e8422b79de02cf6d4b9caf53dd5639be.ppt
- Количество слайдов: 58
 Microarray - Introduction Ka-Lok Ng Asia University
Microarray - Introduction Ka-Lok Ng Asia University
 Topics to be covered • • • Introduction - RNA expression Experimental design, image processing, Microarray databases Data normalization, filter and analysis MATLAB Statistical analysis of gene expression data Clustering methods Time series data (cell cycle) and dynamics programming Gene regulatory networks Gene regulatory networks and protein-protein interaction networks
Topics to be covered • • • Introduction - RNA expression Experimental design, image processing, Microarray databases Data normalization, filter and analysis MATLAB Statistical analysis of gene expression data Clustering methods Time series data (cell cycle) and dynamics programming Gene regulatory networks Gene regulatory networks and protein-protein interaction networks
 • 40% on quiz, classwork, homework, class attendance • Mid-term – 30%,final exam. – 30%, oral presentation
• 40% on quiz, classwork, homework, class attendance • Mid-term – 30%,final exam. – 30%, oral presentation
 References 1. 2. 3. 4. 5. 6. Causton H. , Quackenbush J. , and Brazma A. Microarray Gene Expression Data Analysis. A Beginner’s Guide. Blackwell (2003) Baxevanis A. and Ouellette B. F. Francis. Bioinformatics Ch. 16. J. Wiley (2005) Knudsen S. A Biologist’s Guide to Analysis of DNA Microarray Data. J. Wiley (2002) Benfey P. and Protopapas A. D. Genomics Ch. 5. Prentice Hall (2005). Setubal J. and Meidanis J. Introduction to computational molecular biology. PWS publishing. (1997). A. Gu´enoche (2005). “about the design of oligo-chips”, Discrete Applied Mathematics, v 147(1), pp. 57 -67.
References 1. 2. 3. 4. 5. 6. Causton H. , Quackenbush J. , and Brazma A. Microarray Gene Expression Data Analysis. A Beginner’s Guide. Blackwell (2003) Baxevanis A. and Ouellette B. F. Francis. Bioinformatics Ch. 16. J. Wiley (2005) Knudsen S. A Biologist’s Guide to Analysis of DNA Microarray Data. J. Wiley (2002) Benfey P. and Protopapas A. D. Genomics Ch. 5. Prentice Hall (2005). Setubal J. and Meidanis J. Introduction to computational molecular biology. PWS publishing. (1997). A. Gu´enoche (2005). “about the design of oligo-chips”, Discrete Applied Mathematics, v 147(1), pp. 57 -67.
 Contents • Introduction – the central dogma of molecular biology, applications, data analysis, Microarray slide surface • Printing technologies – spotting, photolithography, ink-jet • Selection of genes for spotting on arrays • Selection of primers for PCR – suffix tree • Microarray application - four different types of brain tumors • Gene co-expression and gene expression profile • Data management
Contents • Introduction – the central dogma of molecular biology, applications, data analysis, Microarray slide surface • Printing technologies – spotting, photolithography, ink-jet • Selection of genes for spotting on arrays • Selection of primers for PCR – suffix tree • Microarray application - four different types of brain tumors • Gene co-expression and gene expression profile • Data management

 Introduction • The last 10 years have brought spectacular achievements in genome sequencing (such as the HGP) • It took >1000 years for science to progress from human anatomy to understand how genomes function) • Even if we assume all the genes have correctly identified, the results represents only sequence • High throughput DNA sequencing technology created a system approach to biology
Introduction • The last 10 years have brought spectacular achievements in genome sequencing (such as the HGP) • It took >1000 years for science to progress from human anatomy to understand how genomes function) • Even if we assume all the genes have correctly identified, the results represents only sequence • High throughput DNA sequencing technology created a system approach to biology
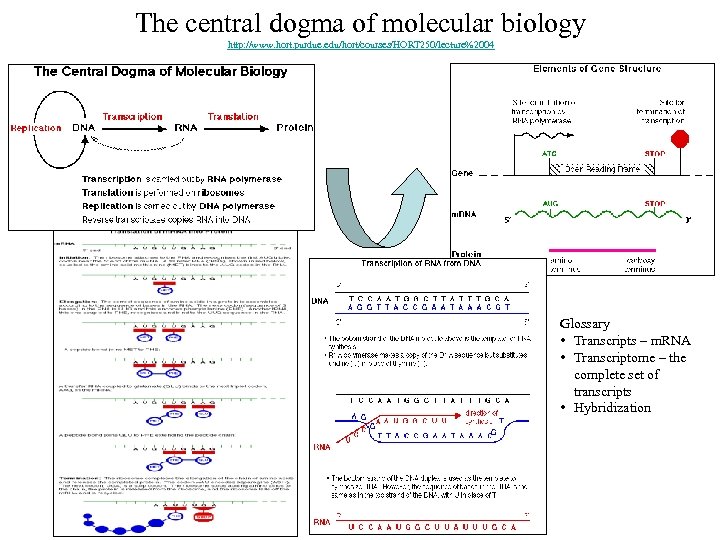 The central dogma of molecular biology http: //www. hort. purdue. edu/hort/courses/HORT 250/lecture%2004 Glossary • Transcripts – m. RNA • Transcriptome – the complete set of transcripts • Hybridization
The central dogma of molecular biology http: //www. hort. purdue. edu/hort/courses/HORT 250/lecture%2004 Glossary • Transcripts – m. RNA • Transcriptome – the complete set of transcripts • Hybridization
 • Microarray technology allow one to identify the genes that are expressed in different cell types, to learn how their expression levels change in different developmental stages or disease states, and to identify the cellular processes in which they participate • Microarray technology provide clues about how genes and gene products interact and their interaction networks Microarray gene expression data analysis • Experimental design data transformations from raw data to gene expression matrices data mining and analysis of gene expression matrices
• Microarray technology allow one to identify the genes that are expressed in different cell types, to learn how their expression levels change in different developmental stages or disease states, and to identify the cellular processes in which they participate • Microarray technology provide clues about how genes and gene products interact and their interaction networks Microarray gene expression data analysis • Experimental design data transformations from raw data to gene expression matrices data mining and analysis of gene expression matrices
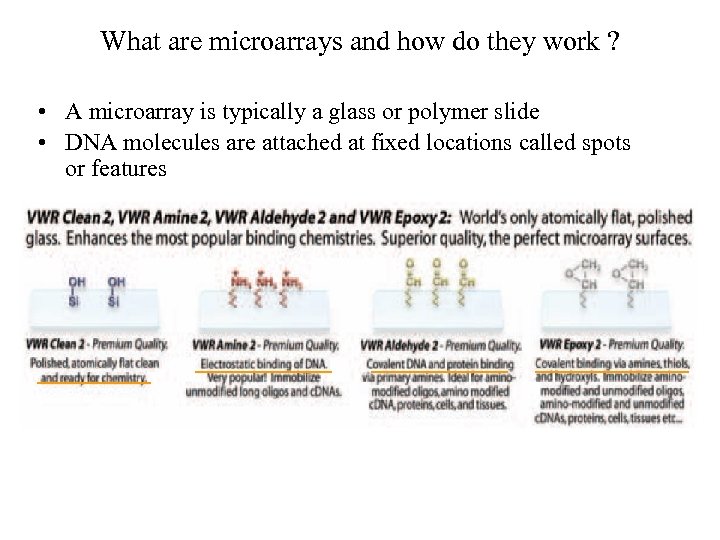 What are microarrays and how do they work ? • A microarray is typically a glass or polymer slide • DNA molecules are attached at fixed locations called spots or features
What are microarrays and how do they work ? • A microarray is typically a glass or polymer slide • DNA molecules are attached at fixed locations called spots or features
 Smooth surface enables even deposition of surface chemistries and perfect spot morphology.
Smooth surface enables even deposition of surface chemistries and perfect spot morphology.
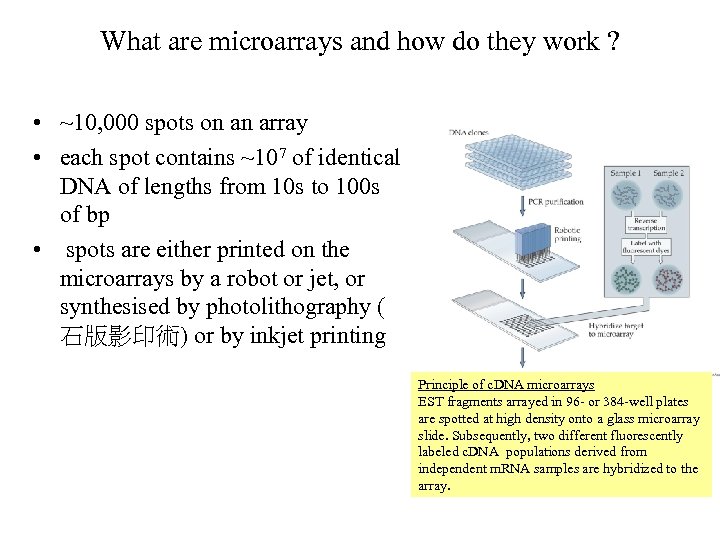 What are microarrays and how do they work ? • ~10, 000 spots on an array • each spot contains ~107 of identical DNA of lengths from 10 s to 100 s of bp • spots are either printed on the microarrays by a robot or jet, or synthesised by photolithography ( 石版影印術) or by inkjet printing Principle of c. DNA microarrays EST fragments arrayed in 96 - or 384 -well plates are spotted at high density onto a glass microarray slide. Subsequently, two different fluorescently labeled c. DNA populations derived from independent m. RNA samples are hybridized to the array.
What are microarrays and how do they work ? • ~10, 000 spots on an array • each spot contains ~107 of identical DNA of lengths from 10 s to 100 s of bp • spots are either printed on the microarrays by a robot or jet, or synthesised by photolithography ( 石版影印術) or by inkjet printing Principle of c. DNA microarrays EST fragments arrayed in 96 - or 384 -well plates are spotted at high density onto a glass microarray slide. Subsequently, two different fluorescently labeled c. DNA populations derived from independent m. RNA samples are hybridized to the array.
