eebaf53c79c186961ef7c02b9408a057.ppt
- Количество слайдов: 48
 Matthew 13: 17 17 For verily I say unto you, That many prophets and righteous men have desired to see those things which ye see, and have not seen them; and to hear those things which ye hear, and have not heard them. © 2001 Timothy G. Standish
Matthew 13: 17 17 For verily I say unto you, That many prophets and righteous men have desired to see those things which ye see, and have not seen them; and to hear those things which ye hear, and have not heard them. © 2001 Timothy G. Standish
 DNA Sequencing Timothy G. Standish, Ph. D. © 2001 Timothy G. Standish
DNA Sequencing Timothy G. Standish, Ph. D. © 2001 Timothy G. Standish
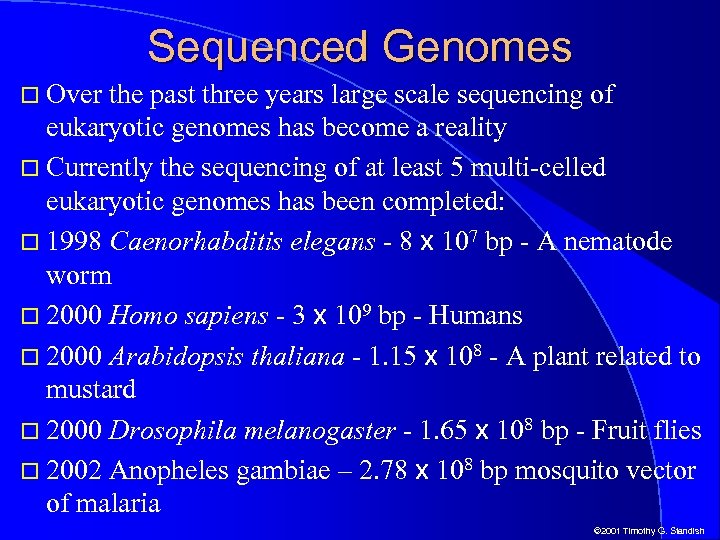 Sequenced Genomes Over the past three years large scale sequencing of eukaryotic genomes has become a reality Currently the sequencing of at least 5 multi-celled eukaryotic genomes has been completed: 1998 Caenorhabditis elegans - 8 x 107 bp - A nematode worm 2000 Homo sapiens - 3 x 109 bp - Humans 2000 Arabidopsis thaliana - 1. 15 x 108 - A plant related to mustard 2000 Drosophila melanogaster - 1. 65 x 108 bp - Fruit flies 2002 Anopheles gambiae – 2. 78 x 108 bp mosquito vector of malaria © 2001 Timothy G. Standish
Sequenced Genomes Over the past three years large scale sequencing of eukaryotic genomes has become a reality Currently the sequencing of at least 5 multi-celled eukaryotic genomes has been completed: 1998 Caenorhabditis elegans - 8 x 107 bp - A nematode worm 2000 Homo sapiens - 3 x 109 bp - Humans 2000 Arabidopsis thaliana - 1. 15 x 108 - A plant related to mustard 2000 Drosophila melanogaster - 1. 65 x 108 bp - Fruit flies 2002 Anopheles gambiae – 2. 78 x 108 bp mosquito vector of malaria © 2001 Timothy G. Standish
 New Technology Rapid sequencing of large complex genomes has been made possible by: Foundational work done over many years and… Dramatic improvement in DNA sequencing technology over the past few years In this presentation we will look at both the basic principles of DNA sequencing and how techniques have been refined to yield the dramatic results we now see © 2001 Timothy G. Standish
New Technology Rapid sequencing of large complex genomes has been made possible by: Foundational work done over many years and… Dramatic improvement in DNA sequencing technology over the past few years In this presentation we will look at both the basic principles of DNA sequencing and how techniques have been refined to yield the dramatic results we now see © 2001 Timothy G. Standish
