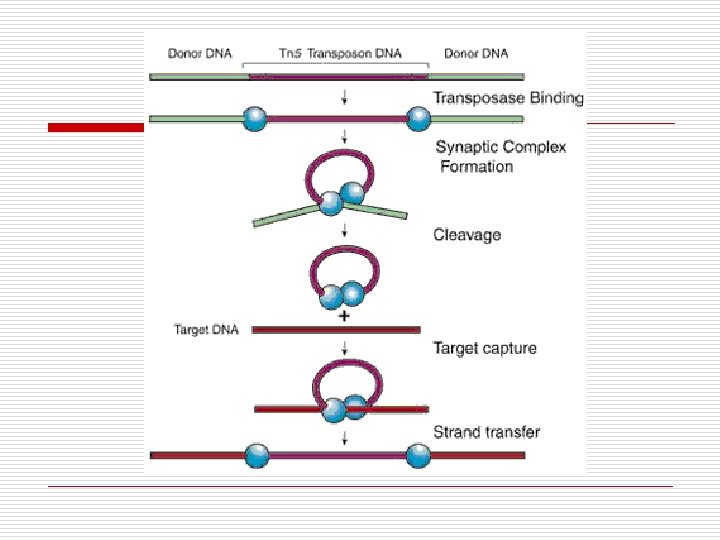
Transposons (Transposable elements) o o o DNA fragments within chromosomal DNA Gene producing enzymes for insertion Types n Simple o n Insertion only Complex o Additional genes n n Jumping genes Move § Within one chromosome § From one chromosome to another
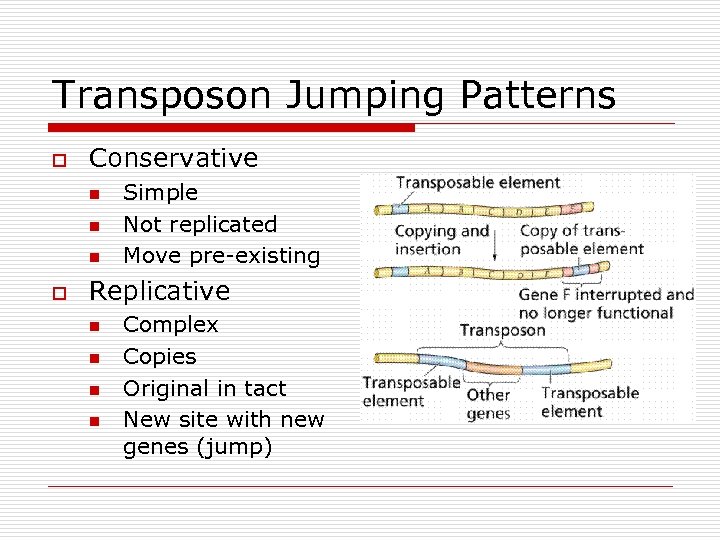
Transposon Jumping Patterns o Conservative n n n o Simple Not replicated Move pre-existing Replicative n n Complex Copies Original in tact New site with new genes (jump)
