9b0095e5cfbbaf14210dcce13cf17641.ppt
- Количество слайдов: 70
 Microarrays, SNPs and Applications DNA m. RNA Protein Eleftherios P. Diamandis MD, Ph. D (ediamandis@mtsinai. on. ca) Website: www. acdclab. org
Microarrays, SNPs and Applications DNA m. RNA Protein Eleftherios P. Diamandis MD, Ph. D (ediamandis@mtsinai. on. ca) Website: www. acdclab. org
 Microarrays What is a microarray? n A microarray is a compact device that contains a large number of well-defined immobilized capture molecules (e. g. synthetic oligos, PCR products, proteins, antibodies) assembled in an addressable format. n You can expose an unknown (test) substance on it and then examine where the molecule was captured. n You can then derive information on identity and amount of captured molecule.
Microarrays What is a microarray? n A microarray is a compact device that contains a large number of well-defined immobilized capture molecules (e. g. synthetic oligos, PCR products, proteins, antibodies) assembled in an addressable format. n You can expose an unknown (test) substance on it and then examine where the molecule was captured. n You can then derive information on identity and amount of captured molecule.
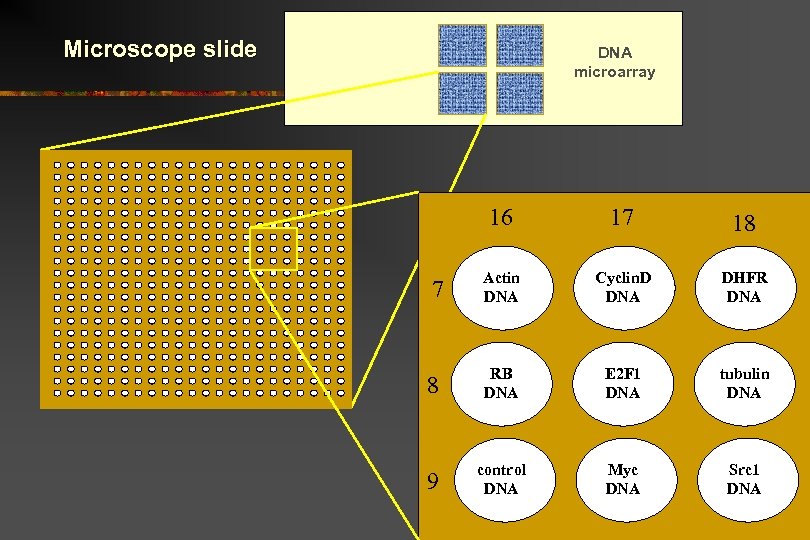 Microscope slide DNA microarray 16 17 18 7 Actin DNA Cyclin. D DNA DHFR DNA 8 RB DNA E 2 F 1 DNA tubulin DNA 9 control DNA Myc DNA Src 1 DNA
Microscope slide DNA microarray 16 17 18 7 Actin DNA Cyclin. D DNA DHFR DNA 8 RB DNA E 2 F 1 DNA tubulin DNA 9 control DNA Myc DNA Src 1 DNA
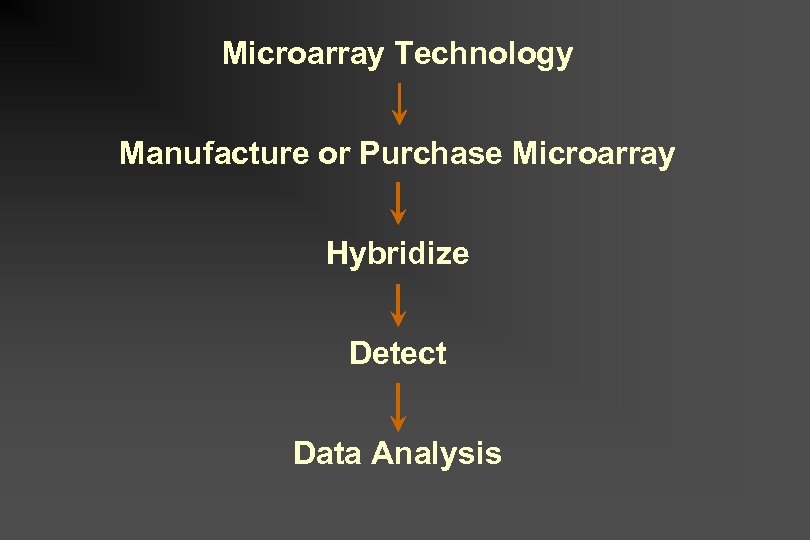 Microarray Technology Manufacture or Purchase Microarray Hybridize Detect Data Analysis
Microarray Technology Manufacture or Purchase Microarray Hybridize Detect Data Analysis
 Advantages of Microarrays n Small volume deposition (n. L) n Minimal wasted reagents n Access many genes / proteins simultaneously n Can be automated n Potentially quantitative
Advantages of Microarrays n Small volume deposition (n. L) n Minimal wasted reagents n Access many genes / proteins simultaneously n Can be automated n Potentially quantitative
 Limitations of Microarrays n Relatively new technology (10 years old) n Still has technical problems (background) n Poor reproducibility between investigators n Still mostly manual procedure n Relatively expensive
Limitations of Microarrays n Relatively new technology (10 years old) n Still has technical problems (background) n Poor reproducibility between investigators n Still mostly manual procedure n Relatively expensive
 Applications of Microarrays n Gene expression patterns n Single nucleotide polymorphism (SNP) detection n Sequence by hybridization / genotyping / mutation detection n Study protein expression (multianalyte assay) n Protein-protein interactions Provides: Massive parallel information
Applications of Microarrays n Gene expression patterns n Single nucleotide polymorphism (SNP) detection n Sequence by hybridization / genotyping / mutation detection n Study protein expression (multianalyte assay) n Protein-protein interactions Provides: Massive parallel information
 If Microarrays Are So Good Why Didn’t We Use Them Before? ? n n Not all genes were available No SNPs known No suitable bioinformatics New proteins now becoming available Microarrays and associated technologies should be regarded as by-products of the Human Genome Initiative, Nanotechnology and Bioinformatics