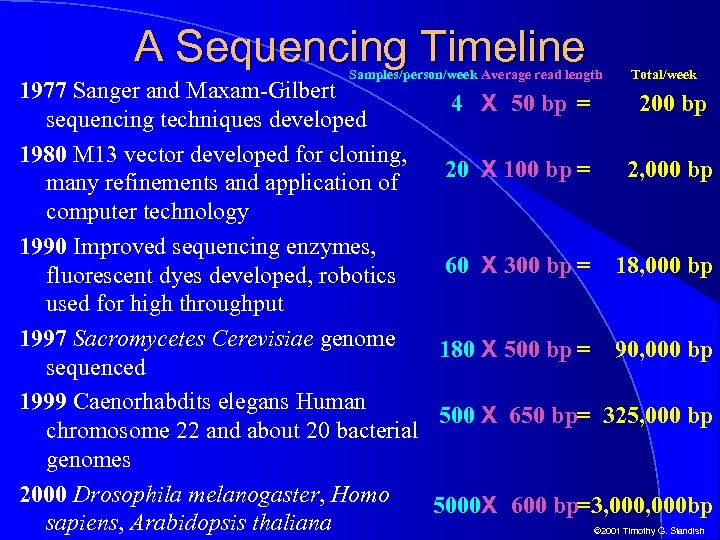 A Sequencing Timeline Samples/person/week Average read length Total/week 1977 Sanger and Maxam-Gilbert 4 X 50 bp = 200 bp sequencing techniques developed 1980 M 13 vector developed for cloning, 20 X 100 bp = 2, 000 bp many refinements and application of computer technology 1990 Improved sequencing enzymes, 60 X 300 bp = 18, 000 bp fluorescent dyes developed, robotics used for high throughput 1997 Sacromycetes Cerevisiae genome 180 X 500 bp = 90, 000 bp sequenced 1999 Caenorhabdits elegans Human 500 X 650 bp= 325, 000 bp chromosome 22 and about 20 bacterial genomes 2000 Drosophila melanogaster, Homo 5000 X 600 bp=3, 000 bp sapiens, Arabidopsis thaliana © 2001 Timothy G. Standish
A Sequencing Timeline Samples/person/week Average read length Total/week 1977 Sanger and Maxam-Gilbert 4 X 50 bp = 200 bp sequencing techniques developed 1980 M 13 vector developed for cloning, 20 X 100 bp = 2, 000 bp many refinements and application of computer technology 1990 Improved sequencing enzymes, 60 X 300 bp = 18, 000 bp fluorescent dyes developed, robotics used for high throughput 1997 Sacromycetes Cerevisiae genome 180 X 500 bp = 90, 000 bp sequenced 1999 Caenorhabdits elegans Human 500 X 650 bp= 325, 000 bp chromosome 22 and about 20 bacterial genomes 2000 Drosophila melanogaster, Homo 5000 X 600 bp=3, 000 bp sapiens, Arabidopsis thaliana © 2001 Timothy G. Standish
 Basic Principles All current practical DNA sequencing techniques can be divided into four major steps: 1. Labeling of DNA so that small quantities can be easily detected, traditionally done by labeling with either P 32 or S 35 2. Generation of fragments for which the specific bases at the 3’ end are known 3. Separation of fragments using gel electrophoresis sensitive ennough to resolve differenced in size of one nucleotide 4. Fragment detection © 2001 Timothy G. Standish
Basic Principles All current practical DNA sequencing techniques can be divided into four major steps: 1. Labeling of DNA so that small quantities can be easily detected, traditionally done by labeling with either P 32 or S 35 2. Generation of fragments for which the specific bases at the 3’ end are known 3. Separation of fragments using gel electrophoresis sensitive ennough to resolve differenced in size of one nucleotide 4. Fragment detection © 2001 Timothy G. Standish
 Outline 1. 2. 3. In this presentation we will look at: The Maxam-Gilbert and Sanger methods of DNA fragment generation Then methods for separation of fragments And finally examine how these techniques have been refined and automated to allow for rapid cheap sequencing of large quantities of DNA © 2001 Timothy G. Standish
Outline 1. 2. 3. In this presentation we will look at: The Maxam-Gilbert and Sanger methods of DNA fragment generation Then methods for separation of fragments And finally examine how these techniques have been refined and automated to allow for rapid cheap sequencing of large quantities of DNA © 2001 Timothy G. Standish