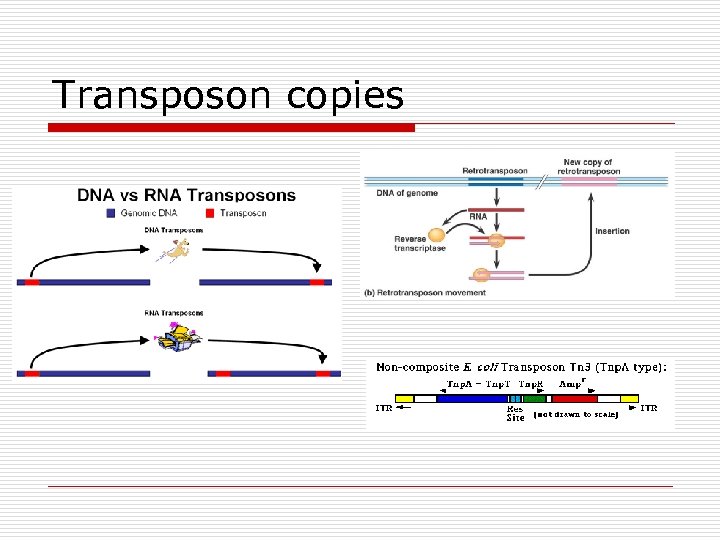
Transposon copies
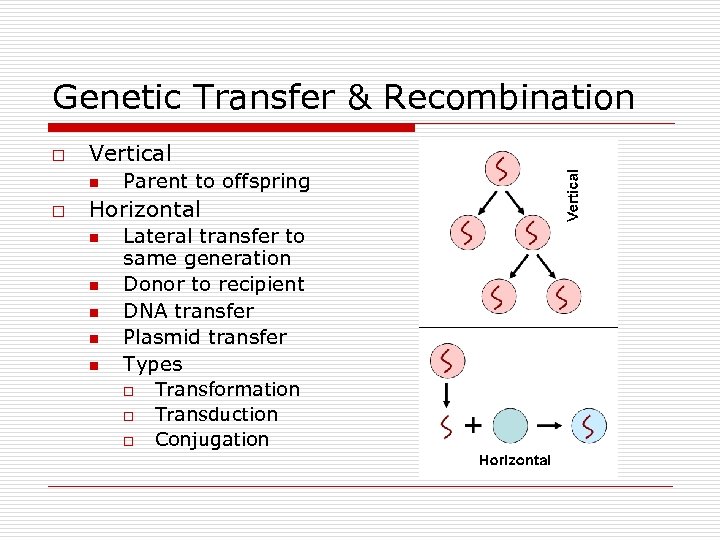
Genetic Transfer & Recombination o Vertical n o Parent to offspring Horizontal n n n Lateral transfer to same generation Donor to recipient DNA transfer Plasmid transfer Types o o o Transformation Transduction Conjugation
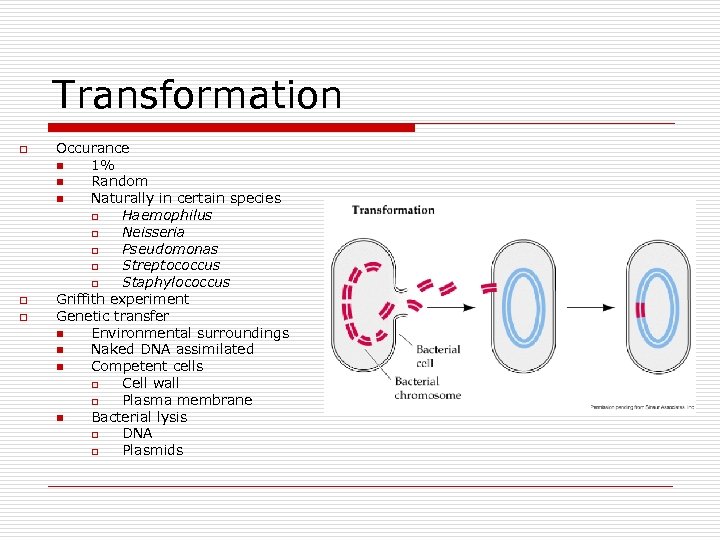