If Microarrays Are So Good Why Didn’t We Use Them Before? ? n n Not all genes were available No SNPs known No suitable bioinformatics New proteins now becoming available Microarrays and associated technologies should be regarded as by-products of the Human Genome Initiative, Nanotechnology and Bioinformatics
 Reviews on Microarrays n A whole issue on Microarray Technology has been published by Nature Genetics, Dec. 2002 (Vol. 32) n Books: n Bowtell D. Sambrook J. DNA Microarrays. Cold Spring Harbor Laboratory Press, 2003 n Schena M. Microarray Analysis. Wiley Liss, 2003
Reviews on Microarrays n A whole issue on Microarray Technology has been published by Nature Genetics, Dec. 2002 (Vol. 32) n Books: n Bowtell D. Sambrook J. DNA Microarrays. Cold Spring Harbor Laboratory Press, 2003 n Schena M. Microarray Analysis. Wiley Liss, 2003
 History 1991 - Photolithographic printing (Affymetrix) 1994 - First c. DNA collections are developed at Stanford. 1995 - Quantitative monitoring of gene expression patterns with a complementary DNA microarray 1996 - Commercialization of arrays (Affymetrix) 1997 - Genome-wide expression monitoring in S. cerevisiae (yeast) 2000 – Portraits/Signatures of cancer 2003 - Introduction to clinical practice 2004 -Whole human genome on one microarray
History 1991 - Photolithographic printing (Affymetrix) 1994 - First c. DNA collections are developed at Stanford. 1995 - Quantitative monitoring of gene expression patterns with a complementary DNA microarray 1996 - Commercialization of arrays (Affymetrix) 1997 - Genome-wide expression monitoring in S. cerevisiae (yeast) 2000 – Portraits/Signatures of cancer 2003 - Introduction to clinical practice 2004 -Whole human genome on one microarray
![Microarray Fabrication Two Major Methods: [a] Affymetrix Photolithography (400, 000 spots in 1. 25 Microarray Fabrication Two Major Methods: [a] Affymetrix Photolithography (400, 000 spots in 1. 25](https://present5.com/presentation/9b0095e5cfbbaf14210dcce13cf17641/image-11.jpg) Microarray Fabrication Two Major Methods: [a] Affymetrix Photolithography (400, 000 spots in 1. 25 x 1. 25 cm area!) [b] Everybody else Mechanical deposition (printing) [0. 5 - 2 n. L] on glass slides, membranes, etc
Microarray Fabrication Two Major Methods: [a] Affymetrix Photolithography (400, 000 spots in 1. 25 x 1. 25 cm area!) [b] Everybody else Mechanical deposition (printing) [0. 5 - 2 n. L] on glass slides, membranes, etc
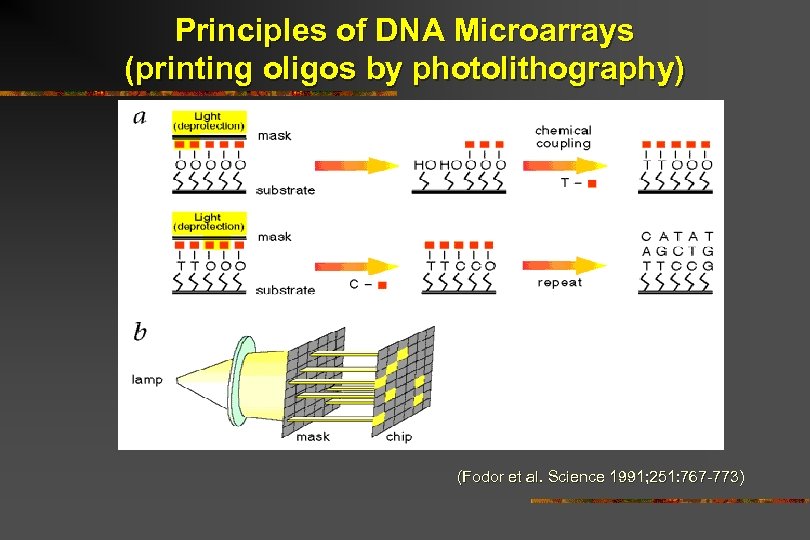 Principles of DNA Microarrays (printing oligos by photolithography) (Fodor et al. Science 1991; 251: 767 -773)
Principles of DNA Microarrays (printing oligos by photolithography) (Fodor et al. Science 1991; 251: 767 -773)
 Microarrays, such as Affymetrix’s Gene. Chip, now include all 50, 000 known human genes. Science, 302: 211, 10 October, 2003
Microarrays, such as Affymetrix’s Gene. Chip, now include all 50, 000 known human genes. Science, 302: 211, 10 October, 2003