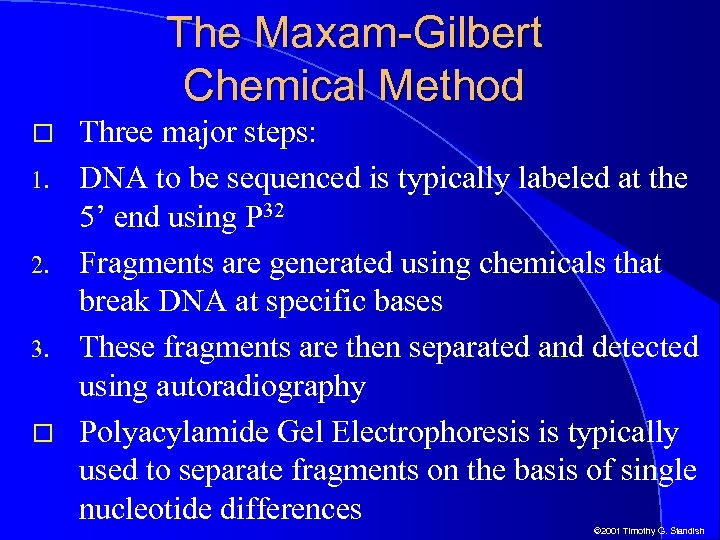 The Maxam-Gilbert Chemical Method 1. 2. 3. Three major steps: DNA to be sequenced is typically labeled at the 5’ end using P 32 Fragments are generated using chemicals that break DNA at specific bases These fragments are then separated and detected using autoradiography Polyacylamide Gel Electrophoresis is typically used to separate fragments on the basis of single nucleotide differences © 2001 Timothy G. Standish
The Maxam-Gilbert Chemical Method 1. 2. 3. Three major steps: DNA to be sequenced is typically labeled at the 5’ end using P 32 Fragments are generated using chemicals that break DNA at specific bases These fragments are then separated and detected using autoradiography Polyacylamide Gel Electrophoresis is typically used to separate fragments on the basis of single nucleotide differences © 2001 Timothy G. Standish
 2 Fragment Generation A number of chemicals will specifically modify the bases in DNA Modified bases can then be removed from the deoxyribose sugar to which they are attached on the sugar-phosphate DNA backbone Piperidine, a volatile secondary amine, is used to cleave the sugar-phosphate back bone of DNA at sites where bases were modified © 2001 Timothy G. Standish
2 Fragment Generation A number of chemicals will specifically modify the bases in DNA Modified bases can then be removed from the deoxyribose sugar to which they are attached on the sugar-phosphate DNA backbone Piperidine, a volatile secondary amine, is used to cleave the sugar-phosphate back bone of DNA at sites where bases were modified © 2001 Timothy G. Standish
 Cleavage at Specific Bases Typically 5 reactions are run: 1. Dimethylsulfate at p. H 8. 0 results in modification of guanine (G) 2. Piperidine formate at p. H 2. 0 breaks glycosidic bonds between deoxyribose and both purines, guanine (G) and adenine (A), by protination of nitrogen atoms 3. Hydrazine (rocket fuel!) opens pyrimidine rings on both pyrimidines, cytosine (C) and thymine (T) 4. Hydrazine in the presence of 1. 5 M Na. Cl only reacts with C 5. 1. 2 N Na. OH at 90 o. C strongly cleaves at A and may also weakly cleave at C © 2001 Timothy G. Standish
Cleavage at Specific Bases Typically 5 reactions are run: 1. Dimethylsulfate at p. H 8. 0 results in modification of guanine (G) 2. Piperidine formate at p. H 2. 0 breaks glycosidic bonds between deoxyribose and both purines, guanine (G) and adenine (A), by protination of nitrogen atoms 3. Hydrazine (rocket fuel!) opens pyrimidine rings on both pyrimidines, cytosine (C) and thymine (T) 4. Hydrazine in the presence of 1. 5 M Na. Cl only reacts with C 5. 1. 2 N Na. OH at 90 o. C strongly cleaves at A and may also weakly cleave at C © 2001 Timothy G. Standish