Transformation o o o Occurance n 1% n Random n Naturally in certain species o Haemophilus o Neisseria o Pseudomonas o Streptococcus o Staphylococcus Griffith experiment Genetic transfer n Environmental surroundings n Naked DNA assimilated n Competent cells o Cell wall o Plasma membrane n Bacterial lysis o DNA o Plasmids
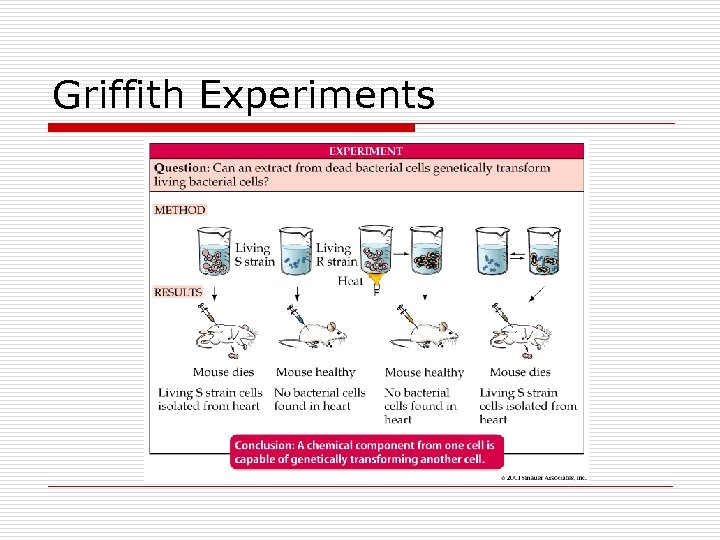
Griffith Experiments
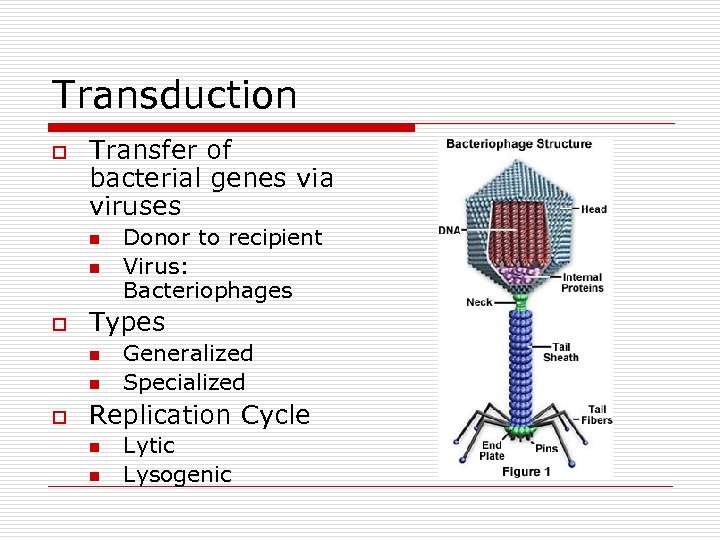
Transduction o Transfer of bacterial genes via viruses n n o Types n n o Donor to recipient Virus: Bacteriophages Generalized Specialized Replication Cycle n n Lytic Lysogenic
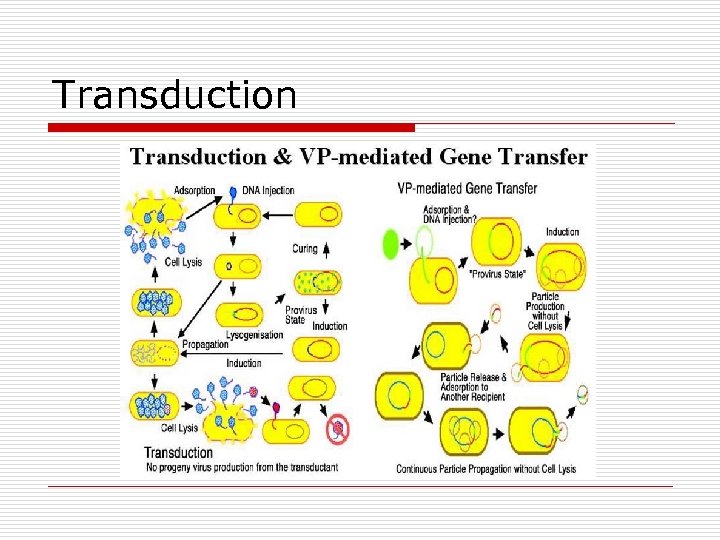
Transduction
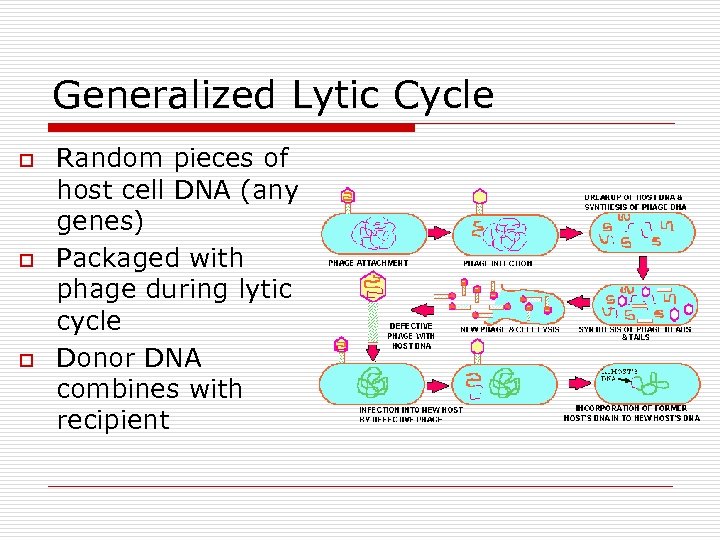
Generalized Lytic Cycle o o o Random pieces of host cell DNA (any genes) Packaged with phage during lytic cycle Donor DNA combines with recipient
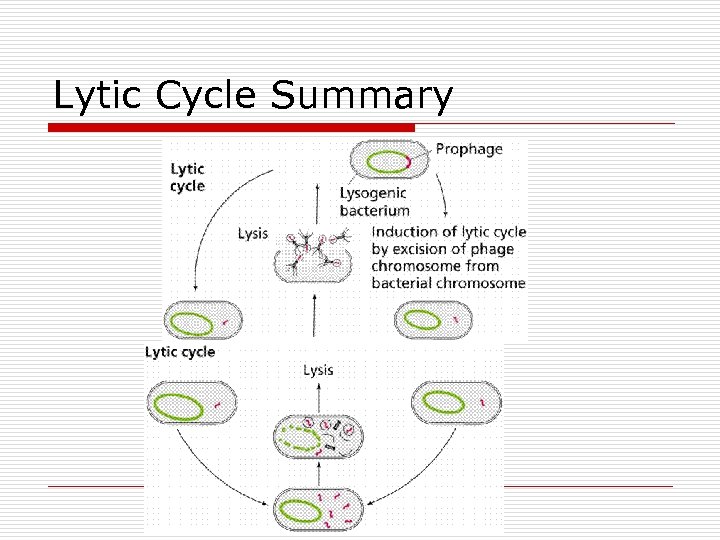
Lytic Cycle Summary
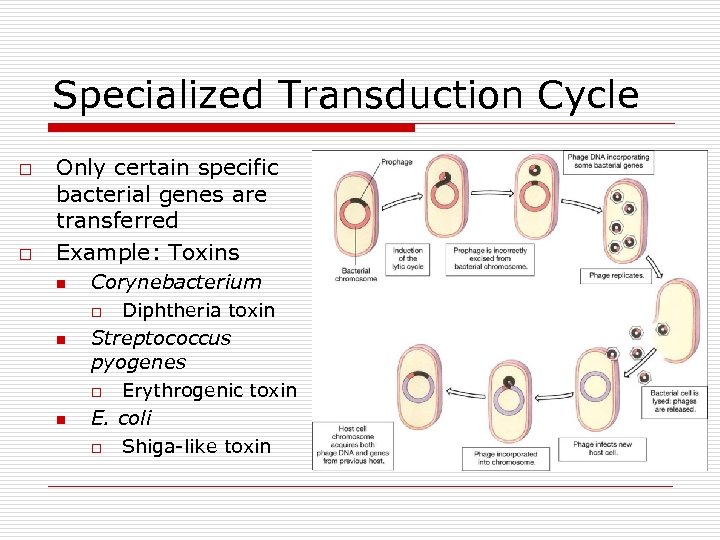