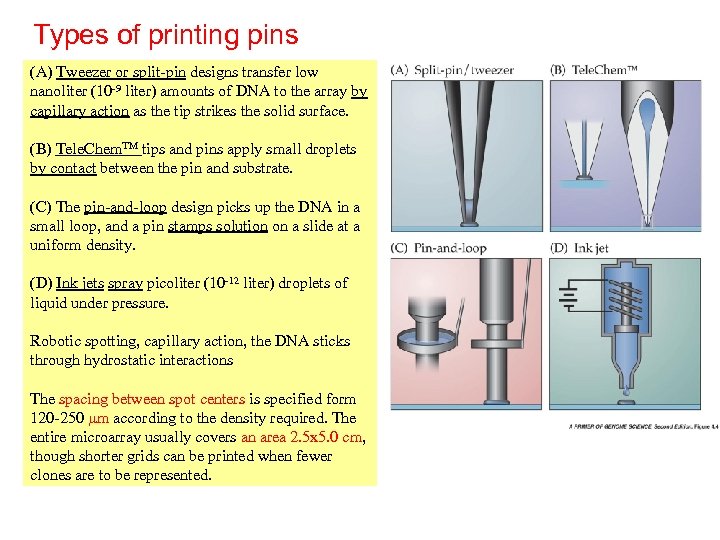 Types of printing pins (A) Tweezer or split-pin designs transfer low nanoliter (10 -9 liter) amounts of DNA to the array by capillary action as the tip strikes the solid surface. (B) Tele. Chem. TM tips and pins apply small droplets by contact between the pin and substrate. (C) The pin-and-loop design picks up the DNA in a small loop, and a pin stamps solution on a slide at a uniform density. (D) Ink jets spray picoliter (10 -12 liter) droplets of liquid under pressure. Robotic spotting, capillary action, the DNA sticks through hydrostatic interactions The spacing between spot centers is specified form 120 -250 mm according to the density required. The entire microarray usually covers an area 2. 5 x 5. 0 cm, though shorter grids can be printed when fewer clones are to be represented.
Types of printing pins (A) Tweezer or split-pin designs transfer low nanoliter (10 -9 liter) amounts of DNA to the array by capillary action as the tip strikes the solid surface. (B) Tele. Chem. TM tips and pins apply small droplets by contact between the pin and substrate. (C) The pin-and-loop design picks up the DNA in a small loop, and a pin stamps solution on a slide at a uniform density. (D) Ink jets spray picoliter (10 -12 liter) droplets of liquid under pressure. Robotic spotting, capillary action, the DNA sticks through hydrostatic interactions The spacing between spot centers is specified form 120 -250 mm according to the density required. The entire microarray usually covers an area 2. 5 x 5. 0 cm, though shorter grids can be printed when fewer clones are to be represented.
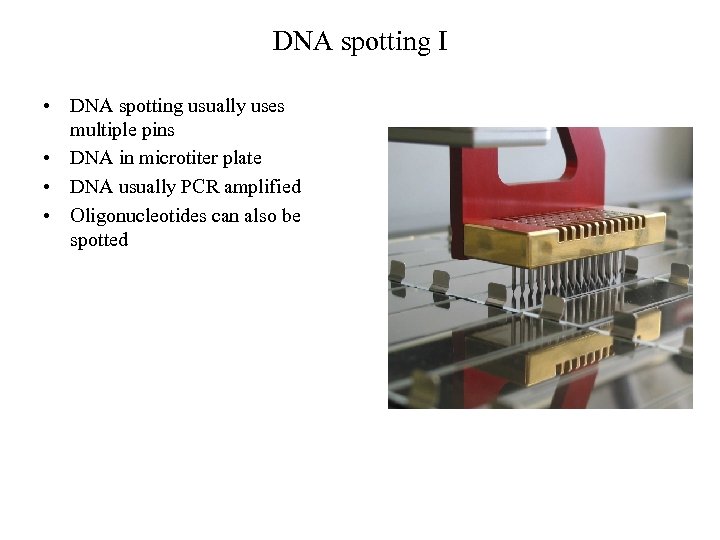 DNA spotting I • DNA spotting usually uses multiple pins • DNA in microtiter plate • DNA usually PCR amplified • Oligonucleotides can also be spotted
DNA spotting I • DNA spotting usually uses multiple pins • DNA in microtiter plate • DNA usually PCR amplified • Oligonucleotides can also be spotted
 Commercial DNA spotter
Commercial DNA spotter
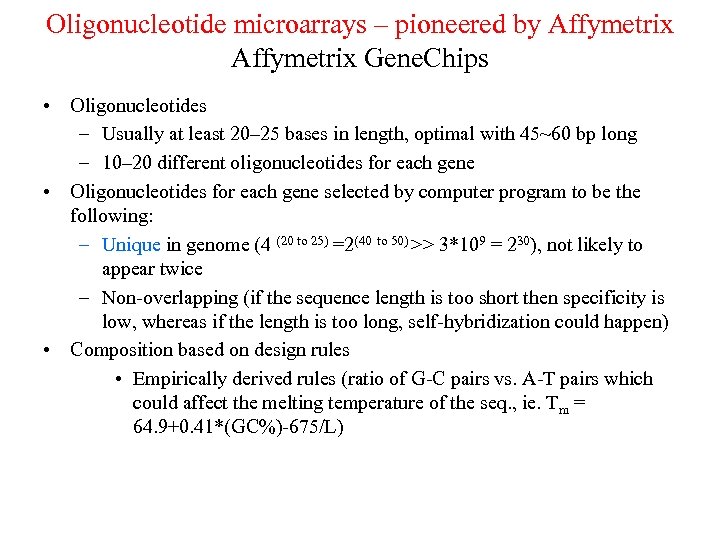 Oligonucleotide microarrays – pioneered by Affymetrix Gene. Chips • Oligonucleotides – Usually at least 20– 25 bases in length, optimal with 45~60 bp long – 10– 20 different oligonucleotides for each gene • Oligonucleotides for each gene selected by computer program to be the following: – Unique in genome (4 (20 to 25) =2(40 to 50) >> 3*109 = 230), not likely to appear twice – Non-overlapping (if the sequence length is too short then specificity is low, whereas if the length is too long, self-hybridization could happen) • Composition based on design rules • Empirically derived rules (ratio of G-C pairs vs. A-T pairs which could affect the melting temperature of the seq. , ie. Tm = 64. 9+0. 41*(GC%)-675/L)
Oligonucleotide microarrays – pioneered by Affymetrix Gene. Chips • Oligonucleotides – Usually at least 20– 25 bases in length, optimal with 45~60 bp long – 10– 20 different oligonucleotides for each gene • Oligonucleotides for each gene selected by computer program to be the following: – Unique in genome (4 (20 to 25) =2(40 to 50) >> 3*109 = 230), not likely to appear twice – Non-overlapping (if the sequence length is too short then specificity is low, whereas if the length is too long, self-hybridization could happen) • Composition based on design rules • Empirically derived rules (ratio of G-C pairs vs. A-T pairs which could affect the melting temperature of the seq. , ie. Tm = 64. 9+0. 41*(GC%)-675/L)
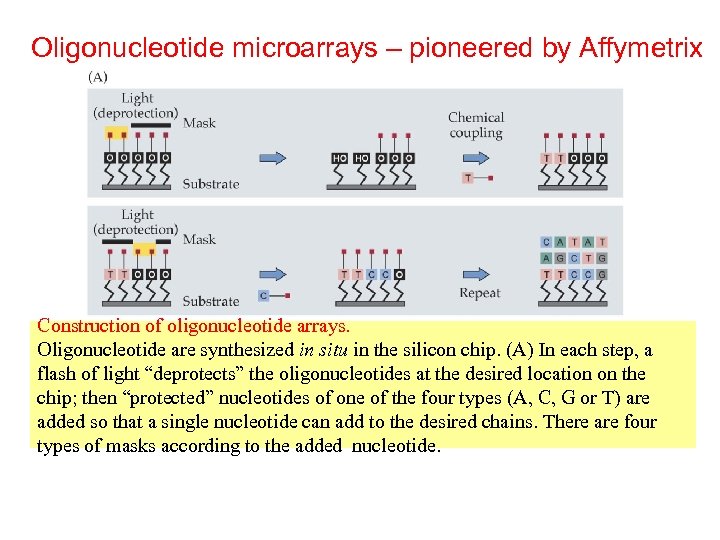 Oligonucleotide microarrays – pioneered by Affymetrix Construction of oligonucleotide arrays. Oligonucleotide are synthesized in situ in the silicon chip. (A) In each step, a flash of light “deprotects” the oligonucleotides at the desired location on the chip; then “protected” nucleotides of one of the four types (A, C, G or T) are added so that a single nucleotide can add to the desired chains. There are four types of masks according to the added nucleotide.
Oligonucleotide microarrays – pioneered by Affymetrix Construction of oligonucleotide arrays. Oligonucleotide are synthesized in situ in the silicon chip. (A) In each step, a flash of light “deprotects” the oligonucleotides at the desired location on the chip; then “protected” nucleotides of one of the four types (A, C, G or T) are added so that a single nucleotide can add to the desired chains. There are four types of masks according to the added nucleotide.
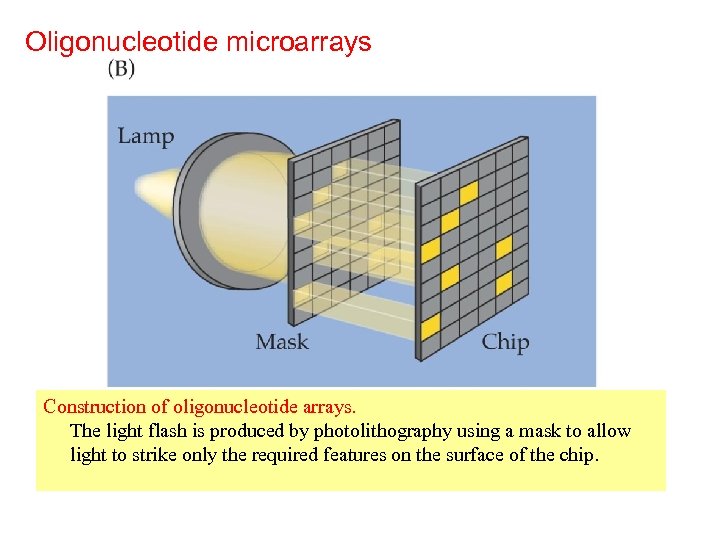 Oligonucleotide microarrays Construction of oligonucleotide arrays. The light flash is produced by photolithography using a mask to allow light to strike only the required features on the surface of the chip.