 Cleavage at Specific Bases The trick in chemical sequencing is to not allow the reactions to go to completion Partial reactions run using the following conditions will result in a series of labeled DNA fragments whose final base is known: Dimethylsulfate at p. H 8. 0 ------> G Piperidine formate at p. H 2. 0 -------> G and A Hydrazine ---------------> C and T Hydrazine in 1. 5 M Na. Cl ------> C 1. 2 N Na. OH at 90 o. C ---------> A and some C © 2001 Timothy G. Standish
Cleavage at Specific Bases The trick in chemical sequencing is to not allow the reactions to go to completion Partial reactions run using the following conditions will result in a series of labeled DNA fragments whose final base is known: Dimethylsulfate at p. H 8. 0 ------> G Piperidine formate at p. H 2. 0 -------> G and A Hydrazine ---------------> C and T Hydrazine in 1. 5 M Na. Cl ------> C 1. 2 N Na. OH at 90 o. C ---------> A and some C © 2001 Timothy G. Standish
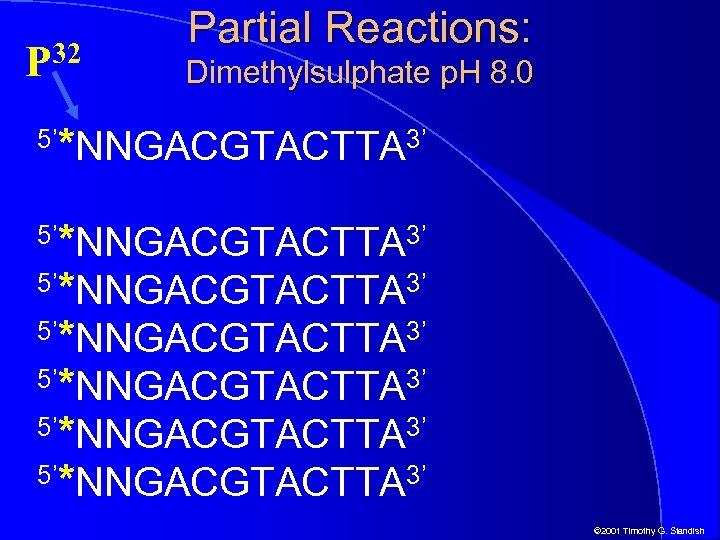 32 P 5’ Partial Reactions: Dimethylsulphate p. H 8. 0 *NNGACGTACTTA 3’ 5’*NNGACGTACTTA 3’ 5’*NNGACGTACTTA 3’ 5’ © 2001 Timothy G. Standish
32 P 5’ Partial Reactions: Dimethylsulphate p. H 8. 0 *NNGACGTACTTA 3’ 5’*NNGACGTACTTA 3’ 5’*NNGACGTACTTA 3’ 5’ © 2001 Timothy G. Standish
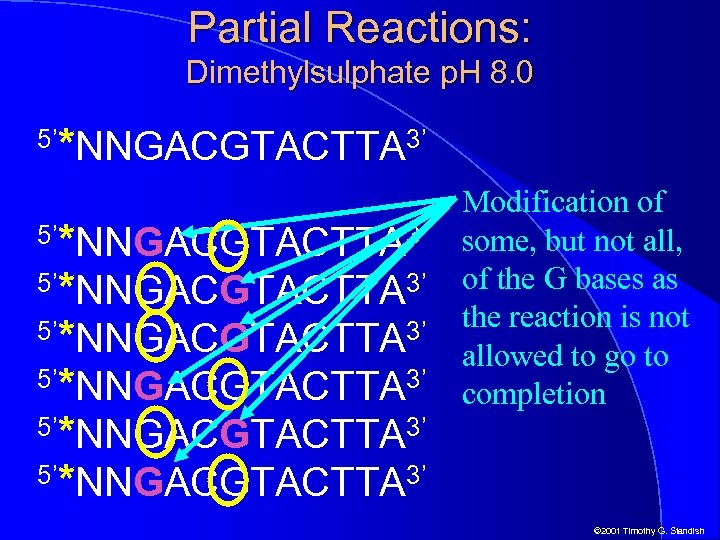 Partial Reactions: Dimethylsulphate p. H 8. 0 5’ *NNGACGTACTTA 3’ 5’*NNGACGTACTTA 3’ 5’*NNGACGTACTTA 3’ 5’ Modification of some, but not all, of the G bases as the reaction is not allowed to go to completion © 2001 Timothy G. Standish
Partial Reactions: Dimethylsulphate p. H 8. 0 5’ *NNGACGTACTTA 3’ 5’*NNGACGTACTTA 3’ 5’*NNGACGTACTTA 3’ 5’ Modification of some, but not all, of the G bases as the reaction is not allowed to go to completion © 2001 Timothy G. Standish
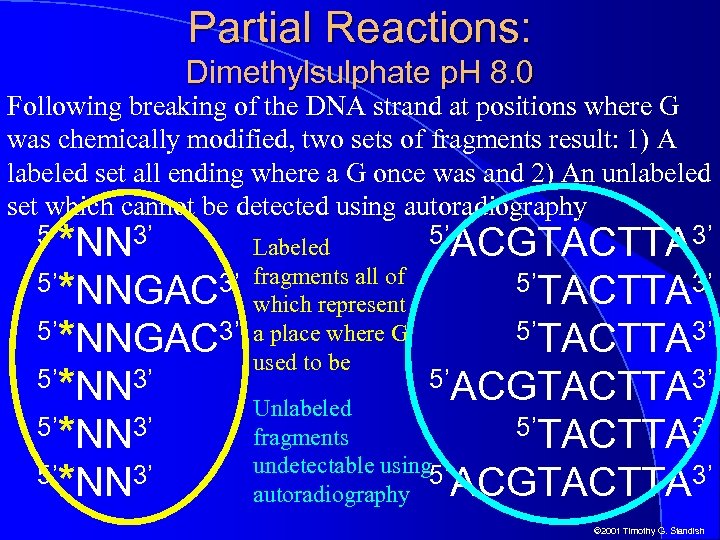 Partial Reactions: Dimethylsulphate p. H 8. 0 Following breaking of the DNA strand at positions where G was chemically modified, two sets of fragments result: 1) A labeled set all ending where a G once was and 2) An unlabeled set which cannot be detected using autoradiography *NN 3’ 5’*NNGAC 3’ 5’*NN 3’ 5’ ACGTACTTA 3’ 5’TACTTA 3’ 5’ACGTACTTA 3’ Unlabeled 5’TACTTA 3’ fragments undetectable using 5’ ACGTACTTA 3’ autoradiography 5’ Labeled fragments all of which represent a place where G used to be © 2001 Timothy G. Standish
Partial Reactions: Dimethylsulphate p. H 8. 0 Following breaking of the DNA strand at positions where G was chemically modified, two sets of fragments result: 1) A labeled set all ending where a G once was and 2) An unlabeled set which cannot be detected using autoradiography *NN 3’ 5’*NNGAC 3’ 5’*NN 3’ 5’ ACGTACTTA 3’ 5’TACTTA 3’ 5’ACGTACTTA 3’ Unlabeled 5’TACTTA 3’ fragments undetectable using 5’ ACGTACTTA 3’ autoradiography 5’ Labeled fragments all of which represent a place where G used to be © 2001 Timothy G. Standish