 Affymetrix Expression Arrays n n They immobilize oligonucleotides (de novo synthesis; 25 mers) For specificity and sensitivity, they array 22 oligos per gene n Latest version covers 50, 000 genes (whole human genome) in one array (Agilent Technologies has the same density array; G 4112 A) n They label-test RNA with biotin and detect with streptavidinfluor conjugates
Affymetrix Expression Arrays n n They immobilize oligonucleotides (de novo synthesis; 25 mers) For specificity and sensitivity, they array 22 oligos per gene n Latest version covers 50, 000 genes (whole human genome) in one array (Agilent Technologies has the same density array; G 4112 A) n They label-test RNA with biotin and detect with streptavidinfluor conjugates
 Preparation of Labeled m. RNA for Hybridization n Use oligo-d. T with a T 7 RNA polymerase promoter for reverse transcription of extracted m. RNA (procedure makes c. DNA) n Use T 7 RNA polymerase and biotin-labeled ribonucleotides for in vitro transcription (produces biotinylated, single-stranded c. RNA) n Alternatively: You can directly label c. RNA with Cy-3 and Cy-5 fluors using T 7 RNA polymerase
Preparation of Labeled m. RNA for Hybridization n Use oligo-d. T with a T 7 RNA polymerase promoter for reverse transcription of extracted m. RNA (procedure makes c. DNA) n Use T 7 RNA polymerase and biotin-labeled ribonucleotides for in vitro transcription (produces biotinylated, single-stranded c. RNA) n Alternatively: You can directly label c. RNA with Cy-3 and Cy-5 fluors using T 7 RNA polymerase
 Microarray Applications Differential Gene Expression
Microarray Applications Differential Gene Expression
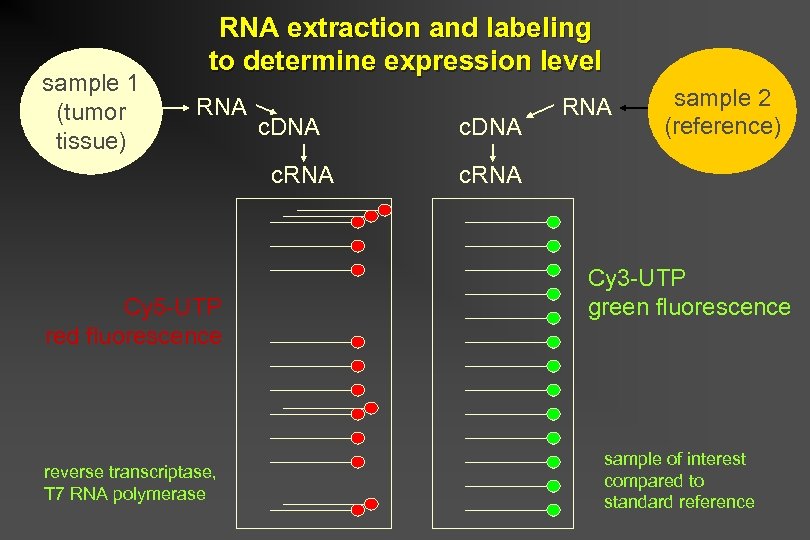 sample 1 (tumor tissue) RNA extraction and labeling to determine expression level RNA c. DNA c. RNA Cy 5 -UTP red fluorescence reverse transcriptase, T 7 RNA polymerase c. DNA RNA sample 2 (reference) c. RNA Cy 3 -UTP green fluorescence sample of interest compared to standard reference
sample 1 (tumor tissue) RNA extraction and labeling to determine expression level RNA c. DNA c. RNA Cy 5 -UTP red fluorescence reverse transcriptase, T 7 RNA polymerase c. DNA RNA sample 2 (reference) c. RNA Cy 3 -UTP green fluorescence sample of interest compared to standard reference
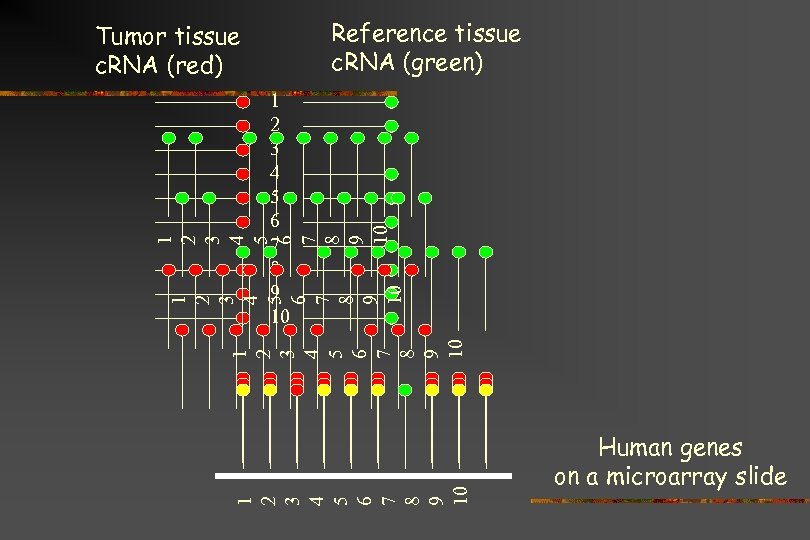 Reference tissue c. RNA (green) Tumor tissue c. RNA (red) 1 2 3 4 5 6 7 8 9 10 1 2 3 4 5 6 7 8 9 10 Human genes on a microarray slide
Reference tissue c. RNA (green) Tumor tissue c. RNA (red) 1 2 3 4 5 6 7 8 9 10 1 2 3 4 5 6 7 8 9 10 Human genes on a microarray slide
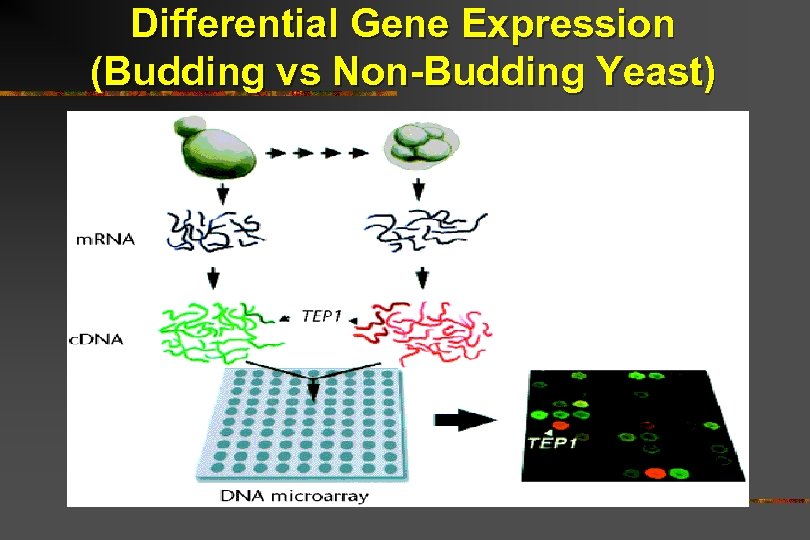 Differential Gene Expression (Budding vs Non-Budding Yeast)
Differential Gene Expression (Budding vs Non-Budding Yeast)
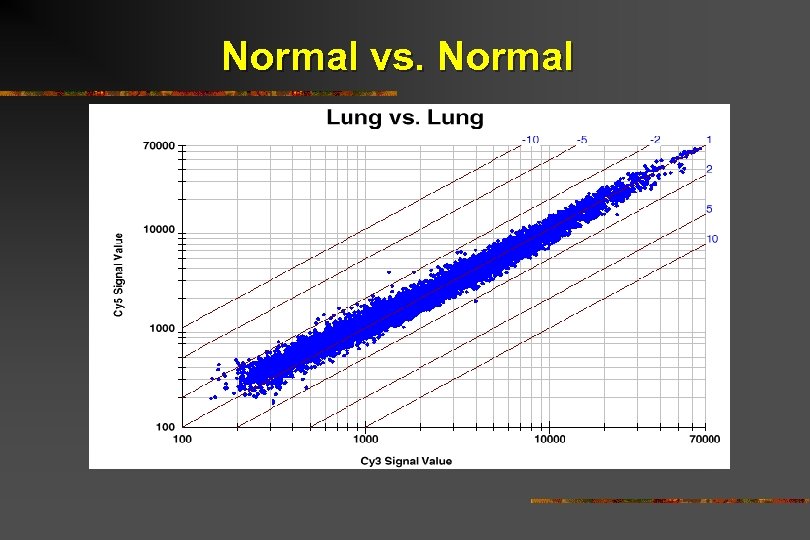 Normal vs. Normal
Normal vs. Normal
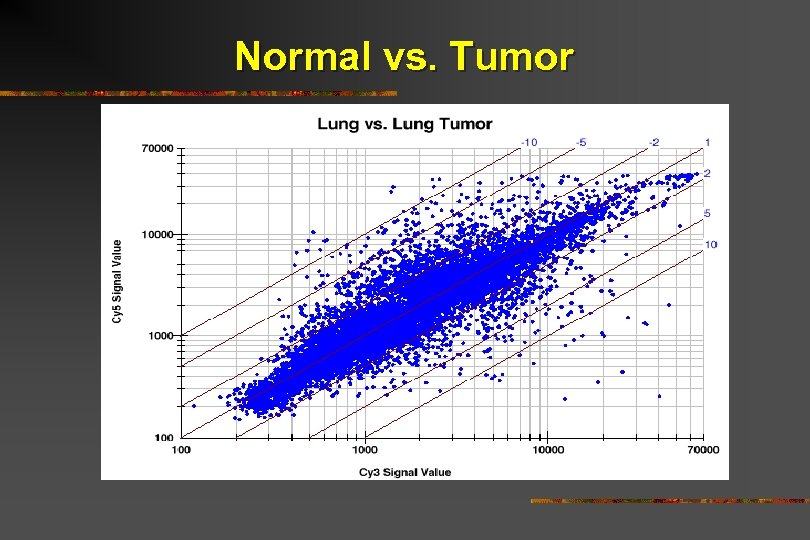 Normal vs. Tumor
Normal vs. Tumor
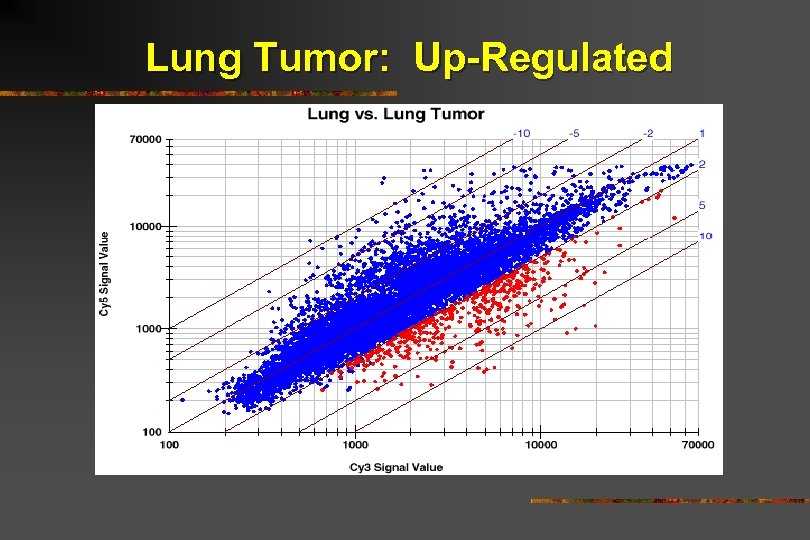 Lung Tumor: Up-Regulated
Lung Tumor: Up-Regulated
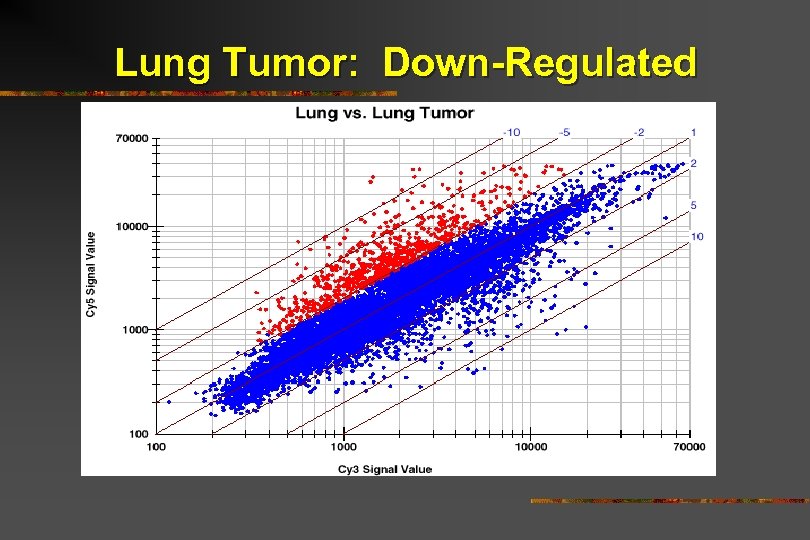 Lung Tumor: Down-Regulated
Lung Tumor: Down-Regulated
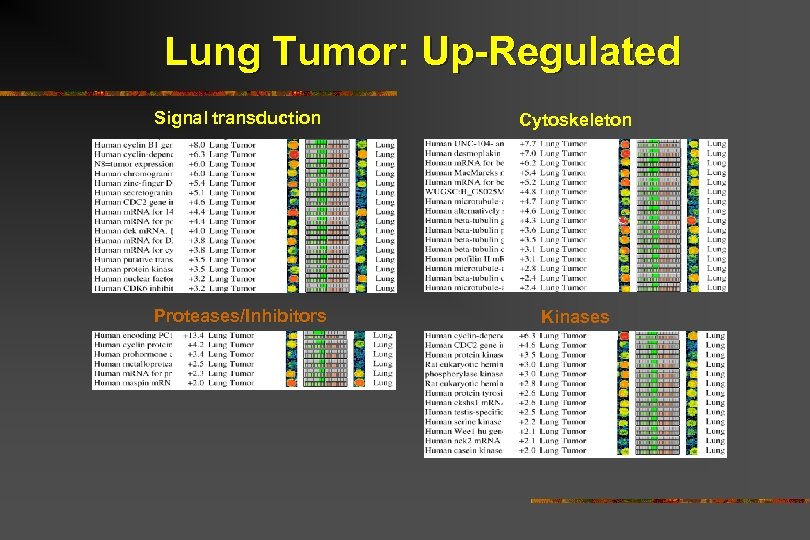 Lung Tumor: Up-Regulated Signal transduction Cytoskeleton Proteases/Inhibitors Kinases
Lung Tumor: Up-Regulated Signal transduction Cytoskeleton Proteases/Inhibitors Kinases
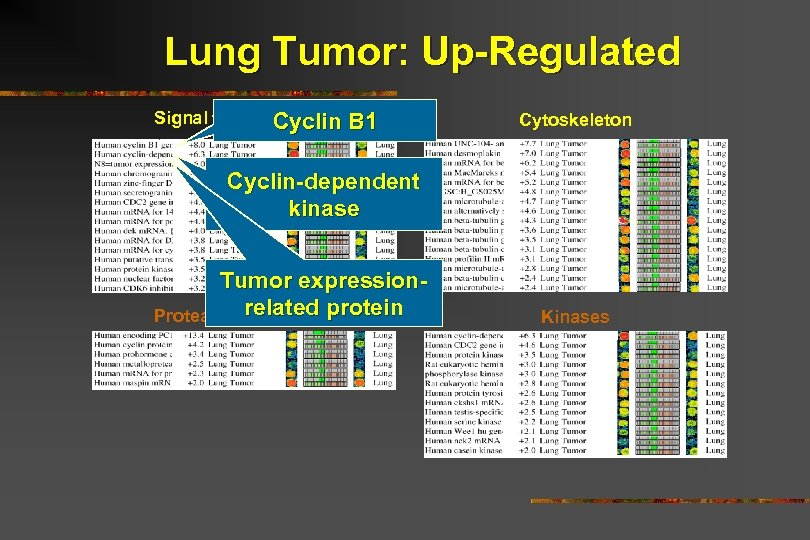 Lung Tumor: Up-Regulated Signal transduction Cyclin B 1 Cytoskeleton Cyclin-dependent kinase Tumor expressionrelated Proteases/Inhibitorsprotein Kinases