Specialized Transduction Cycle o o Only certain specific bacterial genes are transferred Example: Toxins n Corynebacterium o n Streptococcus pyogenes o n Diphtheria toxin Erythrogenic toxin E. coli o Shiga-like toxin
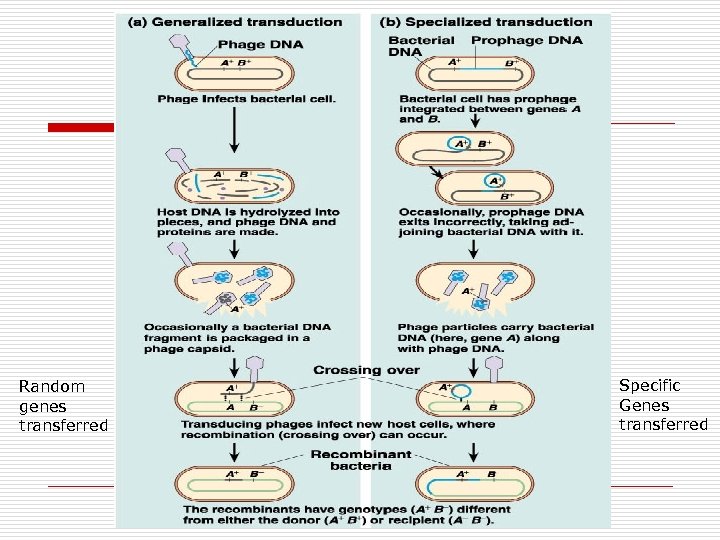
Random genes transferred Specific Genes transferred
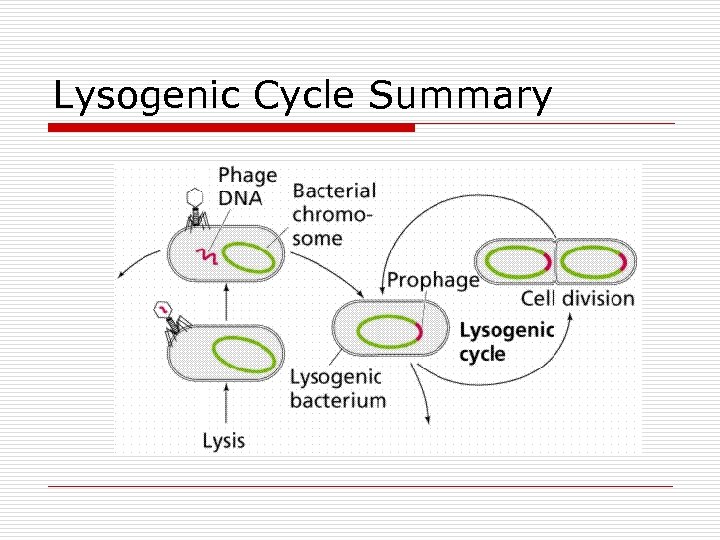
Lysogenic Cycle Summary
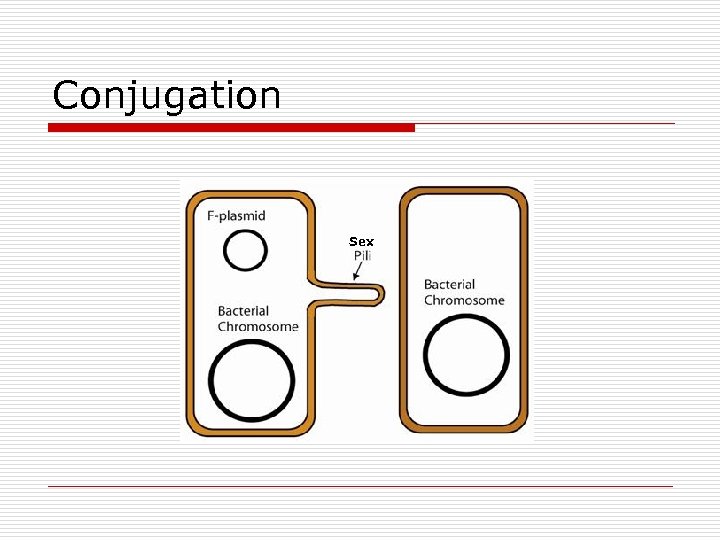
Conjugation Sex
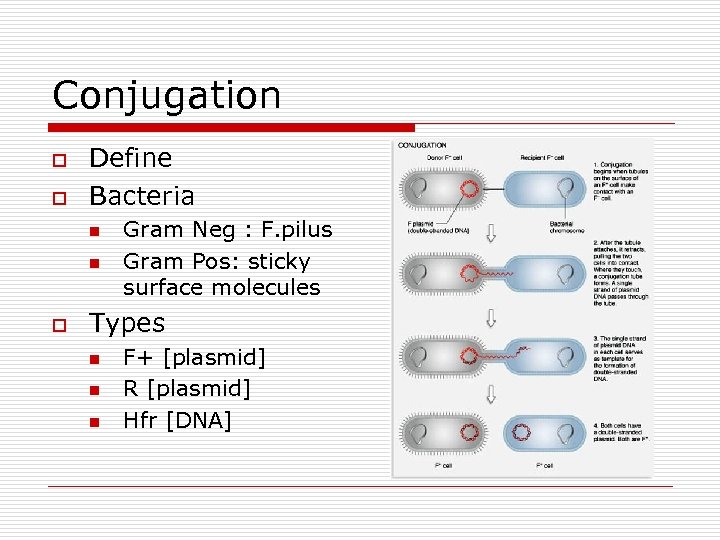