 Partial Reactions: Hydrazine 5’ *NNGACGTACTTA 3’ Some, but not 5’*NNGACGTACTTA 3’ all, C and T 5’*NNGACGTACTTA 3’ bases are 5’*NNGACGTACTTA 3’ modified as the 5’*NNGACGTACTTA 3’ reaction is not 5’*NNGACGTACTTA 3’ allowed to go 5’*NNGACGTACTTA 3’ to completion © 2001 Timothy G. Standish
Partial Reactions: Hydrazine 5’ *NNGACGTACTTA 3’ Some, but not 5’*NNGACGTACTTA 3’ all, C and T 5’*NNGACGTACTTA 3’ bases are 5’*NNGACGTACTTA 3’ modified as the 5’*NNGACGTACTTA 3’ reaction is not 5’*NNGACGTACTTA 3’ allowed to go 5’*NNGACGTACTTA 3’ to completion © 2001 Timothy G. Standish
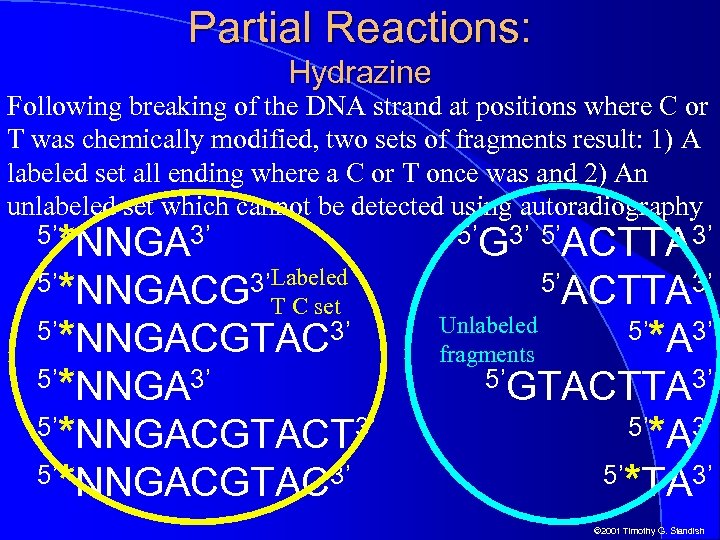 Partial Reactions: Hydrazine Following breaking of the DNA strand at positions where C or T was chemically modified, two sets of fragments result: 1) A labeled set all ending where a C or T once was and 2) An unlabeled set which cannot be detected using autoradiography *NNGA 3’ 5’*NNGACG 3’Labeled T C set 5’*NNGACGTAC 3’ 5’*NNGACGTACT 3’ 5’*NNGACGTAC 3’ 5’ 5’G 3’ 5’ACTTA 3’ Unlabeled fragments 5’ACTTA 3’ 5’ *A 3’ 5’GTACTTA 3’ 5’ 5’ *A 3’ *TA 3’ © 2001 Timothy G. Standish
Partial Reactions: Hydrazine Following breaking of the DNA strand at positions where C or T was chemically modified, two sets of fragments result: 1) A labeled set all ending where a C or T once was and 2) An unlabeled set which cannot be detected using autoradiography *NNGA 3’ 5’*NNGACG 3’Labeled T C set 5’*NNGACGTAC 3’ 5’*NNGACGTACT 3’ 5’*NNGACGTAC 3’ 5’ 5’G 3’ 5’ACTTA 3’ Unlabeled fragments 5’ACTTA 3’ 5’ *A 3’ 5’GTACTTA 3’ 5’ 5’ *A 3’ *TA 3’ © 2001 Timothy G. Standish
 Disadvantages Toxic chemicals Large amounts of radioactivity Sometimes ambiguous and frequently ugly sequencing gels Tricky to read autorads Lack of automated methods © 2001 Timothy G. Standish
Disadvantages Toxic chemicals Large amounts of radioactivity Sometimes ambiguous and frequently ugly sequencing gels Tricky to read autorads Lack of automated methods © 2001 Timothy G. Standish
 Sanger Sequencing The Sanger sequencing method takes advantage of the way that normal DNA replication occurs For DNA to be extended using normal DNA polymerases, a hydroxyl group must be present at the 3’ carbon on deoxyribose Fragments are generated by spiking reactions with small quantities 2’ 3’ dideoxy nucleotides which terminate polymerization whenever they are incorporated into DNA Polymerases used must lack 3’ to 5’ exonuclease proof reading activity for this method to work © 2001 Timothy G. Standish
Sanger Sequencing The Sanger sequencing method takes advantage of the way that normal DNA replication occurs For DNA to be extended using normal DNA polymerases, a hydroxyl group must be present at the 3’ carbon on deoxyribose Fragments are generated by spiking reactions with small quantities 2’ 3’ dideoxy nucleotides which terminate polymerization whenever they are incorporated into DNA Polymerases used must lack 3’ to 5’ exonuclease proof reading activity for this method to work © 2001 Timothy G. Standish