Oligonucleotide microarrays Construction of oligonucleotide arrays. The light flash is produced by photolithography using a mask to allow light to strike only the required features on the surface of the chip.
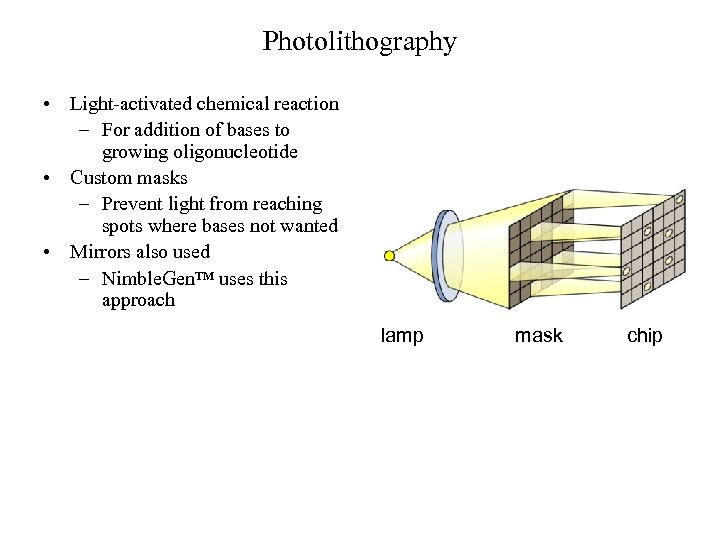 Photolithography • Light-activated chemical reaction – For addition of bases to growing oligonucleotide • Custom masks – Prevent light from reaching spots where bases not wanted • Mirrors also used – Nimble. Gen™ uses this approach lamp mask chip
Photolithography • Light-activated chemical reaction – For addition of bases to growing oligonucleotide • Custom masks – Prevent light from reaching spots where bases not wanted • Mirrors also used – Nimble. Gen™ uses this approach lamp mask chip
 Example: building oligonucleotides by photolithography • Want to add nucleotide G • Mask all other spots on chip • Light shines only where addition of G is desired (light “deprotects” the oligonucleotides at the desired location on the chip) • G added and reacts • Now G is on subset of oligonucleotides light
Example: building oligonucleotides by photolithography • Want to add nucleotide G • Mask all other spots on chip • Light shines only where addition of G is desired (light “deprotects” the oligonucleotides at the desired location on the chip) • G added and reacts • Now G is on subset of oligonucleotides light
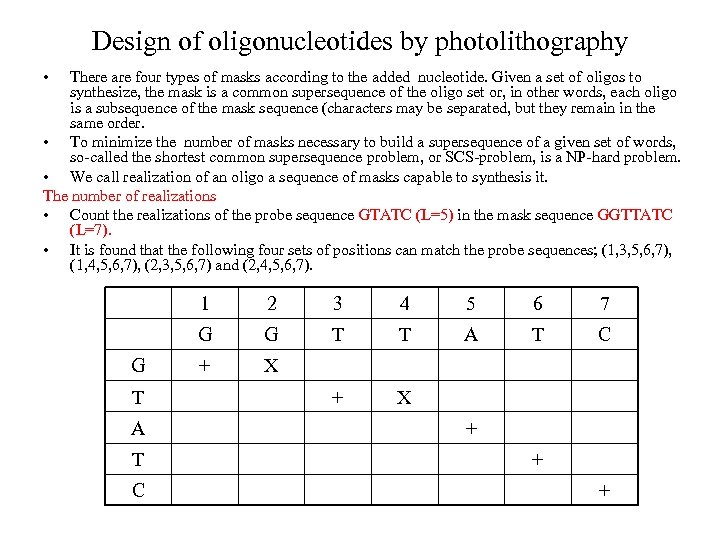 Design of oligonucleotides by photolithography • There are four types of masks according to the added nucleotide. Given a set of oligos to synthesize, the mask is a common supersequence of the oligo set or, in other words, each oligo is a subsequence of the mask sequence (characters may be separated, but they remain in the same order. • To minimize the number of masks necessary to build a supersequence of a given set of words, so-called the shortest common supersequence problem, or SCS-problem, is a NP-hard problem. • We call realization of an oligo a sequence of masks capable to synthesis it. The number of realizations • Count the realizations of the probe sequence GTATC (L=5) in the mask sequence GGTTATC (L=7). • It is found that the following four sets of positions can match the probe sequences; (1, 3, 5, 6, 7), (1, 4, 5, 6, 7), (2, 3, 5, 6, 7) and (2, 4, 5, 6, 7). 1 T A T C 3 4 5 6 7 G G 2 G T T A T C + X + + +
Design of oligonucleotides by photolithography • There are four types of masks according to the added nucleotide. Given a set of oligos to synthesize, the mask is a common supersequence of the oligo set or, in other words, each oligo is a subsequence of the mask sequence (characters may be separated, but they remain in the same order. • To minimize the number of masks necessary to build a supersequence of a given set of words, so-called the shortest common supersequence problem, or SCS-problem, is a NP-hard problem. • We call realization of an oligo a sequence of masks capable to synthesis it. The number of realizations • Count the realizations of the probe sequence GTATC (L=5) in the mask sequence GGTTATC (L=7). • It is found that the following four sets of positions can match the probe sequences; (1, 3, 5, 6, 7), (1, 4, 5, 6, 7), (2, 3, 5, 6, 7) and (2, 4, 5, 6, 7). 1 T A T C 3 4 5 6 7 G G 2 G T T A T C + X + + +
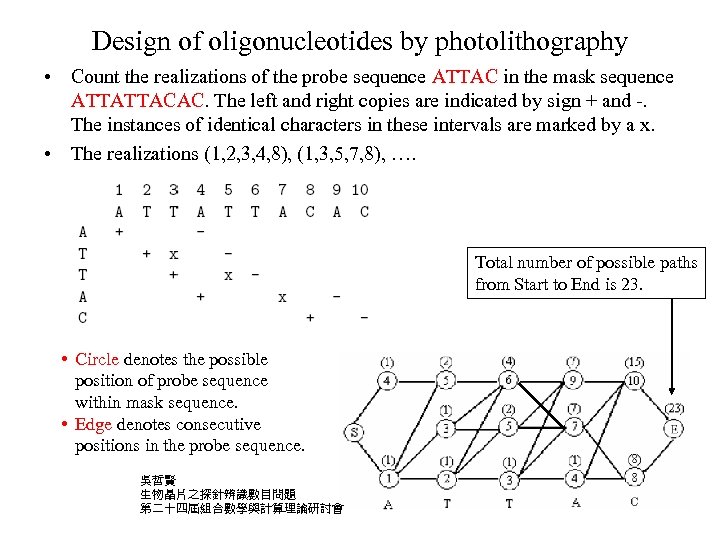 Design of oligonucleotides by photolithography • Count the realizations of the probe sequence ATTAC in the mask sequence ATTATTACAC. The left and right copies are indicated by sign + and -. The instances of identical characters in these intervals are marked by a x. • The realizations (1, 2, 3, 4, 8), (1, 3, 5, 7, 8), …. Total number of possible paths from Start to End is 23. • Circle denotes the possible position of probe sequence within mask sequence. • Edge denotes consecutive positions in the probe sequence. 吳哲賢 生物晶片之探針辨識數目問題 第二十四屆組合數學與計算理論研討會
Design of oligonucleotides by photolithography • Count the realizations of the probe sequence ATTAC in the mask sequence ATTATTACAC. The left and right copies are indicated by sign + and -. The instances of identical characters in these intervals are marked by a x. • The realizations (1, 2, 3, 4, 8), (1, 3, 5, 7, 8), …. Total number of possible paths from Start to End is 23. • Circle denotes the possible position of probe sequence within mask sequence. • Edge denotes consecutive positions in the probe sequence. 吳哲賢 生物晶片之探針辨識數目問題 第二十四屆組合數學與計算理論研討會
 Example: adding a second base • Want to add T • New mask covers spots where T not wanted • Light shines on mask • T added • Continue for all four bases • Need 80 masks for total 20 -mer oligonucleotide light
Example: adding a second base • Want to add T • New mask covers spots where T not wanted • Light shines on mask • T added • Continue for all four bases • Need 80 masks for total 20 -mer oligonucleotide light
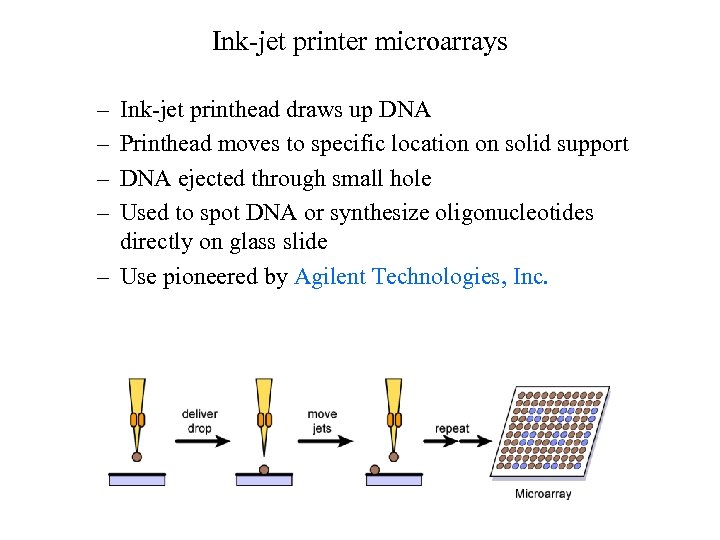 Ink-jet printer microarrays – – Ink-jet printhead draws up DNA Printhead moves to specific location on solid support DNA ejected through small hole Used to spot DNA or synthesize oligonucleotides directly on glass slide – Use pioneered by Agilent Technologies, Inc.