Conjugation o o Define Bacteria n n o Gram Neg : F. pilus Gram Pos: sticky surface molecules Types n n n F+ [plasmid] R [plasmid] Hfr [DNA]
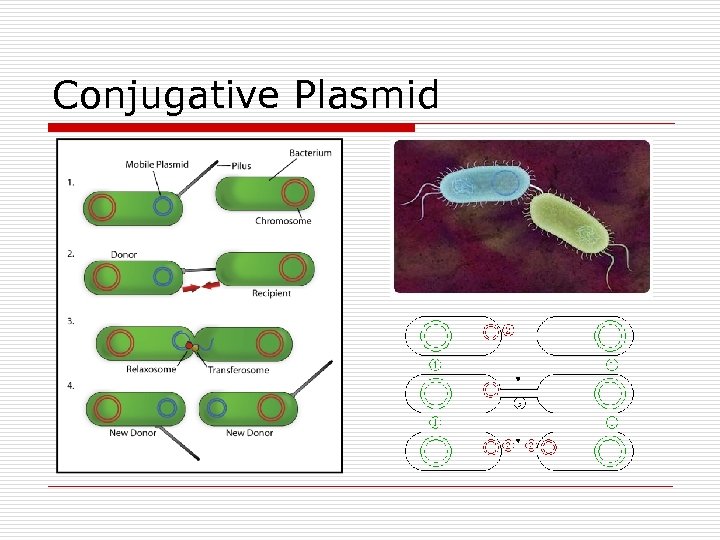
Conjugative Plasmid
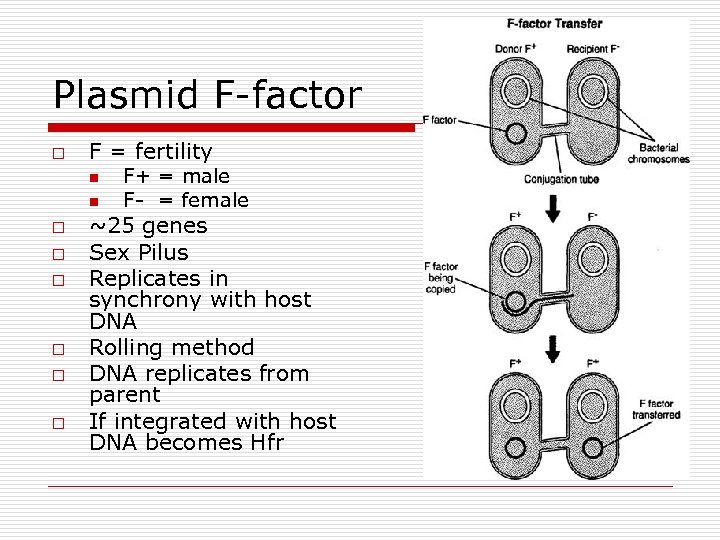
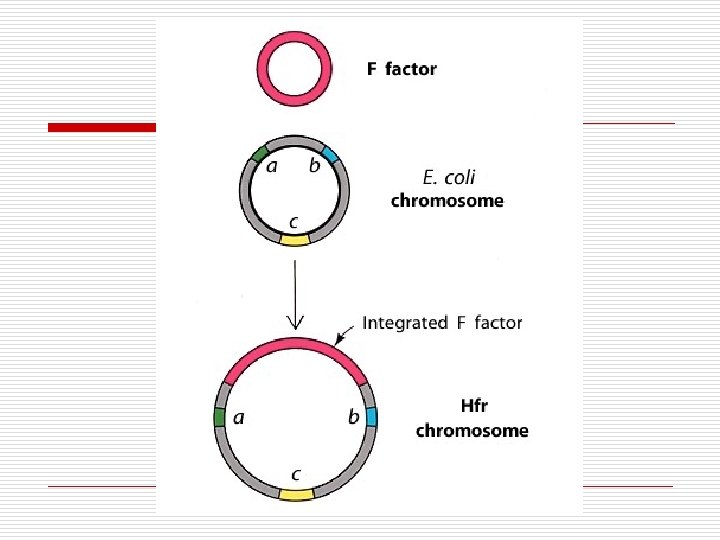
Plasmid F-factor o F = fertility n n o o o F+ = male F- = female ~25 genes Sex Pilus Replicates in synchrony with host DNA Rolling method DNA replicates from parent If integrated with host DNA becomes Hfr