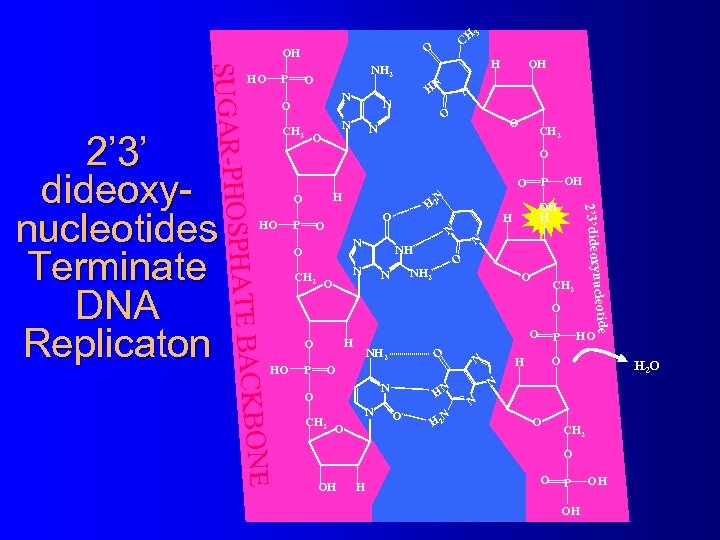
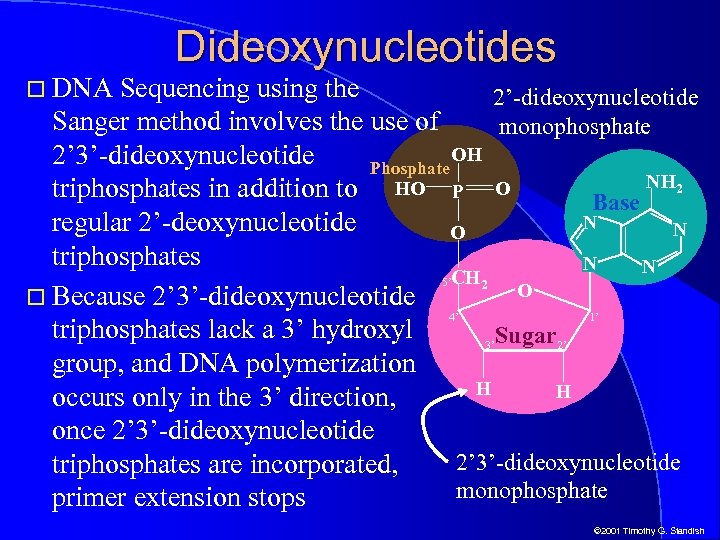 DNA Dideoxynucleotides Sequencing using the 2’-dideoxynucleotide Sanger method involves the use of monophosphate OH 2’ 3’-dideoxynucleotide Phosphate NH 2 triphosphates in addition to HO P O Base N regular 2’-deoxynucleotide N O triphosphates N N CH 2 5’ O Because 2’ 3’-dideoxynucleotide 4’ 1’ triphosphates lack a 3’ hydroxyl 3’ Sugar 2’ group, and DNA polymerization H OH H occurs only in the 3’ direction, once 2’ 3’-dideoxynucleotide triphosphates are incorporated, monophosphate primer extension stops © 2001 Timothy G. Standish
DNA Dideoxynucleotides Sequencing using the 2’-dideoxynucleotide Sanger method involves the use of monophosphate OH 2’ 3’-dideoxynucleotide Phosphate NH 2 triphosphates in addition to HO P O Base N regular 2’-deoxynucleotide N O triphosphates N N CH 2 5’ O Because 2’ 3’-dideoxynucleotide 4’ 1’ triphosphates lack a 3’ hydroxyl 3’ Sugar 2’ group, and DNA polymerization H OH H occurs only in the 3’ direction, once 2’ 3’-dideoxynucleotide triphosphates are incorporated, monophosphate primer extension stops © 2001 Timothy G. Standish
 O OH O O H N N O OH H H N O NH 2 N OH P O O CH 2 O O N O O O H N 2 H H 2 O N N CH 2 O H P HO OH O BONE CH 2 N HN N O O H P NH 2 HO P O HO O H O nucleotide CH 2 NH N CH 2 N H 2 O O O HO P N OH 2’ 3’dideoxy HO HN N N O PHATE BACK 2’ 3’ dideoxynucleotides Terminate DNA Replicaton N CH 2 H NH 2 O 3 E S B A S SUGAR-PHOS P HO CH
O OH O O H N N O OH H H N O NH 2 N OH P O O CH 2 O O N O O O H N 2 H H 2 O N N CH 2 O H P HO OH O BONE CH 2 N HN N O O H P NH 2 HO P O HO O H O nucleotide CH 2 NH N CH 2 N H 2 O O O HO P N OH 2’ 3’dideoxy HO HN N N O PHATE BACK 2’ 3’ dideoxynucleotides Terminate DNA Replicaton N CH 2 H NH 2 O 3 E S B A S SUGAR-PHOS P HO CH
 Making DNA Fragments In Sanger DNA sequencing reactions all the basic components needed to replicate DNA are used 4 reactions are set up, each containing: – – – DNA Polymerase Primer Template to be sequenced d. NTPs A small amount of one dd. NTP dd. ATP, dd. CTP, dd. GTP, dd. TTP As incorporation of dd. NTPs terminates DNA replication, a series of fragments is produced all terminating with the dd. NTP that was added to each reaction © 2001 Timothy G. Standish
Making DNA Fragments In Sanger DNA sequencing reactions all the basic components needed to replicate DNA are used 4 reactions are set up, each containing: – – – DNA Polymerase Primer Template to be sequenced d. NTPs A small amount of one dd. NTP dd. ATP, dd. CTP, dd. GTP, dd. TTP As incorporation of dd. NTPs terminates DNA replication, a series of fragments is produced all terminating with the dd. NTP that was added to each reaction © 2001 Timothy G. Standish