Ink-jet printer microarrays – – Ink-jet printhead draws up DNA Printhead moves to specific location on solid support DNA ejected through small hole Used to spot DNA or synthesize oligonucleotides directly on glass slide – Use pioneered by Agilent Technologies, Inc.
 Comparisons of microarrays Photolithograhy Mechanical printing Ink-jet printing
Comparisons of microarrays Photolithograhy Mechanical printing Ink-jet printing
 Comparison of microarray hybridization • Spotted microarrays – Competitive hybridization • Two labeled c. DNAs hybridized to same slide measure the relative difference between the signal intensity of two targets binding to the same spot of DNA • Affymetrix Gene. Chips – One labeled RNA population per chip – Comparison made between hybridization intensities of same oligonucleotides on different chips
Comparison of microarray hybridization • Spotted microarrays – Competitive hybridization • Two labeled c. DNAs hybridized to same slide measure the relative difference between the signal intensity of two targets binding to the same spot of DNA • Affymetrix Gene. Chips – One labeled RNA population per chip – Comparison made between hybridization intensities of same oligonucleotides on different chips
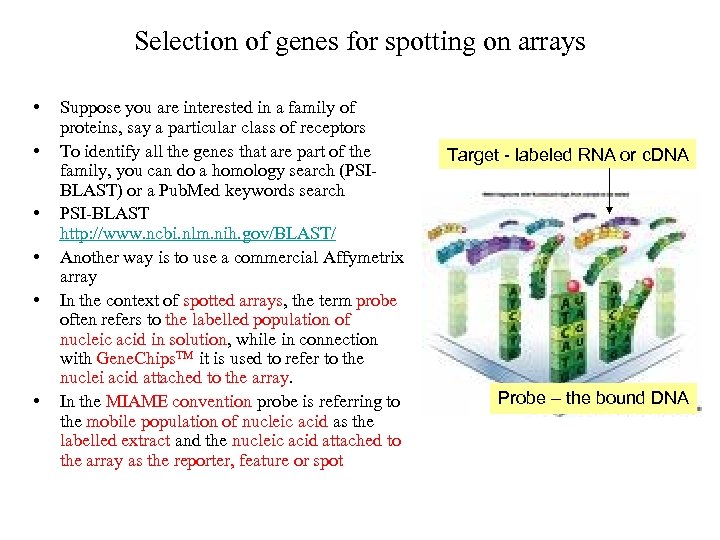 Selection of genes for spotting on arrays • • • Suppose you are interested in a family of proteins, say a particular class of receptors To identify all the genes that are part of the family, you can do a homology search (PSIBLAST) or a Pub. Med keywords search PSI-BLAST http: //www. ncbi. nlm. nih. gov/BLAST/ Another way is to use a commercial Affymetrix array In the context of spotted arrays, the term probe often refers to the labelled population of nucleic acid in solution, while in connection with Gene. Chips. TM it is used to refer to the nuclei acid attached to the array. In the MIAME convention probe is referring to the mobile population of nucleic acid as the labelled extract and the nucleic acid attached to the array as the reporter, feature or spot Target - labeled RNA or c. DNA Probe – the bound DNA
Selection of genes for spotting on arrays • • • Suppose you are interested in a family of proteins, say a particular class of receptors To identify all the genes that are part of the family, you can do a homology search (PSIBLAST) or a Pub. Med keywords search PSI-BLAST http: //www. ncbi. nlm. nih. gov/BLAST/ Another way is to use a commercial Affymetrix array In the context of spotted arrays, the term probe often refers to the labelled population of nucleic acid in solution, while in connection with Gene. Chips. TM it is used to refer to the nuclei acid attached to the array. In the MIAME convention probe is referring to the mobile population of nucleic acid as the labelled extract and the nucleic acid attached to the array as the reporter, feature or spot Target - labeled RNA or c. DNA Probe – the bound DNA
 Selection of regions within genes • Once you have the list of genes you wish to spot on the array • The next question is cross-hybridization • How can you prevent spotting probes (similar) that are complementary to more than one gene (target m. RNA or c. DNA seq. ) if you are working with a gene family with similarities in sequence (such as > 70% similarity) ? – That is a probe could cross-hybridized with different m. RNA – or a gene’s m. RNA could cross-hybridized with different probe nonspecific not a true expression level of the gene under study • Solve this problem by using Probe. Wiz Server • Use Blast to find regions in those genes that are the least homologous to other genes • Probe. Wiz - http: //www. cbs. dtu. dk/services/DNAarray/probewiz. php
Selection of regions within genes • Once you have the list of genes you wish to spot on the array • The next question is cross-hybridization • How can you prevent spotting probes (similar) that are complementary to more than one gene (target m. RNA or c. DNA seq. ) if you are working with a gene family with similarities in sequence (such as > 70% similarity) ? – That is a probe could cross-hybridized with different m. RNA – or a gene’s m. RNA could cross-hybridized with different probe nonspecific not a true expression level of the gene under study • Solve this problem by using Probe. Wiz Server • Use Blast to find regions in those genes that are the least homologous to other genes • Probe. Wiz - http: //www. cbs. dtu. dk/services/DNAarray/probewiz. php
 Selection of primers for PCR • Once those unique regions have been identified, the probe needs to be designed use PCR amplification of a probe • Solve this problem by using Probe. Wiz or Oligo. Array Servers • Probe. Wiz – predicts optimal PCR primer pairs for generation of probes for c. DNA arrays – avoid self-hybridization hairpin structure high specificity • http: //www. cbs. dtu. dk/services/DNAarray/probewiz. php • Oligo. Array – Genome-scale oligonucleotide design for microarrays • http: //berry. engin. umich. edu/oligoarray 2/ • Other option - By using long oligonucleotides (50 to 70 bps) instead of PCR primers • Other complicated issues: alternative splicing, SNP
Selection of primers for PCR • Once those unique regions have been identified, the probe needs to be designed use PCR amplification of a probe • Solve this problem by using Probe. Wiz or Oligo. Array Servers • Probe. Wiz – predicts optimal PCR primer pairs for generation of probes for c. DNA arrays – avoid self-hybridization hairpin structure high specificity • http: //www. cbs. dtu. dk/services/DNAarray/probewiz. php • Oligo. Array – Genome-scale oligonucleotide design for microarrays • http: //berry. engin. umich. edu/oligoarray 2/ • Other option - By using long oligonucleotides (50 to 70 bps) instead of PCR primers • Other complicated issues: alternative splicing, SNP
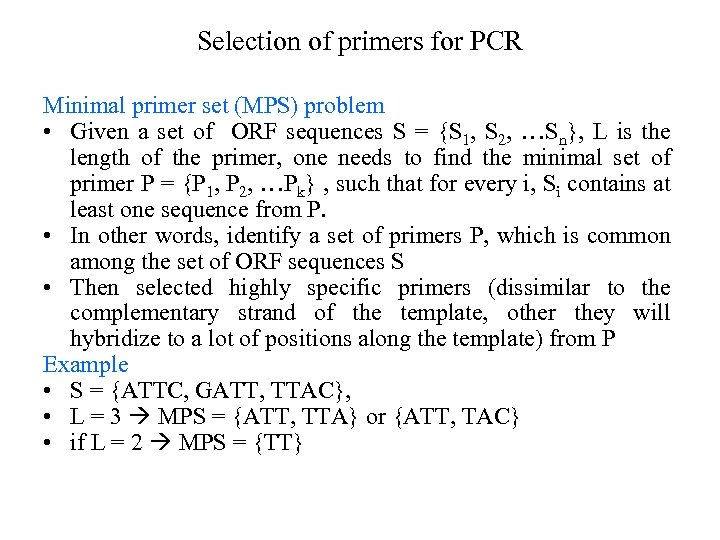 Selection of primers for PCR Minimal primer set (MPS) problem • Given a set of ORF sequences S = {S 1, S 2, …Sn}, L is the length of the primer, one needs to find the minimal set of primer P = {P 1, P 2, …Pk} , such that for every i, Si contains at least one sequence from P. • In other words, identify a set of primers P, which is common among the set of ORF sequences S • Then selected highly specific primers (dissimilar to the complementary strand of the template, other they will hybridize to a lot of positions along the template) from P Example • S = {ATTC, GATT, TTAC}, • L = 3 MPS = {ATT, TTA} or {ATT, TAC} • if L = 2 MPS = {TT}