Lung Tumor: Up-Regulated Signal transduction Cyclin B 1 Cytoskeleton Cyclin-dependent kinase Tumor expressionrelated Proteases/Inhibitorsprotein Kinases
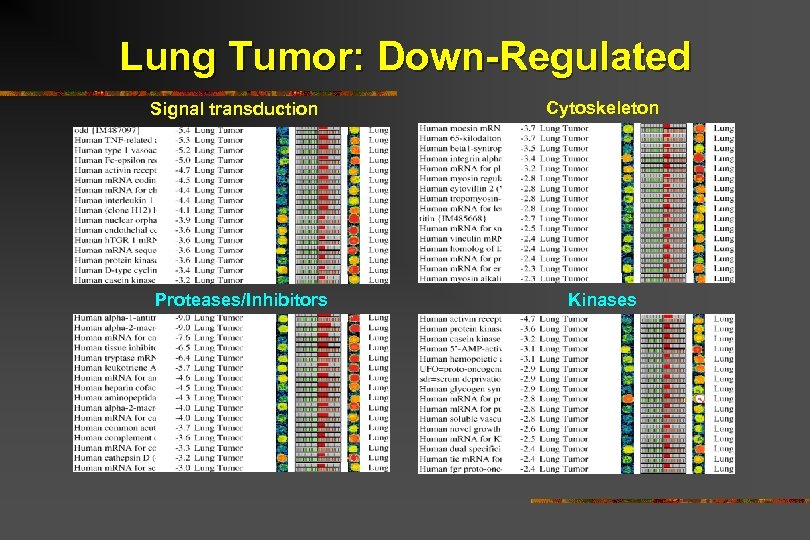 Lung Tumor: Down-Regulated Signal transduction Proteases/Inhibitors Cytoskeleton Kinases
Lung Tumor: Down-Regulated Signal transduction Proteases/Inhibitors Cytoskeleton Kinases
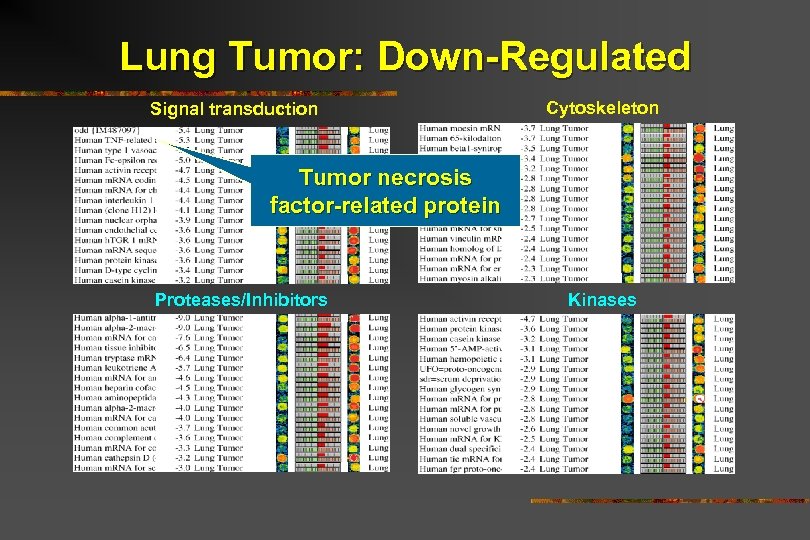 Lung Tumor: Down-Regulated Signal transduction Cytoskeleton Tumor necrosis factor-related protein Proteases/Inhibitors Kinases
Lung Tumor: Down-Regulated Signal transduction Cytoskeleton Tumor necrosis factor-related protein Proteases/Inhibitors Kinases
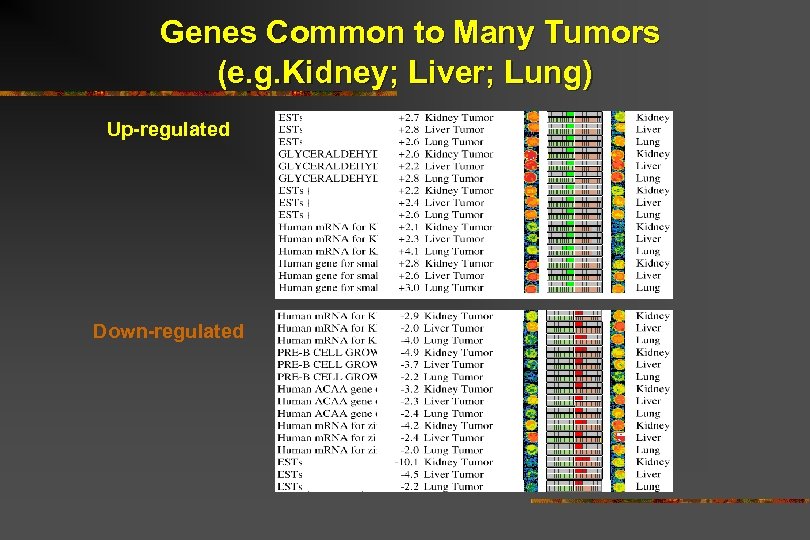 Genes Common to Many Tumors (e. g. Kidney; Liver; Lung) Up-regulated Down-regulated
Genes Common to Many Tumors (e. g. Kidney; Liver; Lung) Up-regulated Down-regulated
 Microarray Applications Whole Organism Biology
Microarray Applications Whole Organism Biology
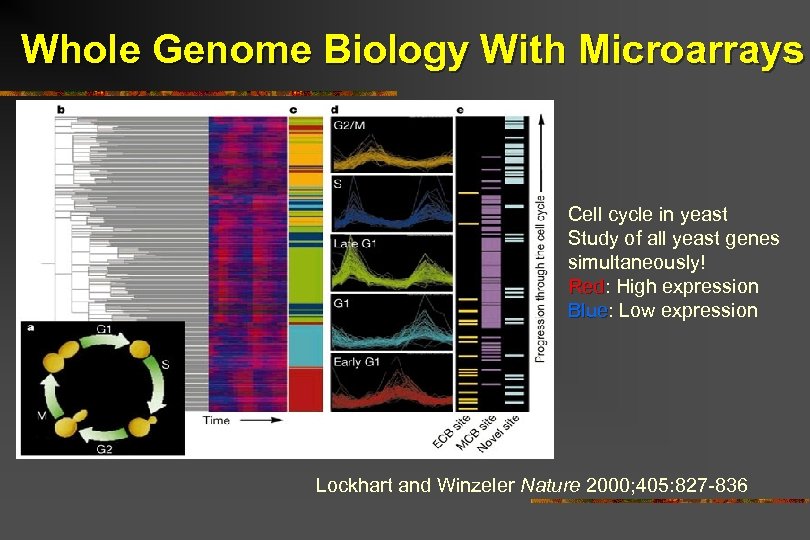 Whole Genome Biology With Microarrays Cell cycle in yeast Study of all yeast genes simultaneously! Red: High expression Red Blue: Low expression Blue Lockhart and Winzeler Nature 2000; 405: 827 -836
Whole Genome Biology With Microarrays Cell cycle in yeast Study of all yeast genes simultaneously! Red: High expression Red Blue: Low expression Blue Lockhart and Winzeler Nature 2000; 405: 827 -836
 Microarray Applications Single Nucleotide Polymorphism (SNP) Analysis
Microarray Applications Single Nucleotide Polymorphism (SNP) Analysis
 Single Nucleotide Polymorphisms (SNP) n DNA variation at one base pair level; found at a frequency of 1 SNP per 1, 000 - 2, 000 bases n A map of 9 x 106 SNPs has been described in humans (by the International SNP map working group (Hap. Map) n 60, 000 SNPs fall within exons; the rest are in introns
Single Nucleotide Polymorphisms (SNP) n DNA variation at one base pair level; found at a frequency of 1 SNP per 1, 000 - 2, 000 bases n A map of 9 x 106 SNPs has been described in humans (by the International SNP map working group (Hap. Map) n 60, 000 SNPs fall within exons; the rest are in introns
 Why Are SNPs Useful? n Human genetic diversity depends on SNPs between individuals (these are our major genetic differences, plus micro/minisatellites) n Specific combinations of alleles (called “Haplotypes”) seem to play a major role in our genetic diversity n How does this genotype affect the phenotype Disease predisposition?
Why Are SNPs Useful? n Human genetic diversity depends on SNPs between individuals (these are our major genetic differences, plus micro/minisatellites) n Specific combinations of alleles (called “Haplotypes”) seem to play a major role in our genetic diversity n How does this genotype affect the phenotype Disease predisposition?
 Why Are SNPs Useful? n Diagnostic Application Determine somebody’s haplotype (sets of SNPs) and assess disease risk. n Be careful: These disease-related haplotypes are not as yet known!
Why Are SNPs Useful? n Diagnostic Application Determine somebody’s haplotype (sets of SNPs) and assess disease risk. n Be careful: These disease-related haplotypes are not as yet known!