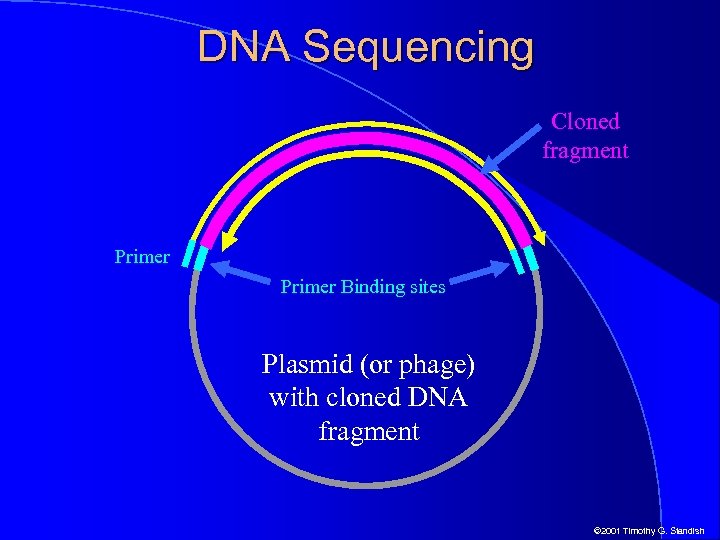 DNA Sequencing Cloned fragment Primer Binding sites Plasmid (or phage) with cloned DNA fragment © 2001 Timothy G. Standish
DNA Sequencing Cloned fragment Primer Binding sites Plasmid (or phage) with cloned DNA fragment © 2001 Timothy G. Standish
 The dd. ATP Reaction Pol. 3’AATAGCATGGTACTGATCTTACGCTAT 5’ Pol. 5’TTATCGTACCATGACTAGA 5’TTATCGTACCATGACTAGATGCGATA Let me Through! Oh come on! Not Again! Agggg…. 5’TTATCGTACCATGACTA 5’TTATCGTACCATGACTAGATGCGATA © 2001 Timothy G. Standish
The dd. ATP Reaction Pol. 3’AATAGCATGGTACTGATCTTACGCTAT 5’ Pol. 5’TTATCGTACCATGACTAGA 5’TTATCGTACCATGACTAGATGCGATA Let me Through! Oh come on! Not Again! Agggg…. 5’TTATCGTACCATGACTA 5’TTATCGTACCATGACTAGATGCGATA © 2001 Timothy G. Standish
 Separation of DNA Fragments All current practical sequencing methods rely on separation of DNA fragments in such a way that differences in length of a single base can be resolved This is typically done using polyacrylamide gel electrophoresis © 2001 Timothy G. Standish
Separation of DNA Fragments All current practical sequencing methods rely on separation of DNA fragments in such a way that differences in length of a single base can be resolved This is typically done using polyacrylamide gel electrophoresis © 2001 Timothy G. Standish
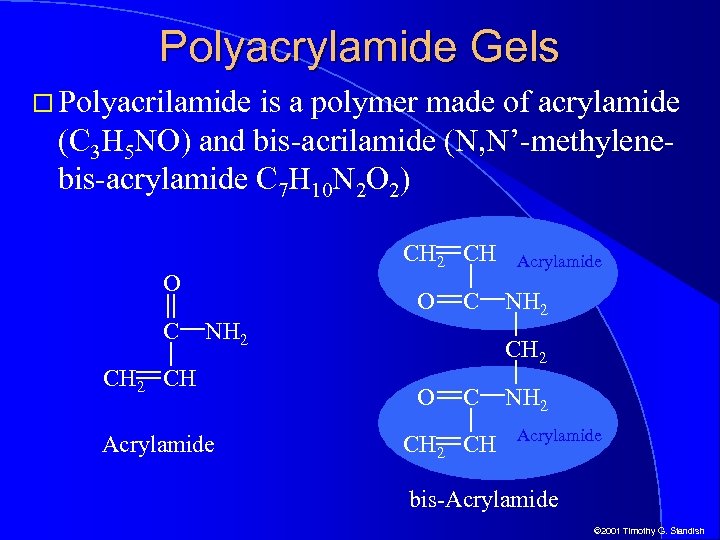 Polyacrylamide Gels Polyacrilamide is a polymer made of acrylamide (C 3 H 5 NO) and bis-acrilamide (N, N’-methylenebis-acrylamide C 7 H 10 N 2 O 2) CH 2 CH O C NH 2 CH Acrylamide NH 2 CH 2 O C CH 2 CH NH 2 Acrylamide bis-Acrylamide © 2001 Timothy G. Standish
Polyacrylamide Gels Polyacrilamide is a polymer made of acrylamide (C 3 H 5 NO) and bis-acrilamide (N, N’-methylenebis-acrylamide C 7 H 10 N 2 O 2) CH 2 CH O C NH 2 CH Acrylamide NH 2 CH 2 O C CH 2 CH NH 2 Acrylamide bis-Acrylamide © 2001 Timothy G. Standish
 Polyacrylamide Gels Acrylamide polymerizes in the presence of free radicals typically supplied by ammonium persulfate O C CH 2 CH O NH 2 CH SO 4 -. © 2001 Timothy G. Standish
Polyacrylamide Gels Acrylamide polymerizes in the presence of free radicals typically supplied by ammonium persulfate O C CH 2 CH O NH 2 CH SO 4 -. © 2001 Timothy G. Standish
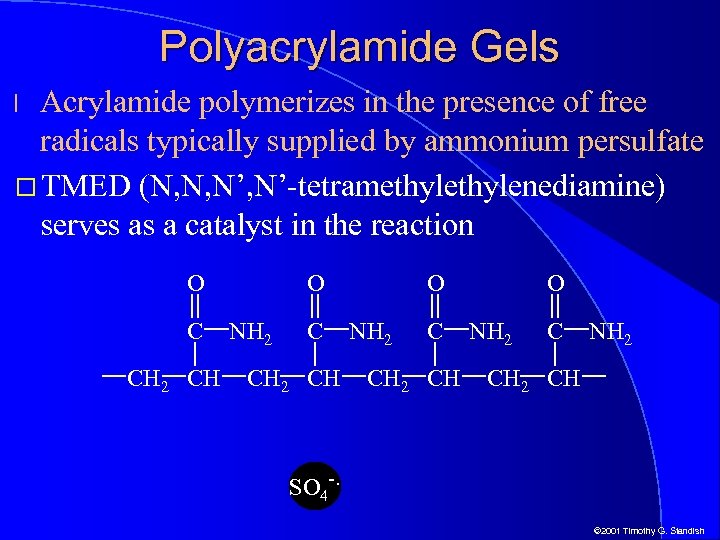 Polyacrylamide Gels Acrylamide polymerizes in the presence of free radicals typically supplied by ammonium persulfate TMED (N, N, N’-tetramethylenediamine) serves as a catalyst in the reaction l O C CH 2 CH O NH 2 CH 2 CH SO 4 -. © 2001 Timothy G. Standish