Selection of primers for PCR Minimal primer set (MPS) problem • Given a set of ORF sequences S = {S 1, S 2, …Sn}, L is the length of the primer, one needs to find the minimal set of primer P = {P 1, P 2, …Pk} , such that for every i, Si contains at least one sequence from P. • In other words, identify a set of primers P, which is common among the set of ORF sequences S • Then selected highly specific primers (dissimilar to the complementary strand of the template, other they will hybridize to a lot of positions along the template) from P Example • S = {ATTC, GATT, TTAC}, • L = 3 MPS = {ATT, TTA} or {ATT, TAC} • if L = 2 MPS = {TT}
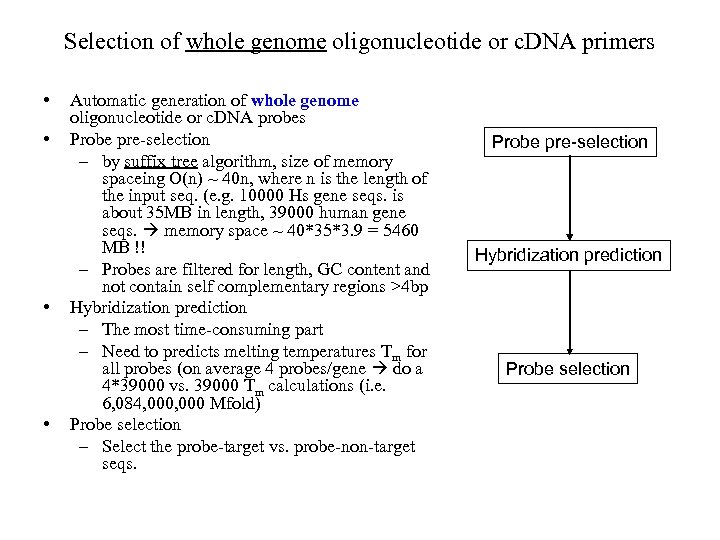 Selection of whole genome oligonucleotide or c. DNA primers • • Automatic generation of whole genome oligonucleotide or c. DNA probes Probe pre-selection – by suffix tree algorithm, size of memory spaceing O(n) ~ 40 n, where n is the length of the input seq. (e. g. 10000 Hs gene seqs. is about 35 MB in length, 39000 human gene seqs. memory space ~ 40*35*3. 9 = 5460 MB !! – Probes are filtered for length, GC content and not contain self complementary regions >4 bp Hybridization prediction – The most time-consuming part – Need to predicts melting temperatures Tm for all probes (on average 4 probes/gene do a 4*39000 vs. 39000 Tm calculations (i. e. 6, 084, 000 Mfold) Probe selection – Select the probe-target vs. probe-non-target seqs. Probe pre-selection Hybridization prediction Probe selection
Selection of whole genome oligonucleotide or c. DNA primers • • Automatic generation of whole genome oligonucleotide or c. DNA probes Probe pre-selection – by suffix tree algorithm, size of memory spaceing O(n) ~ 40 n, where n is the length of the input seq. (e. g. 10000 Hs gene seqs. is about 35 MB in length, 39000 human gene seqs. memory space ~ 40*35*3. 9 = 5460 MB !! – Probes are filtered for length, GC content and not contain self complementary regions >4 bp Hybridization prediction – The most time-consuming part – Need to predicts melting temperatures Tm for all probes (on average 4 probes/gene do a 4*39000 vs. 39000 Tm calculations (i. e. 6, 084, 000 Mfold) Probe selection – Select the probe-target vs. probe-non-target seqs. Probe pre-selection Hybridization prediction Probe selection
 Suffix tree - Basic notation • Concatenation (串聯) of two strings s and t is denoted by st and is formed by appending all characters of t after s, in the order they appear in t, for instance, if s =GGCTA and t=CAAC, then st=GGCTACAAC. The length of st is |s|+|t|. • A prefix of s is any substring of s of the form s[1…. j] for 0≦j≦ |s|. It is admit j=0 and define s[1…. 0] as being the empty string, which is a prefix of s as well. Note that t is a prefix of s if and only if there is another string u such that s=tu. Sometimes one needs to refer to the prefix of s with exactly k characters, with 0≦k≦|s|, and we use the notation prefix(s, k) to denote this string. • prefix(s, 3) ATT is a prefix of ATTCGATTTTAC • A suffix of s is a substring of the form s[i…. |s|] for a certain i such that 1≦i≦ |s|+1. one admit i=|s|+1, in which case s[|s|+1…. |s|] denotes the empty string. A string t is a suffix of s if and only if there is another string u such that s=ut. The notation suffix(s, k) denotes the unique suffix of s with k characters, for 0≦k≦|s|. • suffix(s, 3) TAC is a suffix of ATTCGATTTTAC
Suffix tree - Basic notation • Concatenation (串聯) of two strings s and t is denoted by st and is formed by appending all characters of t after s, in the order they appear in t, for instance, if s =GGCTA and t=CAAC, then st=GGCTACAAC. The length of st is |s|+|t|. • A prefix of s is any substring of s of the form s[1…. j] for 0≦j≦ |s|. It is admit j=0 and define s[1…. 0] as being the empty string, which is a prefix of s as well. Note that t is a prefix of s if and only if there is another string u such that s=tu. Sometimes one needs to refer to the prefix of s with exactly k characters, with 0≦k≦|s|, and we use the notation prefix(s, k) to denote this string. • prefix(s, 3) ATT is a prefix of ATTCGATTTTAC • A suffix of s is a substring of the form s[i…. |s|] for a certain i such that 1≦i≦ |s|+1. one admit i=|s|+1, in which case s[|s|+1…. |s|] denotes the empty string. A string t is a suffix of s if and only if there is another string u such that s=ut. The notation suffix(s, k) denotes the unique suffix of s with k characters, for 0≦k≦|s|. • suffix(s, 3) TAC is a suffix of ATTCGATTTTAC
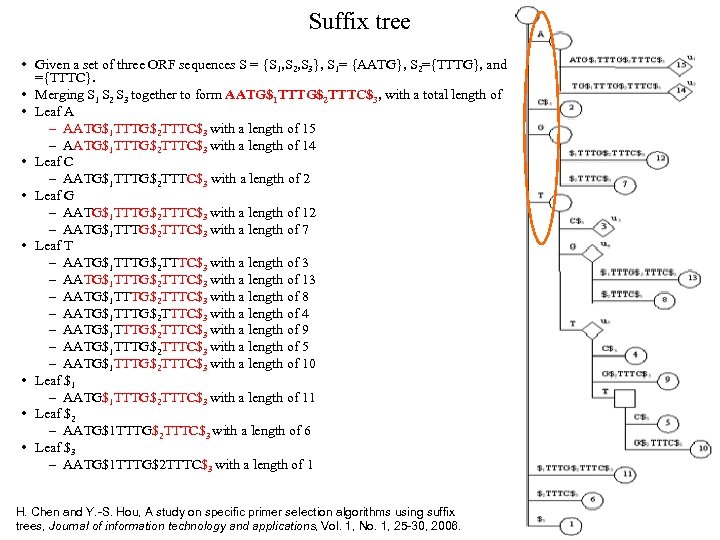 Suffix tree • Given a set of three ORF sequences S = {S 1, S 2, S 3}, S 1= {AATG}, S 2={TTTG}, and S 3 ={TTTC}. • Merging S 1 S 2 S 3 together to form AATG$1 TTTG$2 TTTC$3, with a total length of 15. • Leaf A – AATG$1 TTTG$2 TTTC$3 with a length of 15 – AATG$1 TTTG$2 TTTC$3 with a length of 14 • Leaf C – AATG$1 TTTG$2 TTTC$3 with a length of 2 • Leaf G – AATG$1 TTTG$2 TTTC$3 with a length of 12 – AATG$1 TTTG$2 TTTC$3 with a length of 7 • Leaf T – AATG$1 TTTG$2 TTTC$3 with a length of 3 – AATG$1 TTTG$2 TTTC$3 with a length of 13 – AATG$1 TTTG$2 TTTC$3 with a length of 8 – AATG$1 TTTG$2 TTTC$3 with a length of 4 – AATG$1 TTTG$2 TTTC$3 with a length of 9 – AATG$1 TTTG$2 TTTC$3 with a length of 5 – AATG$1 TTTG$2 TTTC$3 with a length of 10 • Leaf $1 – AATG$1 TTTG$2 TTTC$3 with a length of 11 • Leaf $2 – AATG$1 TTTG$2 TTTC$3 with a length of 6 • Leaf $3 – AATG$1 TTTG$2 TTTC$3 with a length of 1 H. Chen and Y. -S. Hou, A study on specific primer selection algorithms using suffix trees, Journal of information technology and applications, Vol. 1, No. 1, 25 -30, 2006.
Suffix tree • Given a set of three ORF sequences S = {S 1, S 2, S 3}, S 1= {AATG}, S 2={TTTG}, and S 3 ={TTTC}. • Merging S 1 S 2 S 3 together to form AATG$1 TTTG$2 TTTC$3, with a total length of 15. • Leaf A – AATG$1 TTTG$2 TTTC$3 with a length of 15 – AATG$1 TTTG$2 TTTC$3 with a length of 14 • Leaf C – AATG$1 TTTG$2 TTTC$3 with a length of 2 • Leaf G – AATG$1 TTTG$2 TTTC$3 with a length of 12 – AATG$1 TTTG$2 TTTC$3 with a length of 7 • Leaf T – AATG$1 TTTG$2 TTTC$3 with a length of 3 – AATG$1 TTTG$2 TTTC$3 with a length of 13 – AATG$1 TTTG$2 TTTC$3 with a length of 8 – AATG$1 TTTG$2 TTTC$3 with a length of 4 – AATG$1 TTTG$2 TTTC$3 with a length of 9 – AATG$1 TTTG$2 TTTC$3 with a length of 5 – AATG$1 TTTG$2 TTTC$3 with a length of 10 • Leaf $1 – AATG$1 TTTG$2 TTTC$3 with a length of 11 • Leaf $2 – AATG$1 TTTG$2 TTTC$3 with a length of 6 • Leaf $3 – AATG$1 TTTG$2 TTTC$3 with a length of 1 H. Chen and Y. -S. Hou, A study on specific primer selection algorithms using suffix trees, Journal of information technology and applications, Vol. 1, No. 1, 25 -30, 2006.
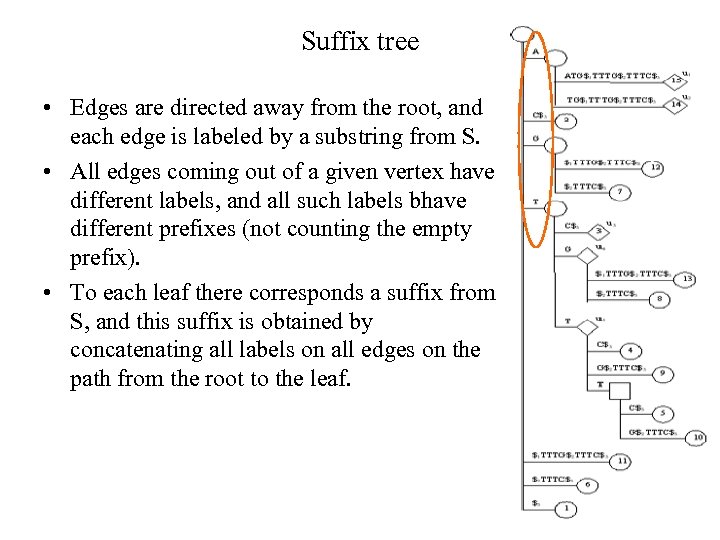 Suffix tree • Edges are directed away from the root, and each edge is labeled by a substring from S. • All edges coming out of a given vertex have different labels, and all such labels bhave different prefixes (not counting the empty prefix). • To each leaf there corresponds a suffix from S, and this suffix is obtained by concatenating all labels on all edges on the path from the root to the leaf.