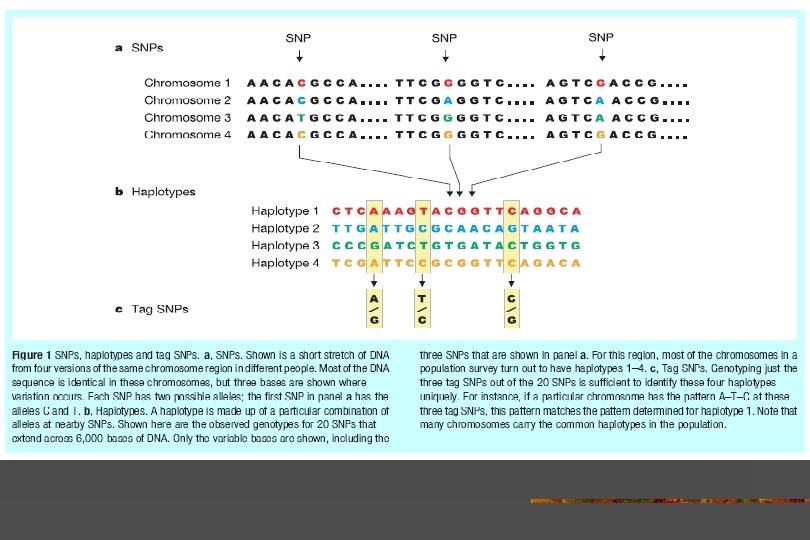
 Nature 2003 426: 789 -796
Nature 2003 426: 789 -796
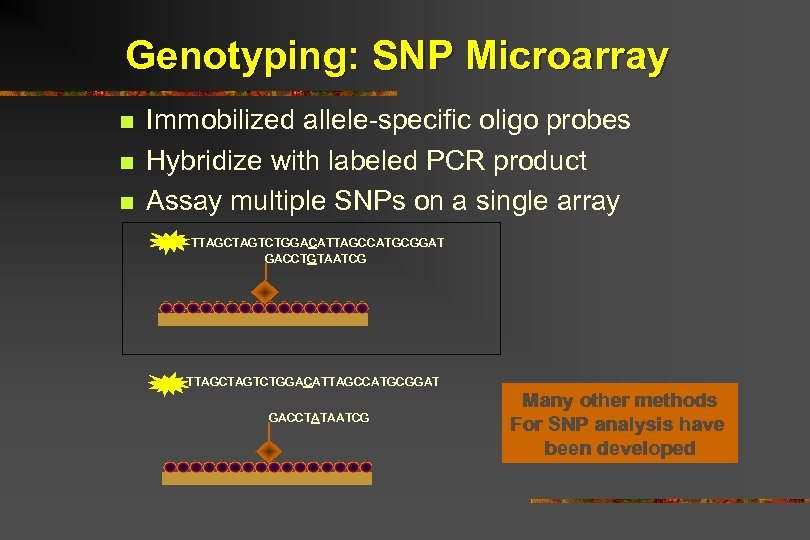 Genotyping: SNP Microarray n n n Immobilized allele-specific oligo probes Hybridize with labeled PCR product Assay multiple SNPs on a single array TTAGCTAGTCTGGACATTAGCCATGCGGAT GACCTGTAATCG TTAGCTAGTCTGGACATTAGCCATGCGGAT GACCTATAATCG Many other methods For SNP analysis have been developed
Genotyping: SNP Microarray n n n Immobilized allele-specific oligo probes Hybridize with labeled PCR product Assay multiple SNPs on a single array TTAGCTAGTCTGGACATTAGCCATGCGGAT GACCTGTAATCG TTAGCTAGTCTGGACATTAGCCATGCGGAT GACCTATAATCG Many other methods For SNP analysis have been developed
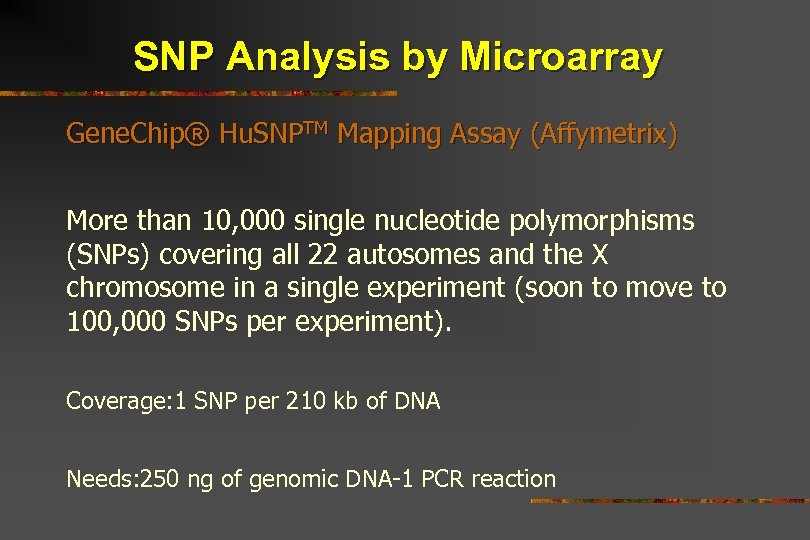 SNP Analysis by Microarray Gene. Chip® Hu. SNPTM Mapping Assay (Affymetrix) More than 10, 000 single nucleotide polymorphisms (SNPs) covering all 22 autosomes and the X chromosome in a single experiment (soon to move to 100, 000 SNPs per experiment). Coverage: 1 SNP per 210 kb of DNA Needs: 250 ng of genomic DNA-1 PCR reaction
SNP Analysis by Microarray Gene. Chip® Hu. SNPTM Mapping Assay (Affymetrix) More than 10, 000 single nucleotide polymorphisms (SNPs) covering all 22 autosomes and the X chromosome in a single experiment (soon to move to 100, 000 SNPs per experiment). Coverage: 1 SNP per 210 kb of DNA Needs: 250 ng of genomic DNA-1 PCR reaction
 Commercial Microarray for Clinical Use (Pharmacogenomics) Roche Product CYP 450 Genotyping (drug metabolizing system) FDA Confusion Class 1 medical device? (no PMA) Class 2 or 3 medical device? (requires pre-market approval) From: Nature Biotechnology 2003 21: 959 -60
Commercial Microarray for Clinical Use (Pharmacogenomics) Roche Product CYP 450 Genotyping (drug metabolizing system) FDA Confusion Class 1 medical device? (no PMA) Class 2 or 3 medical device? (requires pre-market approval) From: Nature Biotechnology 2003 21: 959 -60
 “The US government has blocked the sale of a new kind of DNA diagnostic test, putting up an unexpected barrier to the marketing of technology to distinguish genetic differences in how patients metabolize certain drugs. ” Science 2003 302: 1134
“The US government has blocked the sale of a new kind of DNA diagnostic test, putting up an unexpected barrier to the marketing of technology to distinguish genetic differences in how patients metabolize certain drugs. ” Science 2003 302: 1134
 SNP Detection by Mass Spectrometry n High throughput detection of SNPs can be achieved by mass spectrometry n SNP Center in Toronto (PMH) runs a Sequenom Mass Spectrometry system
SNP Detection by Mass Spectrometry n High throughput detection of SNPs can be achieved by mass spectrometry n SNP Center in Toronto (PMH) runs a Sequenom Mass Spectrometry system
 Microarray Applications Sequencing by Hybridization
Microarray Applications Sequencing by Hybridization
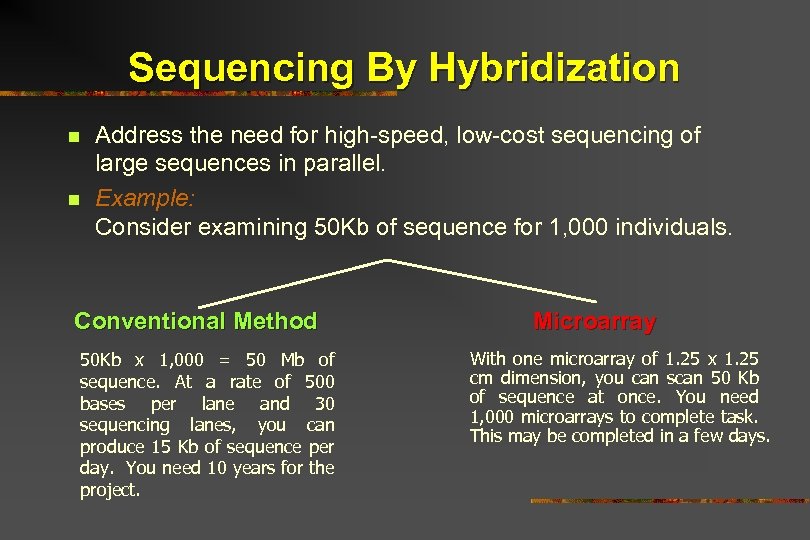 Sequencing By Hybridization n n Address the need for high-speed, low-cost sequencing of large sequences in parallel. Example: Consider examining 50 Kb of sequence for 1, 000 individuals. Conventional Method 50 Kb x 1, 000 = 50 Mb of sequence. At a rate of 500 bases per lane and 30 sequencing lanes, you can produce 15 Kb of sequence per day. You need 10 years for the project. Microarray With one microarray of 1. 25 x 1. 25 cm dimension, you can scan 50 Kb of sequence at once. You need 1, 000 microarrays to complete task. This may be completed in a few days.
Sequencing By Hybridization n n Address the need for high-speed, low-cost sequencing of large sequences in parallel. Example: Consider examining 50 Kb of sequence for 1, 000 individuals. Conventional Method 50 Kb x 1, 000 = 50 Mb of sequence. At a rate of 500 bases per lane and 30 sequencing lanes, you can produce 15 Kb of sequence per day. You need 10 years for the project. Microarray With one microarray of 1. 25 x 1. 25 cm dimension, you can scan 50 Kb of sequence at once. You need 1, 000 microarrays to complete task. This may be completed in a few days.