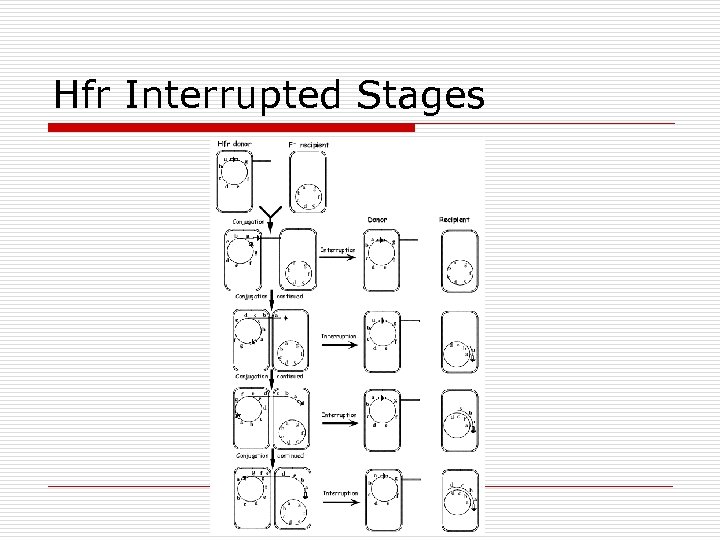
Hfr Interrupted Stages
Other Plasmids o R factors n n n o Bacteriocin factors n n o Resistance to AB ~10 genes Different bacterial species share Specific protein toxins Kill other cells of same or similar species Virulence n Pathogenicity o o o Structures Enzymes Toxins
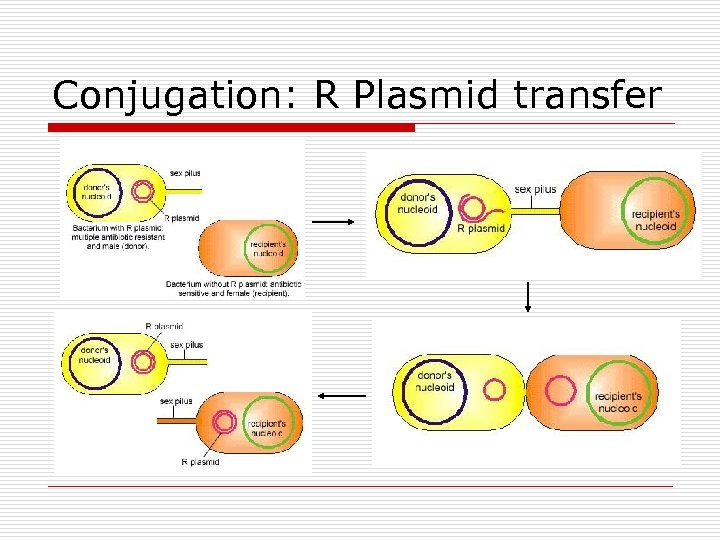
Conjugation: R Plasmid transfer
Genetic Recombination o General n n n o Site Specific n n o o Homologous chromosomes Any location