Suffix tree • Edges are directed away from the root, and each edge is labeled by a substring from S. • All edges coming out of a given vertex have different labels, and all such labels bhave different prefixes (not counting the empty prefix). • To each leaf there corresponds a suffix from S, and this suffix is obtained by concatenating all labels on all edges on the path from the root to the leaf.
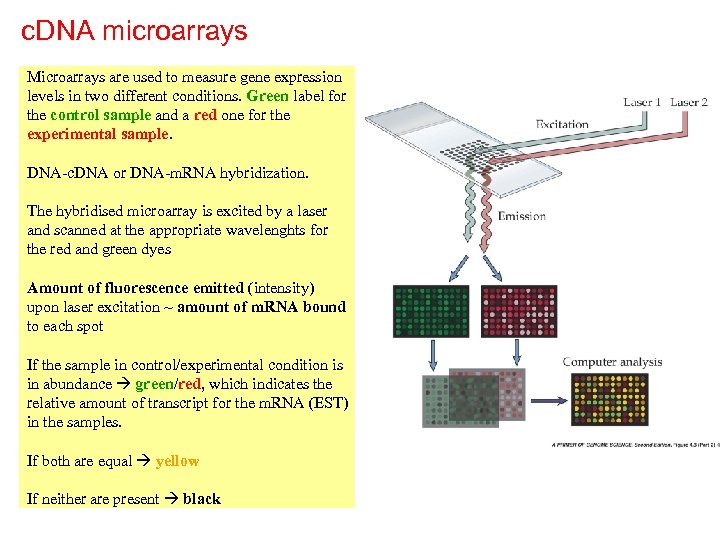 c. DNA microarrays Microarrays are used to measure gene expression levels in two different conditions. Green label for the control sample and a red one for the experimental sample. DNA-c. DNA or DNA-m. RNA hybridization. The hybridised microarray is excited by a laser and scanned at the appropriate wavelenghts for the red and green dyes Amount of fluorescence emitted (intensity) upon laser excitation ~ amount of m. RNA bound to each spot If the sample in control/experimental condition is in abundance green/red, which indicates the relative amount of transcript for the m. RNA (EST) in the samples. If both are equal yellow If neither are present black
c. DNA microarrays Microarrays are used to measure gene expression levels in two different conditions. Green label for the control sample and a red one for the experimental sample. DNA-c. DNA or DNA-m. RNA hybridization. The hybridised microarray is excited by a laser and scanned at the appropriate wavelenghts for the red and green dyes Amount of fluorescence emitted (intensity) upon laser excitation ~ amount of m. RNA bound to each spot If the sample in control/experimental condition is in abundance green/red, which indicates the relative amount of transcript for the m. RNA (EST) in the samples. If both are equal yellow If neither are present black
 Scanning of microarrays • Confocal laser scanning microscopy • Laser beam excites each spot of DNA • Amount of fluorescence detected • Different lasers used for different wavelengths – Cy 3 – Cy 5 laser detection
Scanning of microarrays • Confocal laser scanning microscopy • Laser beam excites each spot of DNA • Amount of fluorescence detected • Different lasers used for different wavelengths – Cy 3 – Cy 5 laser detection
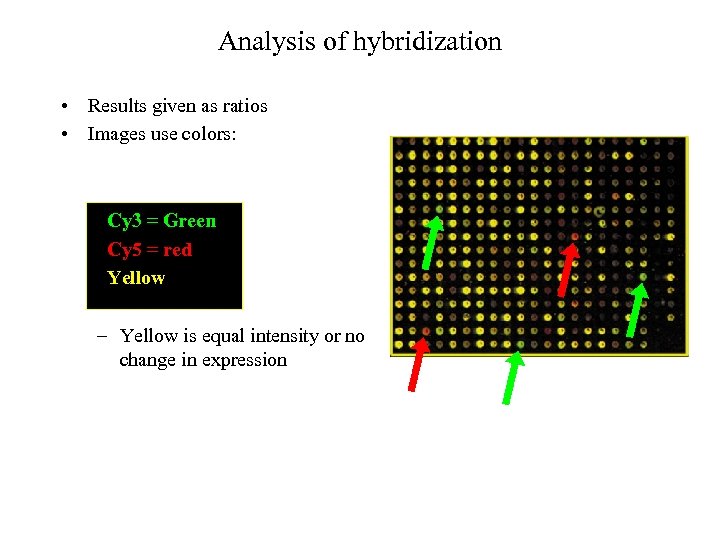 Analysis of hybridization • Results given as ratios • Images use colors: Cy 3 = Green Cy 5 = red Yellow – Yellow is equal intensity or no change in expression
Analysis of hybridization • Results given as ratios • Images use colors: Cy 3 = Green Cy 5 = red Yellow – Yellow is equal intensity or no change in expression
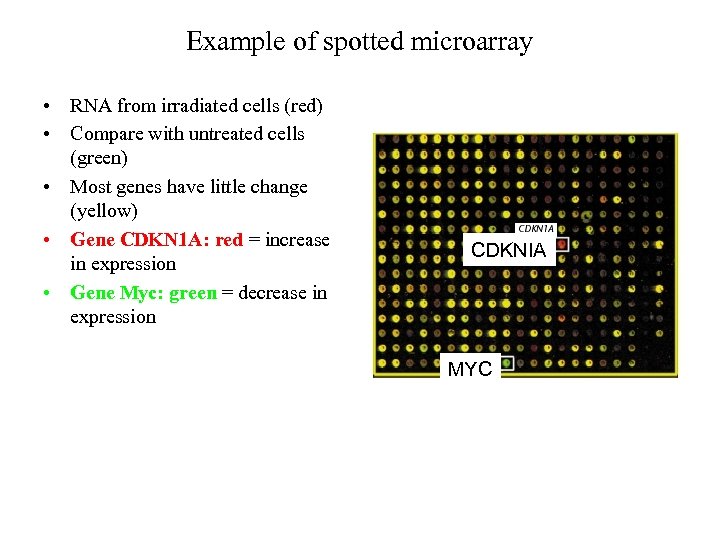 Example of spotted microarray • RNA from irradiated cells (red) • Compare with untreated cells (green) • Most genes have little change (yellow) • Gene CDKN 1 A: red = increase in expression • Gene Myc: green = decrease in expression CDKNIA MYC
Example of spotted microarray • RNA from irradiated cells (red) • Compare with untreated cells (green) • Most genes have little change (yellow) • Gene CDKN 1 A: red = increase in expression • Gene Myc: green = decrease in expression CDKNIA MYC
 Visualizing the hybridized target on a microarray can be performed by using either a confocal detector or a charge couple detector (CCD) camera. Microarray images produced with a pin-and-loop arrayer. (A) Two common undesirable features are indicated, namely high local background (arrow head) and scratches (two arrows) that would suggest “flagging” of the associated spots. (B) A close-up of a portion of the array demonstrates the uniformity of relative hybridization within each spot and differences in the red: green ratio of reach clone.
Visualizing the hybridized target on a microarray can be performed by using either a confocal detector or a charge couple detector (CCD) camera. Microarray images produced with a pin-and-loop arrayer. (A) Two common undesirable features are indicated, namely high local background (arrow head) and scratches (two arrows) that would suggest “flagging” of the associated spots. (B) A close-up of a portion of the array demonstrates the uniformity of relative hybridization within each spot and differences in the red: green ratio of reach clone.