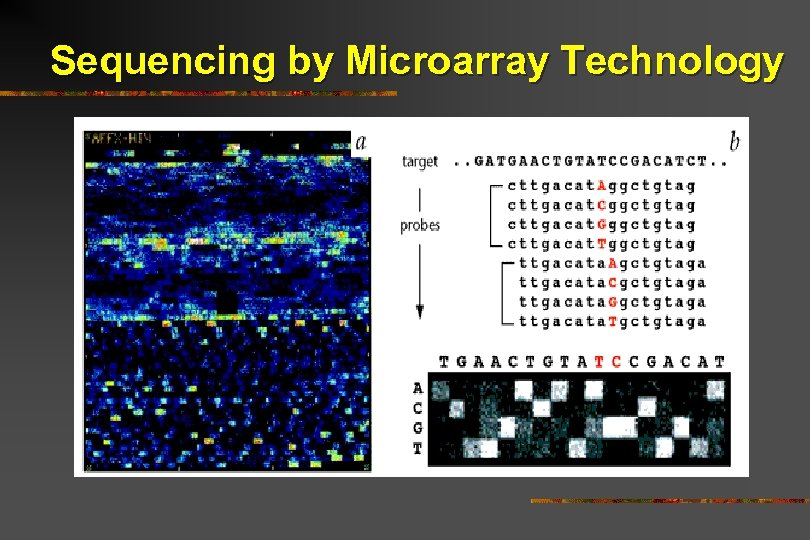 Sequencing by Microarray Technology
Sequencing by Microarray Technology
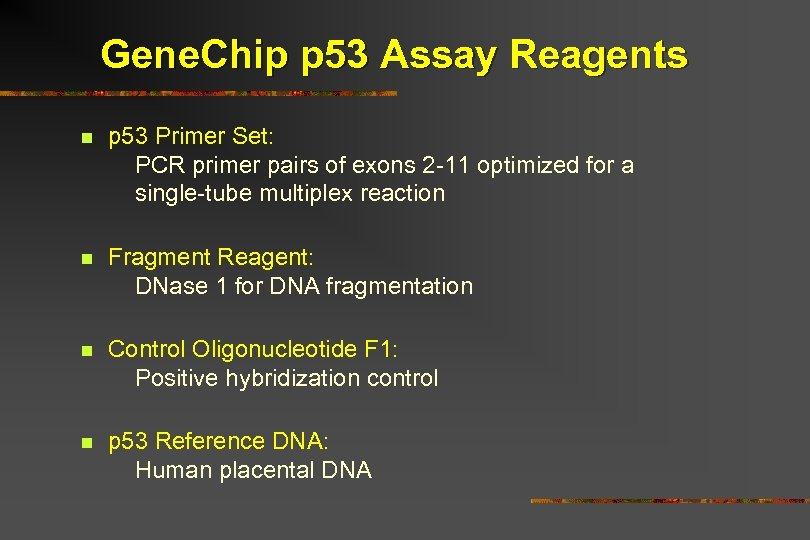 Gene. Chip p 53 Assay Reagents n p 53 Primer Set: PCR primer pairs of exons 2 -11 optimized for a single-tube multiplex reaction n Fragment Reagent: DNase 1 for DNA fragmentation n Control Oligonucleotide F 1: Positive hybridization control n p 53 Reference DNA: Human placental DNA
Gene. Chip p 53 Assay Reagents n p 53 Primer Set: PCR primer pairs of exons 2 -11 optimized for a single-tube multiplex reaction n Fragment Reagent: DNase 1 for DNA fragmentation n Control Oligonucleotide F 1: Positive hybridization control n p 53 Reference DNA: Human placental DNA
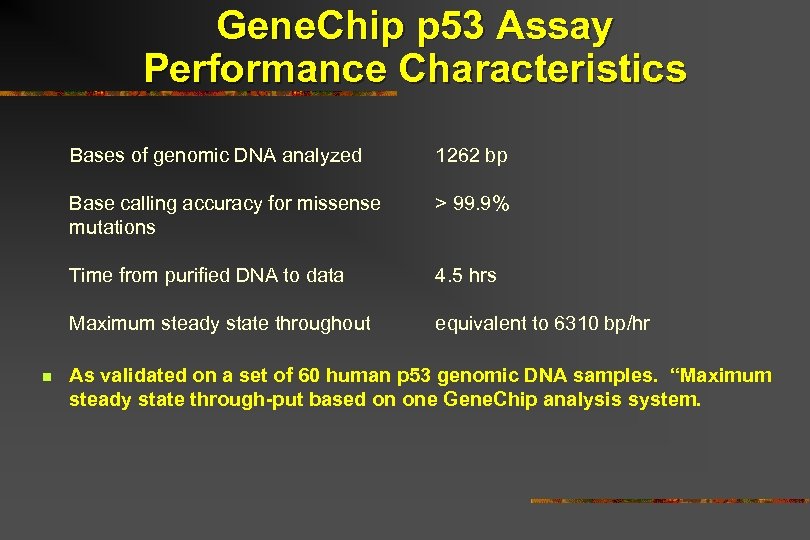 Gene. Chip p 53 Assay Performance Characteristics Bases of genomic DNA analyzed Base calling accuracy for missense mutations > 99. 9% Time from purified DNA to data 4. 5 hrs Maximum steady state throughout n 1262 bp equivalent to 6310 bp/hr As validated on a set of 60 human p 53 genomic DNA samples. “Maximum steady state through-put based on one Gene. Chip analysis system.
Gene. Chip p 53 Assay Performance Characteristics Bases of genomic DNA analyzed Base calling accuracy for missense mutations > 99. 9% Time from purified DNA to data 4. 5 hrs Maximum steady state throughout n 1262 bp equivalent to 6310 bp/hr As validated on a set of 60 human p 53 genomic DNA samples. “Maximum steady state through-put based on one Gene. Chip analysis system.
 Microarray Applications-Non Human - Chips Avaliable Now (2004) n Pathogens (detection of Bird-Flu Virus strains) n Smallpox (bioterrorism) n Malaria (Plasmodium anopheles) n Zebrafish/Xenopus laevis (model organisms) n SARS Virus sequencing
Microarray Applications-Non Human - Chips Avaliable Now (2004) n Pathogens (detection of Bird-Flu Virus strains) n Smallpox (bioterrorism) n Malaria (Plasmodium anopheles) n Zebrafish/Xenopus laevis (model organisms) n SARS Virus sequencing
 Microarray Applications n Food Expert-ID (available by Bio-Merieux; 2004) n DNA chip can verify quickly the animal species composition and the authenticity of raw or processed food animal feed n By providing multi-species identification, Food. Expert-ID will help to improve safety of food for human and animal consumption, thereby contributing to consumer health protection
Microarray Applications n Food Expert-ID (available by Bio-Merieux; 2004) n DNA chip can verify quickly the animal species composition and the authenticity of raw or processed food animal feed n By providing multi-species identification, Food. Expert-ID will help to improve safety of food for human and animal consumption, thereby contributing to consumer health protection
 Microarray Applications Protein Microarrays
Microarray Applications Protein Microarrays
 Protein Microarrays n Protein microarrays are lagging behind DNA microarrays n Same idea but immobilized elements are proteins instead of nucleic acids n Number of elements (proteins) on current protein microarrays are limited (approx. 500) n Antibodies for high density microarrays have limitations (crossreactivities) n Aptamers or engineered antibodies/proteins may be viable alternatives (Aptamers: RNAs that bind proteins with high specificity and affinity)
Protein Microarrays n Protein microarrays are lagging behind DNA microarrays n Same idea but immobilized elements are proteins instead of nucleic acids n Number of elements (proteins) on current protein microarrays are limited (approx. 500) n Antibodies for high density microarrays have limitations (crossreactivities) n Aptamers or engineered antibodies/proteins may be viable alternatives (Aptamers: RNAs that bind proteins with high specificity and affinity)
 Applications Screening for: n Small molecule targets n Post-translational modifications n Protein-protein interactions n Protein-DNA interactions n Enzyme assays n Epitope mapping
Applications Screening for: n Small molecule targets n Post-translational modifications n Protein-protein interactions n Protein-DNA interactions n Enzyme assays n Epitope mapping
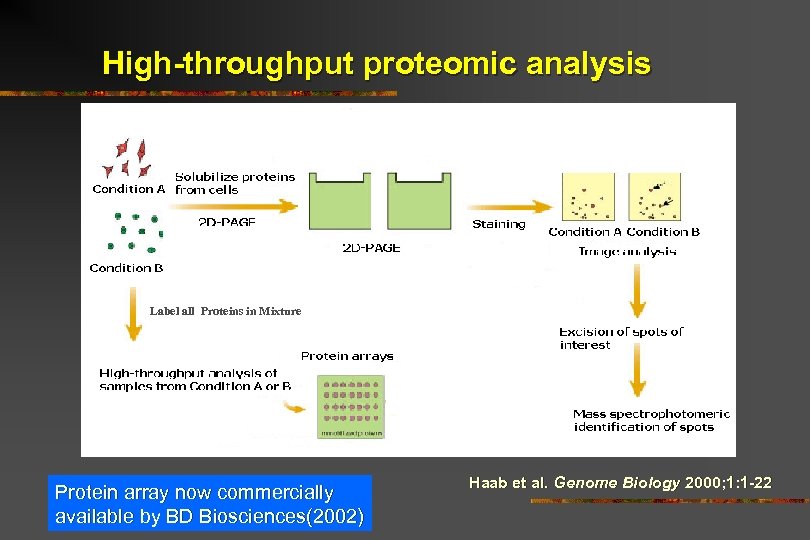 High-throughput proteomic analysis Label all Proteins in Mixture Protein array now commercially available by BD Biosciences(2002) Haab et al. Genome Biology 2000; 1: 1 -22
High-throughput proteomic analysis Label all Proteins in Mixture Protein array now commercially available by BD Biosciences(2002) Haab et al. Genome Biology 2000; 1: 1 -22
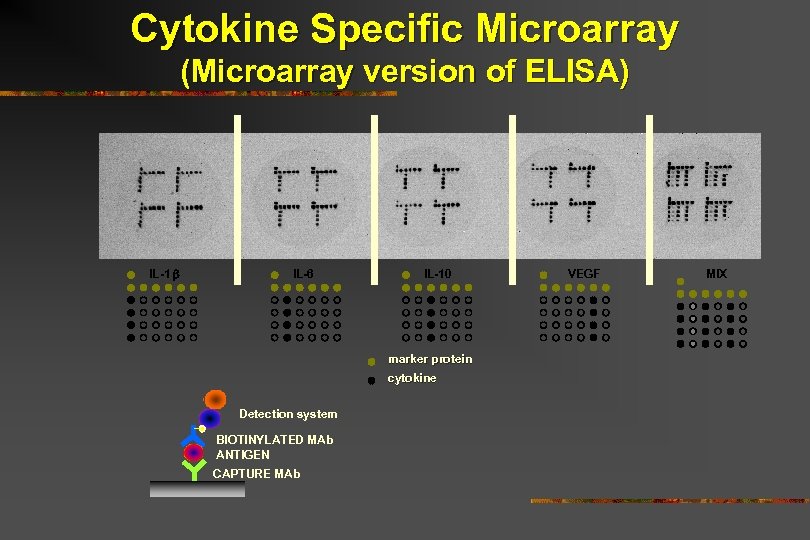 Cytokine Specific Microarray (Microarray version of ELISA) IL-1 IL-6 IL-10 marker protein cytokine Detection system BIOTINYLATED MAb ANTIGEN CAPTURE MAb VEGF MIX