DNA breakage and repair Non-homologous Viral genomes in bacterial chromosomes Replicative Health and Food industries
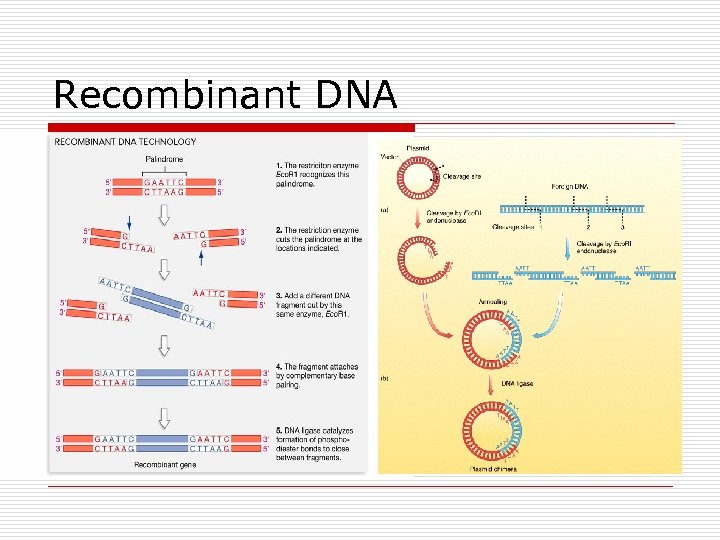
Recombinant DNA
Genetic Engineering o Use n n o Plasmids Recombinant DNA Applications n Therapeutic o o n n Hormones Enzymes Vaccines Gene therapy Agricultural Scientific o o Southern Blot ELISA tests
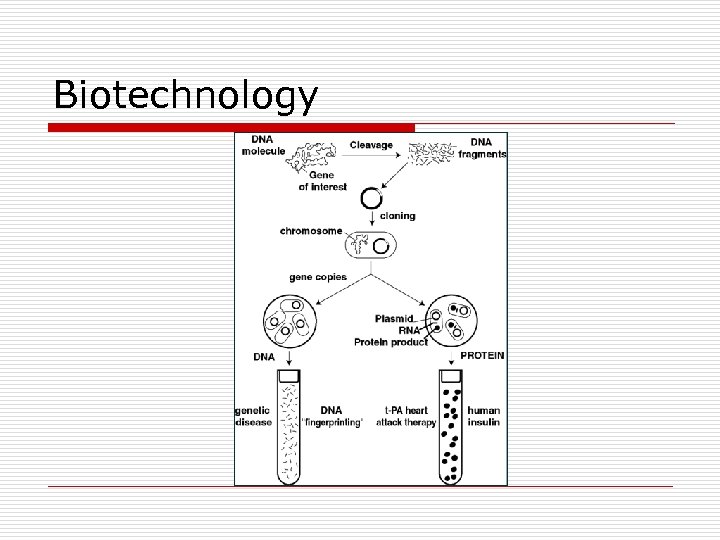
Biotechnology

Questions?
bb1c5db3ddb07121950540a035f41cb8.ppt