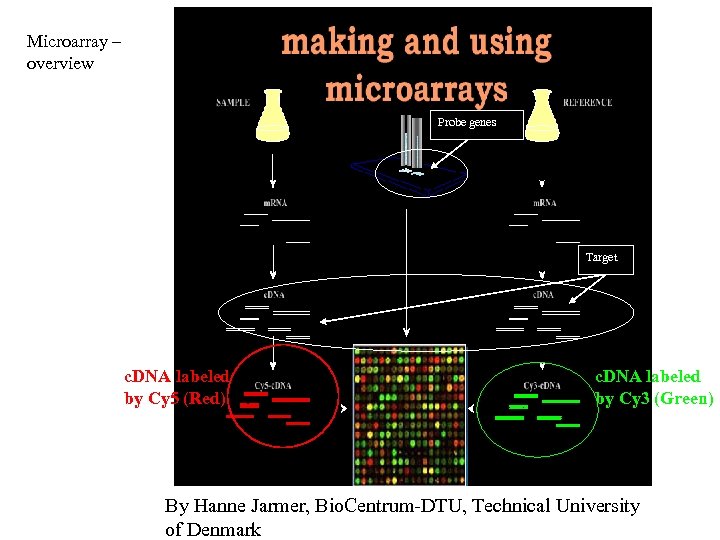 Microarray – overview Probe genes Target c. DNA labeled by Cy 5 (Red) c. DNA labeled by Cy 3 (Green) By Hanne Jarmer, Bio. Centrum-DTU, Technical University of Denmark
Microarray – overview Probe genes Target c. DNA labeled by Cy 5 (Red) c. DNA labeled by Cy 3 (Green) By Hanne Jarmer, Bio. Centrum-DTU, Technical University of Denmark
 What can we learn from the microarray data ? (1) Microarray permits an integrated approach to biology, in which genetic regulation can be examined allows us to build a gene network (2) Classification of disease, diagnosis, prognostic (judgment of the likely or expected development of a disease) prediction and pharmaceutical applications
What can we learn from the microarray data ? (1) Microarray permits an integrated approach to biology, in which genetic regulation can be examined allows us to build a gene network (2) Classification of disease, diagnosis, prognostic (judgment of the likely or expected development of a disease) prediction and pharmaceutical applications
 Co-expression of gene expression • Co-expressed genes involved in common processes clustering of genes Examples • Genes required for nutrition and stress responses • Genes whose products encode components of metabolic pathways • Genes encoding subunits of multi-subunit complexes such as the ribosome, the proteasome and the nucleosome are coordinately expressed • Ribosome - site of cellular protein synthesis • Proteasome - large multi-enzyme complexes that digest proteins • Nucleosome – A length of DNA consisting of about 140 base pairs makes two turns around the histone core thus forming a nucleosome. • Animation - http: //www. johnkyrk. com/index. html
Co-expression of gene expression • Co-expressed genes involved in common processes clustering of genes Examples • Genes required for nutrition and stress responses • Genes whose products encode components of metabolic pathways • Genes encoding subunits of multi-subunit complexes such as the ribosome, the proteasome and the nucleosome are coordinately expressed • Ribosome - site of cellular protein synthesis • Proteasome - large multi-enzyme complexes that digest proteins • Nucleosome – A length of DNA consisting of about 140 base pairs makes two turns around the histone core thus forming a nucleosome. • Animation - http: //www. johnkyrk. com/index. html
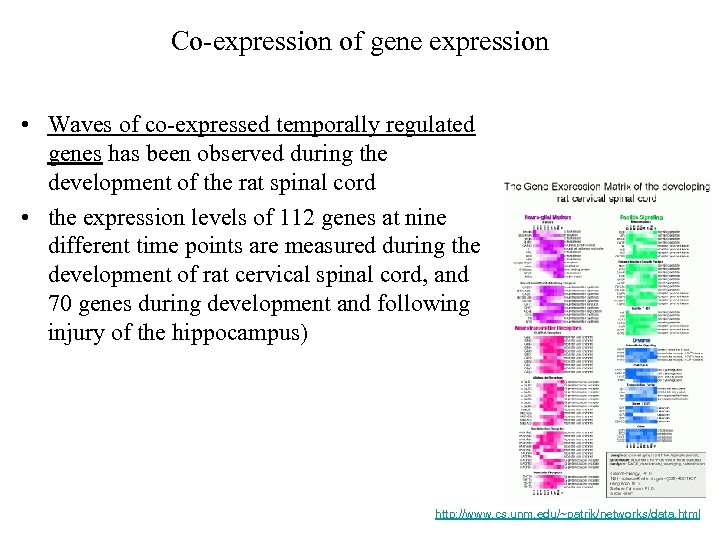 Co-expression of gene expression • Waves of co-expressed temporally regulated genes has been observed during the development of the rat spinal cord • the expression levels of 112 genes at nine different time points are measured during the development of rat cervical spinal cord, and 70 genes during development and following injury of the hippocampus) http: //www. cs. unm. edu/~patrik/networks/data. html
Co-expression of gene expression • Waves of co-expressed temporally regulated genes has been observed during the development of the rat spinal cord • the expression levels of 112 genes at nine different time points are measured during the development of rat cervical spinal cord, and 70 genes during development and following injury of the hippocampus) http: //www. cs. unm. edu/~patrik/networks/data. html
 Gene expression profile and phenotype • Profile or so-called signature • the combination of the m. RNAs (representing a subset of the total genotype) being expressed by the cell [Thomas A. Houpt, Nutrition, 827 (2000)] • Can be thought of a s a precise molecular definition of the cell in a specific state • Expression profile is a way to describe a phenotype, and can be used to characterize a wide variety of samples • Example • human cancer cell lines treated with 70000 agents independently or in combinations have been used to link drug activity with its mode of action • genes and putative drug targets
Gene expression profile and phenotype • Profile or so-called signature • the combination of the m. RNAs (representing a subset of the total genotype) being expressed by the cell [Thomas A. Houpt, Nutrition, 827 (2000)] • Can be thought of a s a precise molecular definition of the cell in a specific state • Expression profile is a way to describe a phenotype, and can be used to characterize a wide variety of samples • Example • human cancer cell lines treated with 70000 agents independently or in combinations have been used to link drug activity with its mode of action • genes and putative drug targets
 Affymetrix Gene. Chip experiment • RNA from four different types of brain tumors extracted • Extracted RNA hybridized to Gene. Chips containing approximately 6, 800 human genes • Identified gene expression profiles specific to each type of tumor
Affymetrix Gene. Chip experiment • RNA from four different types of brain tumors extracted • Extracted RNA hybridized to Gene. Chips containing approximately 6, 800 human genes • Identified gene expression profiles specific to each type of tumor
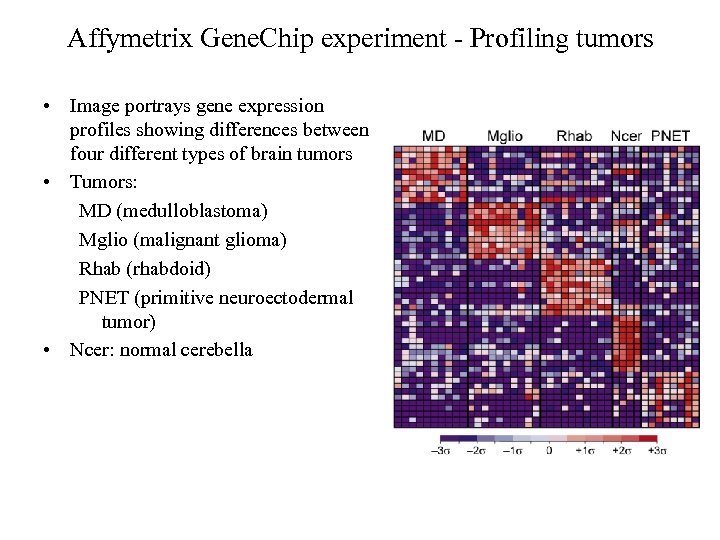 Affymetrix Gene. Chip experiment - Profiling tumors • Image portrays gene expression profiles showing differences between four different types of brain tumors • Tumors: MD (medulloblastoma) Mglio (malignant glioma) Rhab (rhabdoid) PNET (primitive neuroectodermal tumor) • Ncer: normal cerebella
Affymetrix Gene. Chip experiment - Profiling tumors • Image portrays gene expression profiles showing differences between four different types of brain tumors • Tumors: MD (medulloblastoma) Mglio (malignant glioma) Rhab (rhabdoid) PNET (primitive neuroectodermal tumor) • Ncer: normal cerebella
 • Gene expression differences for medulloblastoma correlated with response to chemotherapy • Those who failed to respond had a different profile from survivors • Can use this approach to determine which tumors are likely to respond to different treatment 60 different samples Affymetrix Gene. Chip experiment - Cancer diagnosis by microarray
• Gene expression differences for medulloblastoma correlated with response to chemotherapy • Those who failed to respond had a different profile from survivors • Can use this approach to determine which tumors are likely to respond to different treatment 60 different samples Affymetrix Gene. Chip experiment - Cancer diagnosis by microarray
 Microarray data generation, processing and analysis Two parts 1. Material processing and data collection 2. Information processing Five steps - Material processing and data collection • Array fabrication • Preparation of the biological samples to be studied • Extraction and labeling of the RNA from the samples • Hybridization of the labeled extracts to the array • Scanning of the hybridized array
Microarray data generation, processing and analysis Two parts 1. Material processing and data collection 2. Information processing Five steps - Material processing and data collection • Array fabrication • Preparation of the biological samples to be studied • Extraction and labeling of the RNA from the samples • Hybridization of the labeled extracts to the array • Scanning of the hybridized array
 Microarray data generation, processing and analysis Four steps - Information processing Image analysis • Image quantitation – locating the spots and measuring their fluorescence intensities • Data normalization and integration – construction of the gene expression matrix from sets of spot • Gene expression data analysis and Data analysis mining – finding differentially http: //www. mathworks. com/company/pressroom/i clustering mage_library/biotech. html expressed genes or clusters of similarly expressed genes • Generation from these analyses of new hypotheses about the underlying biological processes stimulates new hypotheses that in turn should be tested in follow-up experiments
Microarray data generation, processing and analysis Four steps - Information processing Image analysis • Image quantitation – locating the spots and measuring their fluorescence intensities • Data normalization and integration – construction of the gene expression matrix from sets of spot • Gene expression data analysis and Data analysis mining – finding differentially http: //www. mathworks. com/company/pressroom/i clustering mage_library/biotech. html expressed genes or clusters of similarly expressed genes • Generation from these analyses of new hypotheses about the underlying biological processes stimulates new hypotheses that in turn should be tested in follow-up experiments
 Microarray data processing and analysis http: //www. ebi. ac. uk/microarray/biology_intro. html Microarray experimental raw data (image data) spot quantitation matrices (row = spot on array, column = quantitation of that spot, i. e. mean, median, background) gene expression matrix data analysis (clustering or classification (SVD or PCA, see http: //public. lanl. gov/mewall/kluwer 2002. html))
Microarray data processing and analysis http: //www. ebi. ac. uk/microarray/biology_intro. html Microarray experimental raw data (image data) spot quantitation matrices (row = spot on array, column = quantitation of that spot, i. e. mean, median, background) gene expression matrix data analysis (clustering or classification (SVD or PCA, see http: //public. lanl. gov/mewall/kluwer 2002. html))
 Microarray data processing and analysis • Clustering – unsupervised method, i. e. do not assign some prior knowledge about function to the genes and/or samples • Class prediction (classification) – supervised method, i. e. assign some prior knowledge about function to the genes and/or samples • Next, the reverse engineering of gene regulatory networks based on the hypothesis that genes have similar expression profiles under a variety of conditions are likely to be regulated by common mechanisms • Cluster of genes some of these promoter sequences are obtained may contain a ‘signal’, e. g. a specific seq. pattern relevant to gene regulation • Application of different algorithms, or different parameters (such as distance measures), or different data filtering methods produce different results !! • What happen ? Well, it reflects the fact that cells typically carry out multiple processes simultaneously via multiple interacting pathways Future research directions – data analysis method, quality or reliability of data in the next generation of microarrays, where each spot is printed or synthesised multiple times estimate the measurement reliability using the standard deviation between the individual measurements data mining
Microarray data processing and analysis • Clustering – unsupervised method, i. e. do not assign some prior knowledge about function to the genes and/or samples • Class prediction (classification) – supervised method, i. e. assign some prior knowledge about function to the genes and/or samples • Next, the reverse engineering of gene regulatory networks based on the hypothesis that genes have similar expression profiles under a variety of conditions are likely to be regulated by common mechanisms • Cluster of genes some of these promoter sequences are obtained may contain a ‘signal’, e. g. a specific seq. pattern relevant to gene regulation • Application of different algorithms, or different parameters (such as distance measures), or different data filtering methods produce different results !! • What happen ? Well, it reflects the fact that cells typically carry out multiple processes simultaneously via multiple interacting pathways Future research directions – data analysis method, quality or reliability of data in the next generation of microarrays, where each spot is printed or synthesised multiple times estimate the measurement reliability using the standard deviation between the individual measurements data mining
 Microarray data management • Microarray database consists of three major parts – the gene expression matrix, gene annotation, and sample annotation • No established standards for microarray experiments or raw data processing • No standard ways for measuring gene expression levels
Microarray data management • Microarray database consists of three major parts – the gene expression matrix, gene annotation, and sample annotation • No established standards for microarray experiments or raw data processing • No standard ways for measuring gene expression levels
 Microarray data management • Microarray Gene Expression Data Society (MGED), http: //www. mged. org • Has developed recommendations for the Minimum Information About a Microarray Experiment (MIAME) that attempt to define the set of information sufficient to interpret the experiment, and the experiment, unambiguously, and to enable verification of the data • A set of guidelines for the describing an experiment, and the guidelines are translated into protocols enabling the electronic exchange of data in a standard format • The MIAME standard has been adopted and supported by the EBI Array. Express database, NCBI GEO and the CIBEX database at the DDBJ • Members of MGED joins with Rosetta Inpharmatics lead to the development of the microarray gene expression object model (MAGE-OM) and an XML-based extensible markup language (MAGE-ML) • MAGE is now built into a wide range of free available software, including BASE, Bio. Conductor, and TM 4.