Cytokine Specific Microarray (Microarray version of ELISA) IL-1 IL-6 IL-10 marker protein cytokine Detection system BIOTINYLATED MAb ANTIGEN CAPTURE MAb VEGF MIX
 Competing High Throughput Protein Technologies Bead-Based Technologies n Luminex-flow cytometry n Illumina-bead chips Microfluidics n Zyomyx Mass spectrometry n Ciphergen-protein chips
Competing High Throughput Protein Technologies Bead-Based Technologies n Luminex-flow cytometry n Illumina-bead chips Microfluidics n Zyomyx Mass spectrometry n Ciphergen-protein chips
 Microarray Clinical Applications Cancer Diagnostics
Microarray Clinical Applications Cancer Diagnostics
 Molecular Portraits of Cancer Rationale: The phenotypic diversity of breast and other tumors might be accompanied by a corresponding diversity in gene expression patterns that can be captured by using c. DNA microarrays Then Systematic investigation of gene expression patterns in human tumors might provide the basis of an improved taxonomy of breast cancers Perou et al. Nature 2000; 406: 747 -752
Molecular Portraits of Cancer Rationale: The phenotypic diversity of breast and other tumors might be accompanied by a corresponding diversity in gene expression patterns that can be captured by using c. DNA microarrays Then Systematic investigation of gene expression patterns in human tumors might provide the basis of an improved taxonomy of breast cancers Perou et al. Nature 2000; 406: 747 -752
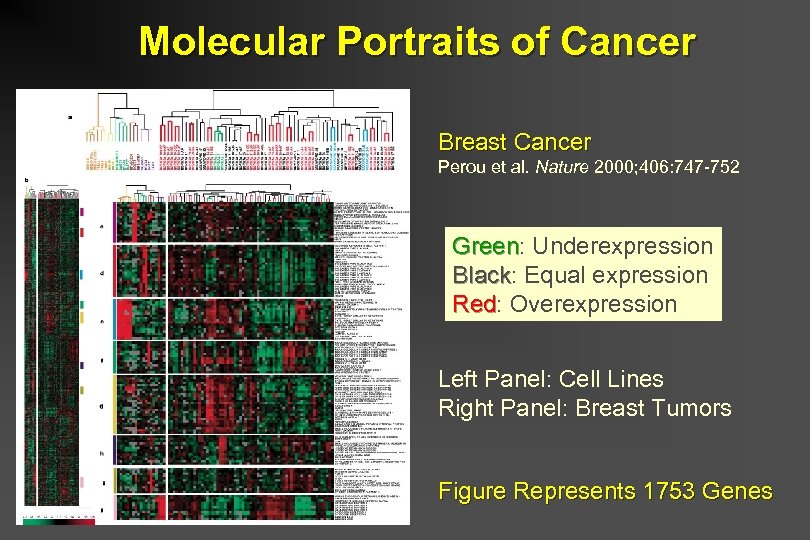 Molecular Portraits of Cancer Breast Cancer Perou et al. Nature 2000; 406: 747 -752 Green: Underexpression Green Black: Equal expression Black Red: Overexpression Red Left Panel: Cell Lines Right Panel: Breast Tumors Figure Represents 1753 Genes
Molecular Portraits of Cancer Breast Cancer Perou et al. Nature 2000; 406: 747 -752 Green: Underexpression Green Black: Equal expression Black Red: Overexpression Red Left Panel: Cell Lines Right Panel: Breast Tumors Figure Represents 1753 Genes
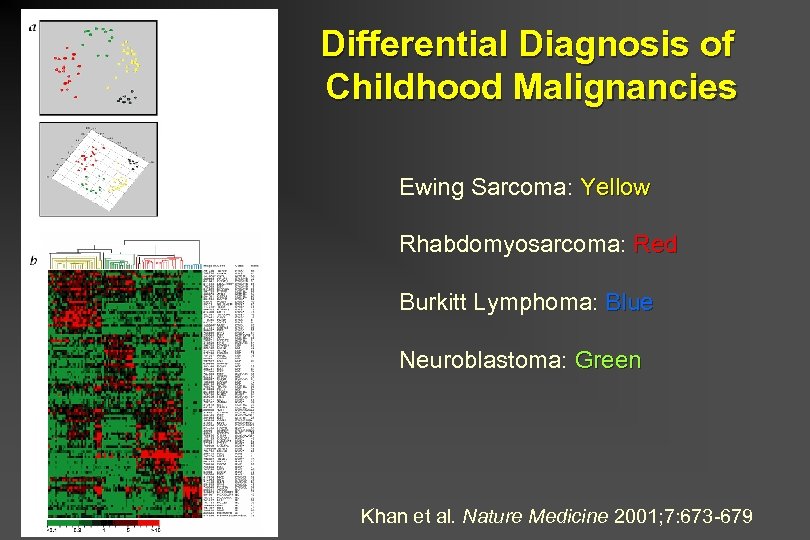 Differential Diagnosis of Childhood Malignancies Ewing Sarcoma: Yellow Rhabdomyosarcoma: Red Burkitt Lymphoma: Blue Neuroblastoma: Green Khan et al. Nature Medicine 2001; 7: 673 -679
Differential Diagnosis of Childhood Malignancies Ewing Sarcoma: Yellow Rhabdomyosarcoma: Red Burkitt Lymphoma: Blue Neuroblastoma: Green Khan et al. Nature Medicine 2001; 7: 673 -679
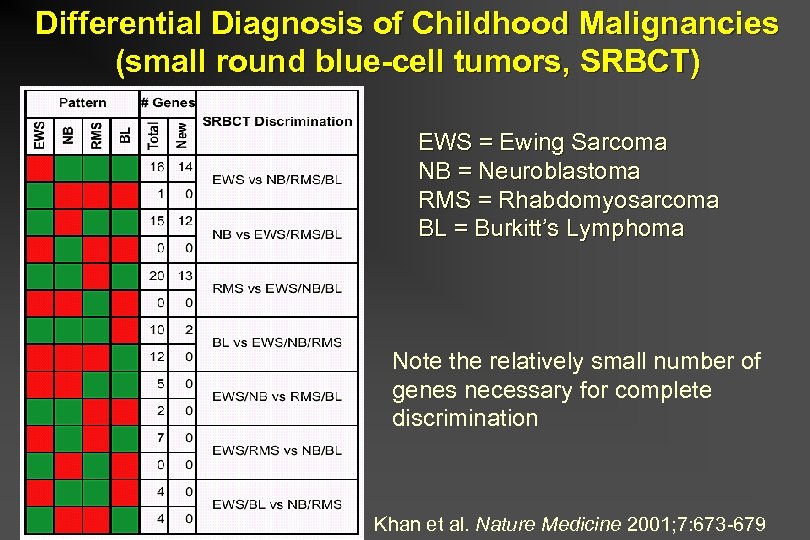 Differential Diagnosis of Childhood Malignancies (small round blue-cell tumors, SRBCT) EWS = Ewing Sarcoma NB = Neuroblastoma RMS = Rhabdomyosarcoma BL = Burkitt’s Lymphoma Note the relatively small number of genes necessary for complete discrimination Khan et al. Nature Medicine 2001; 7: 673 -679
Differential Diagnosis of Childhood Malignancies (small round blue-cell tumors, SRBCT) EWS = Ewing Sarcoma NB = Neuroblastoma RMS = Rhabdomyosarcoma BL = Burkitt’s Lymphoma Note the relatively small number of genes necessary for complete discrimination Khan et al. Nature Medicine 2001; 7: 673 -679
 Microarray Milestone: June 2003 Question: Can microarray profiling be used in clinical practice? Prognosis/Prediction of therapy/Selection of patients who should be treated aggressively? Nature 2002; 415: 530 -536 NEJM 2002; 347: 1999 -2009 Van’t Veer and colleagues are using microarray profiling as a routine tool for breast cancer management (administration of adjuvant chemotherapy after surgery). Their profile is based on expression of 70 genes
Microarray Milestone: June 2003 Question: Can microarray profiling be used in clinical practice? Prognosis/Prediction of therapy/Selection of patients who should be treated aggressively? Nature 2002; 415: 530 -536 NEJM 2002; 347: 1999 -2009 Van’t Veer and colleagues are using microarray profiling as a routine tool for breast cancer management (administration of adjuvant chemotherapy after surgery). Their profile is based on expression of 70 genes
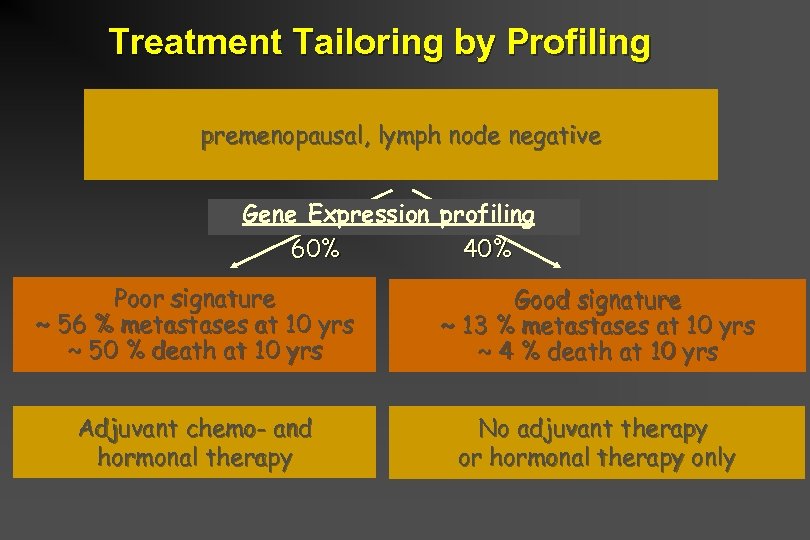 Treatment Tailoring by Profiling premenopausal, lymph node negative Gene Expression profiling 60% 40% Poor signature ~ 56 % metastases at 10 yrs ~ 50 % death at 10 yrs Good signature ~ 13 % metastases at 10 yrs ~ 4 % death at 10 yrs Adjuvant chemo- and hormonal therapy No adjuvant therapy or hormonal therapy only
Treatment Tailoring by Profiling premenopausal, lymph node negative Gene Expression profiling 60% 40% Poor signature ~ 56 % metastases at 10 yrs ~ 50 % death at 10 yrs Good signature ~ 13 % metastases at 10 yrs ~ 4 % death at 10 yrs Adjuvant chemo- and hormonal therapy No adjuvant therapy or hormonal therapy only
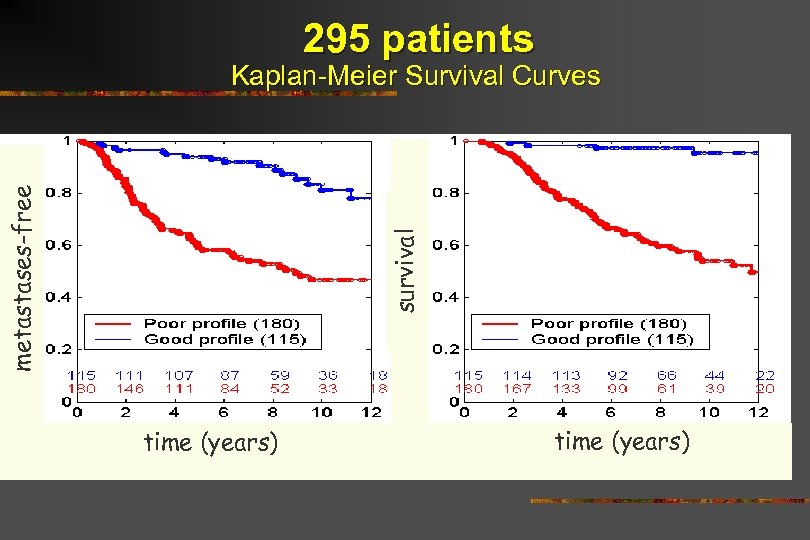 295 patients survival metastases-free Kaplan-Meier Survival Curves time (years)
295 patients survival metastases-free Kaplan-Meier Survival Curves time (years)
 Profiling in Clinical Practice n Metastatic potential is an early and inherent ability rather than late and acquired n Predictive power of prognostic signature confirmed in validation series n Prognostic profile outperforms clinical parameters n ~30 -40% reduction of unnecessary treatment and avoidance of undertreatment (LN 0 and LN+)
Profiling in Clinical Practice n Metastatic potential is an early and inherent ability rather than late and acquired n Predictive power of prognostic signature confirmed in validation series n Prognostic profile outperforms clinical parameters n ~30 -40% reduction of unnecessary treatment and avoidance of undertreatment (LN 0 and LN+)
 Therapeutic Implications n Who to treat: n Prognostic profile as diagnostic tool n n Prognostic profile implemented in clinical trials n n improvement of accurate selection for adjuvant therapy (less under- and over-treatment) reduction in number of patients & costs (select only patients that are at metastatic risk) How to treat: n Predictive profile for drug response n selection of patients who benefit
Therapeutic Implications n Who to treat: n Prognostic profile as diagnostic tool n n Prognostic profile implemented in clinical trials n n improvement of accurate selection for adjuvant therapy (less under- and over-treatment) reduction in number of patients & costs (select only patients that are at metastatic risk) How to treat: n Predictive profile for drug response n selection of patients who benefit
 Commercial Clashes n n n Oncotype DX by “Genomic Health Inc”, Redwood City, CA A prognostic test for breast cancer metastasis based on profiling 250 genes; 16 genes as a group have predictive value; $3, 400 per test 215, 000 breast cancer cases per year (potential market value > $500 million!) No validation of test; No FDA approval Test has no value for predicting response to treatment Science 2004; 303: 1754 -5
Commercial Clashes n n n Oncotype DX by “Genomic Health Inc”, Redwood City, CA A prognostic test for breast cancer metastasis based on profiling 250 genes; 16 genes as a group have predictive value; $3, 400 per test 215, 000 breast cancer cases per year (potential market value > $500 million!) No validation of test; No FDA approval Test has no value for predicting response to treatment Science 2004; 303: 1754 -5
 Commercial Clashes n n n Mammaprint marketed by Agendia, Amsterdam, The Netherlands Based on L. Van’t Veer publications Test costs Euro 1650; based on 70 gene signature Prospective trials underway Celera and Arcturus developing similar tests (prognosis/prediction of therapy) Science 2004; 303: 1754 -5
Commercial Clashes n n n Mammaprint marketed by Agendia, Amsterdam, The Netherlands Based on L. Van’t Veer publications Test costs Euro 1650; based on 70 gene signature Prospective trials underway Celera and Arcturus developing similar tests (prognosis/prediction of therapy) Science 2004; 303: 1754 -5
 Tissue Microarrays n Printing on a slide tiny amounts of tissue n Array many patients in one slide (e. g. 500) n Process all at once (e. g. immunohistochemistry) n Works with archival tissue (paraffin blocks)
Tissue Microarrays n Printing on a slide tiny amounts of tissue n Array many patients in one slide (e. g. 500) n Process all at once (e. g. immunohistochemistry) n Works with archival tissue (paraffin blocks)
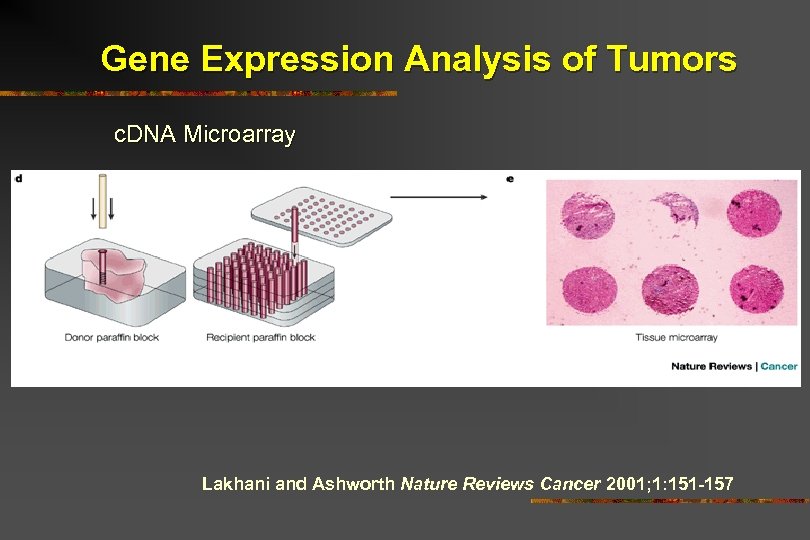 Gene Expression Analysis of Tumors c. DNA Microarray Lakhani and Ashworth Nature Reviews Cancer 2001; 1: 151 -157
Gene Expression Analysis of Tumors c. DNA Microarray Lakhani and Ashworth Nature Reviews Cancer 2001; 1: 151 -157
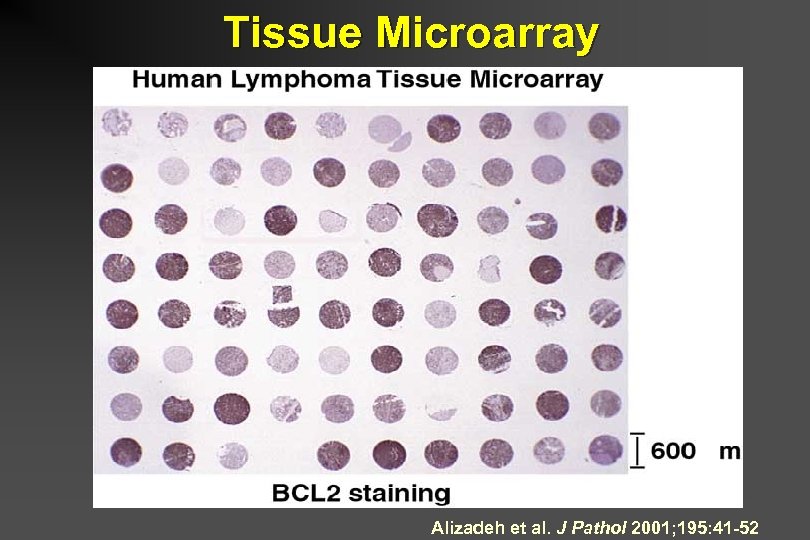 Tissue Microarray Alizadeh et al. J Pathol 2001; 195: 41 -52
Tissue Microarray Alizadeh et al. J Pathol 2001; 195: 41 -52
 Microarray Future: Conclusions n Differential gene experssion studies will continue(robusness) n Inexpensive, high-throughput, genome-wide scans for clinical applications n Protein microarrays are now being deployed (may take over) n Focus on biology and improved technology n SNP analysis-Disease predisposition n Pharmacogenomics n Diagnostics-Multiparametric analysis n Replacement of single-gene experiments(paradigm shift)
Microarray Future: Conclusions n Differential gene experssion studies will continue(robusness) n Inexpensive, high-throughput, genome-wide scans for clinical applications n Protein microarrays are now being deployed (may take over) n Focus on biology and improved technology n SNP analysis-Disease predisposition n Pharmacogenomics n Diagnostics-Multiparametric analysis n Replacement of single-gene experiments(paradigm shift)