Microarray data management • Microarray Gene Expression Data Society (MGED), http: //www. mged. org • Has developed recommendations for the Minimum Information About a Microarray Experiment (MIAME) that attempt to define the set of information sufficient to interpret the experiment, and the experiment, unambiguously, and to enable verification of the data • A set of guidelines for the describing an experiment, and the guidelines are translated into protocols enabling the electronic exchange of data in a standard format • The MIAME standard has been adopted and supported by the EBI Array. Express database, NCBI GEO and the CIBEX database at the DDBJ • Members of MGED joins with Rosetta Inpharmatics lead to the development of the microarray gene expression object model (MAGE-OM) and an XML-based extensible markup language (MAGE-ML) • MAGE is now built into a wide range of free available software, including BASE, Bio. Conductor, and TM 4.
 Microarray image processing • Labeled probe transform the fluorescence intensity transcript abundance most of these steps are done by software provided with commercial scanners • Image processing essentially involved four steps (1) image acquisition, (2) spot location, (3) computation of spot intensities, and (4) data reporting (1) image acquisition • Raw image of a microarray scan a 16 -bit image file of the intensity of fluorescence associated with each pixel a number between 0 and 65536 (i. e. 216). • Higher resolution use a 32 -bit image file (i. e. 0 ~ 4*109) • However the sources of experimental error are greater than the image resolution ! • Gain on the laser – too high intensity spots will converge on the same upper value, if the gain is too low information at the low end is lost in the background • Dyes (Cy 3 and Cy 5) quench with time, and different rates, it is not a good idea to repeatedly scan the same array (2) spot location • Spot location achieved by laying a grid over the image that places a square or circle around each spot • Always imperfections in the spacing of spots must be re-centered by deforming the grid so as to maximize the coverage of the spots by the circles
Microarray image processing • Labeled probe transform the fluorescence intensity transcript abundance most of these steps are done by software provided with commercial scanners • Image processing essentially involved four steps (1) image acquisition, (2) spot location, (3) computation of spot intensities, and (4) data reporting (1) image acquisition • Raw image of a microarray scan a 16 -bit image file of the intensity of fluorescence associated with each pixel a number between 0 and 65536 (i. e. 216). • Higher resolution use a 32 -bit image file (i. e. 0 ~ 4*109) • However the sources of experimental error are greater than the image resolution ! • Gain on the laser – too high intensity spots will converge on the same upper value, if the gain is too low information at the low end is lost in the background • Dyes (Cy 3 and Cy 5) quench with time, and different rates, it is not a good idea to repeatedly scan the same array (2) spot location • Spot location achieved by laying a grid over the image that places a square or circle around each spot • Always imperfections in the spacing of spots must be re-centered by deforming the grid so as to maximize the coverage of the spots by the circles
 Microarray image processing (3) computation of spot intensities • Spot intensities = mean intensity for each pixel within the circle surrounding a spot – mean (median) intensity of the background pixels immediately surrounding the spot (4) data reporting • Data is usually reported as a tab-delimited text file linkage of the data to genome databases • Microarray data or protocols are built on XML-based languages that allow storage and retrieval from public databases
Microarray image processing (3) computation of spot intensities • Spot intensities = mean intensity for each pixel within the circle surrounding a spot – mean (median) intensity of the background pixels immediately surrounding the spot (4) data reporting • Data is usually reported as a tab-delimited text file linkage of the data to genome databases • Microarray data or protocols are built on XML-based languages that allow storage and retrieval from public databases
 Summary • A comparison between c. DNA and oligonucleotides arrays • Patterns of gene expression – Deduce gene function based on patterns of expression vs. uses patterns of gene expression as a biomarker to classify samples
Summary • A comparison between c. DNA and oligonucleotides arrays • Patterns of gene expression – Deduce gene function based on patterns of expression vs. uses patterns of gene expression as a biomarker to classify samples
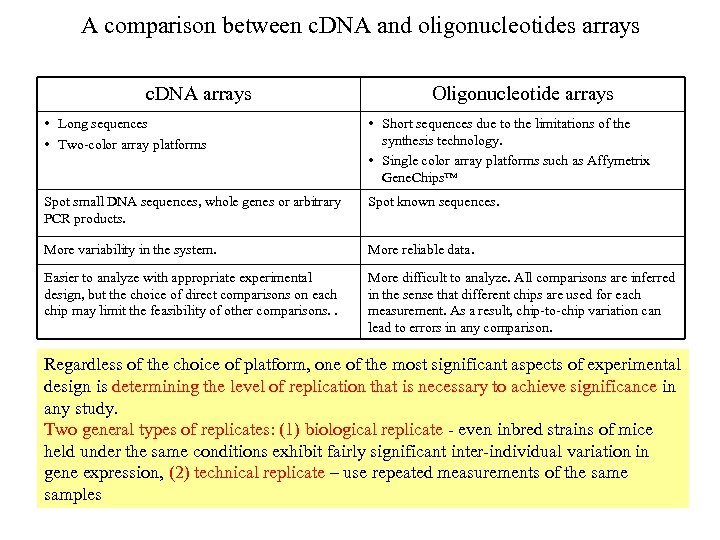 A comparison between c. DNA and oligonucleotides arrays c. DNA arrays Oligonucleotide arrays • Long sequences • Two-color array platforms • Short sequences due to the limitations of the synthesis technology. • Single color array platforms such as Affymetrix Gene. Chips™ Spot small DNA sequences, whole genes or arbitrary PCR products. Spot known sequences. More variability in the system. More reliable data. Easier to analyze with appropriate experimental design, but the choice of direct comparisons on each chip may limit the feasibility of other comparisons. . More difficult to analyze. All comparisons are inferred in the sense that different chips are used for each measurement. As a result, chip-to-chip variation can lead to errors in any comparison. Regardless of the choice of platform, one of the most significant aspects of experimental design is determining the level of replication that is necessary to achieve significance in any study. Two general types of replicates: (1) biological replicate - even inbred strains of mice held under the same conditions exhibit fairly significant inter-individual variation in gene expression, (2) technical replicate – use repeated measurements of the samples
A comparison between c. DNA and oligonucleotides arrays c. DNA arrays Oligonucleotide arrays • Long sequences • Two-color array platforms • Short sequences due to the limitations of the synthesis technology. • Single color array platforms such as Affymetrix Gene. Chips™ Spot small DNA sequences, whole genes or arbitrary PCR products. Spot known sequences. More variability in the system. More reliable data. Easier to analyze with appropriate experimental design, but the choice of direct comparisons on each chip may limit the feasibility of other comparisons. . More difficult to analyze. All comparisons are inferred in the sense that different chips are used for each measurement. As a result, chip-to-chip variation can lead to errors in any comparison. Regardless of the choice of platform, one of the most significant aspects of experimental design is determining the level of replication that is necessary to achieve significance in any study. Two general types of replicates: (1) biological replicate - even inbred strains of mice held under the same conditions exhibit fairly significant inter-individual variation in gene expression, (2) technical replicate – use repeated measurements of the samples
 Patterns of gene expression Deduce gene function based on patterns of expression Uses patterns of gene expression as a biomarker to classify samples Gene function Infer gene function by monitoring changes in expression resulting from experimental perturbations. • Search for genes exhibiting patterns of expression that differentiate the various groups • If the transcriptional differences between groups can be validated, these expression patterns can then be used as “biomarkers” in classifying other experimental subjects. Disadvantage Even simple changes can often produce a large number of transcriptional changes and these may be difficult to link to the underlying biological perturbation. Disadvantage In applications such as these, it is not essential that the genes themselves be linked causally to the underlying disease or other phenomenon that separates the classes. • Functional studies and searches for biomarkers are not mutually exclusive. Ultimately the most useful and informative biomarkers are likely those that can be linked causally to a disease or outcome. • Northerns are generally used to test a hypothesis based on biology. • Microarrays generate hypotheses that should be tested to validate them.
Patterns of gene expression Deduce gene function based on patterns of expression Uses patterns of gene expression as a biomarker to classify samples Gene function Infer gene function by monitoring changes in expression resulting from experimental perturbations. • Search for genes exhibiting patterns of expression that differentiate the various groups • If the transcriptional differences between groups can be validated, these expression patterns can then be used as “biomarkers” in classifying other experimental subjects. Disadvantage Even simple changes can often produce a large number of transcriptional changes and these may be difficult to link to the underlying biological perturbation. Disadvantage In applications such as these, it is not essential that the genes themselves be linked causally to the underlying disease or other phenomenon that separates the classes. • Functional studies and searches for biomarkers are not mutually exclusive. Ultimately the most useful and informative biomarkers are likely those that can be linked causally to a disease or outcome. • Northerns are generally used to test a hypothesis based on biology. • Microarrays generate hypotheses that should be tested to validate them.