Polyacrylamide Gels Acrylamide polymerizes in the presence of free radicals typically supplied by ammonium persulfate TMED (N, N, N’-tetramethylenediamine) serves as a catalyst in the reaction l O C CH 2 CH O NH 2 CH 2 CH SO 4 -. © 2001 Timothy G. Standish
 Polyacrylamide Gels bis-Acrylamide polymerizes along with acrylamide forming cross-links between acrylamide chains O C CH 2 CH O NH 2 C CH 2 CH O C CH 2 CH NH 2 CH 2 O O O NH 2 C CH 2 CH O NH 2 CH bis-Acrylamide © 2001 Timothy G. Standish
Polyacrylamide Gels bis-Acrylamide polymerizes along with acrylamide forming cross-links between acrylamide chains O C CH 2 CH O NH 2 C CH 2 CH O C CH 2 CH NH 2 CH 2 O O O NH 2 C CH 2 CH O NH 2 CH bis-Acrylamide © 2001 Timothy G. Standish
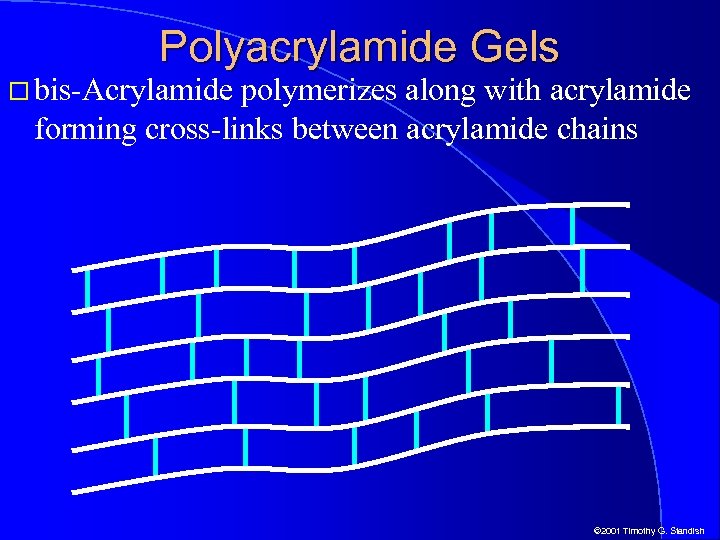 Polyacrylamide Gels bis-Acrylamide polymerizes along with acrylamide forming cross-links between acrylamide chains © 2001 Timothy G. Standish
Polyacrylamide Gels bis-Acrylamide polymerizes along with acrylamide forming cross-links between acrylamide chains © 2001 Timothy G. Standish
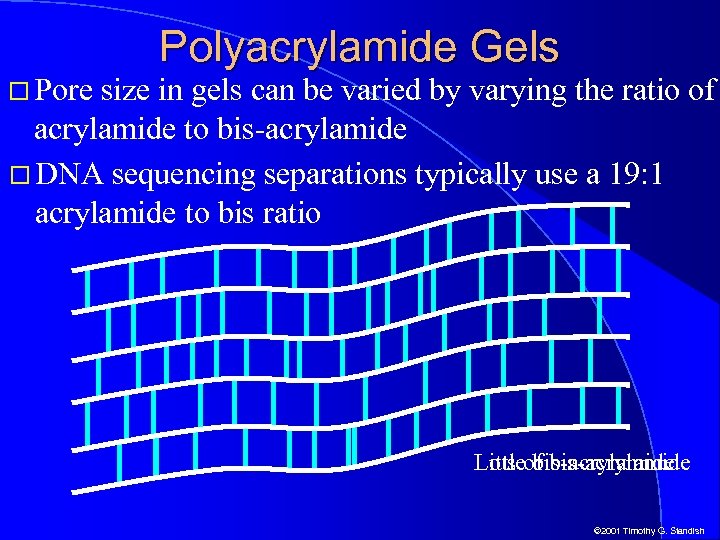 Pore Polyacrylamide Gels size in gels can be varied by varying the ratio of acrylamide to bis-acrylamide DNA sequencing separations typically use a 19: 1 acrylamide to bis ratio Little bis-acrylamide Lots of bis-acrylamide © 2001 Timothy G. Standish
Pore Polyacrylamide Gels size in gels can be varied by varying the ratio of acrylamide to bis-acrylamide DNA sequencing separations typically use a 19: 1 acrylamide to bis ratio Little bis-acrylamide Lots of bis-acrylamide © 2001 Timothy G. Standish
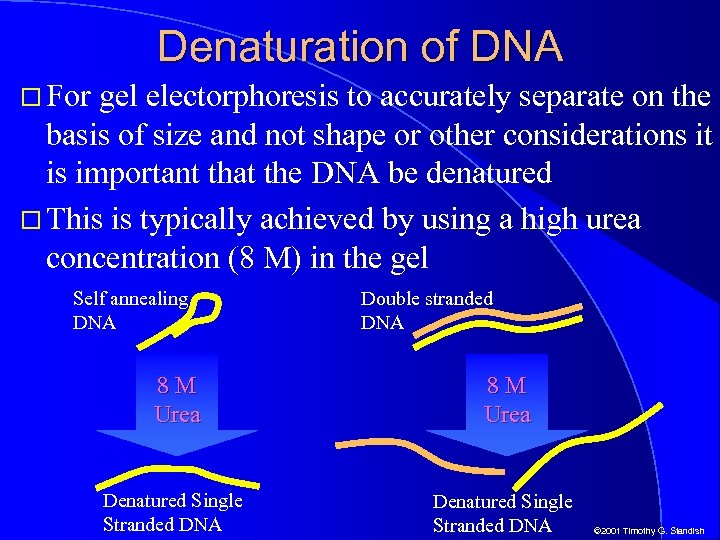 Denaturation of DNA For gel electorphoresis to accurately separate on the basis of size and not shape or other considerations it is important that the DNA be denatured This is typically achieved by using a high urea concentration (8 M) in the gel Self annealing DNA Double stranded DNA 8 M Urea Denatured Single Stranded DNA © 2001 Timothy G. Standish
Denaturation of DNA For gel electorphoresis to accurately separate on the basis of size and not shape or other considerations it is important that the DNA be denatured This is typically achieved by using a high urea concentration (8 M) in the gel Self annealing DNA Double stranded DNA 8 M Urea Denatured Single Stranded DNA © 2001 Timothy G. Standish
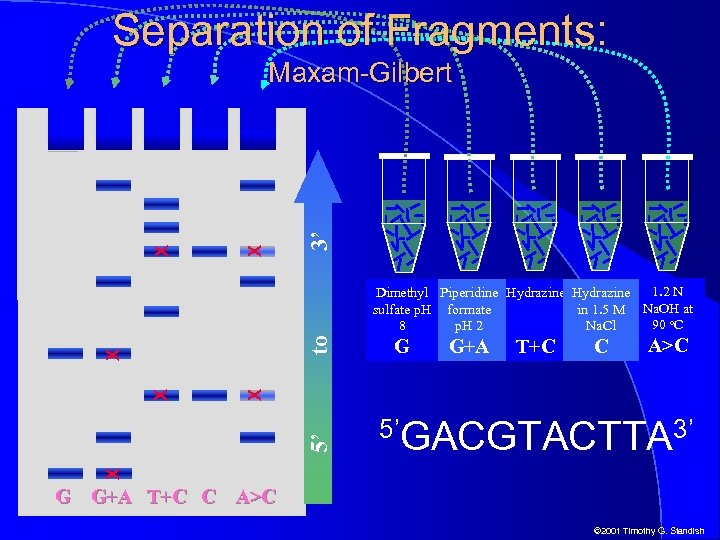 Separation of Fragments: 3’ X X Maxam-Gilbert G G+A T+C C A>C 5’GACGTACTTA 3’ X 5’ X X X to 1. 2 N Dimethyl Piperidine Hydrazine sulfate p. H formate in 1. 5 M Na. OH at 90 o. C 8 p. H 2 Na. Cl G G+A T+C C A>C © 2001 Timothy G. Standish
Separation of Fragments: 3’ X X Maxam-Gilbert G G+A T+C C A>C 5’GACGTACTTA 3’ X 5’ X X X to 1. 2 N Dimethyl Piperidine Hydrazine sulfate p. H formate in 1. 5 M Na. OH at 90 o. C 8 p. H 2 Na. Cl G G+A T+C C A>C © 2001 Timothy G. Standish
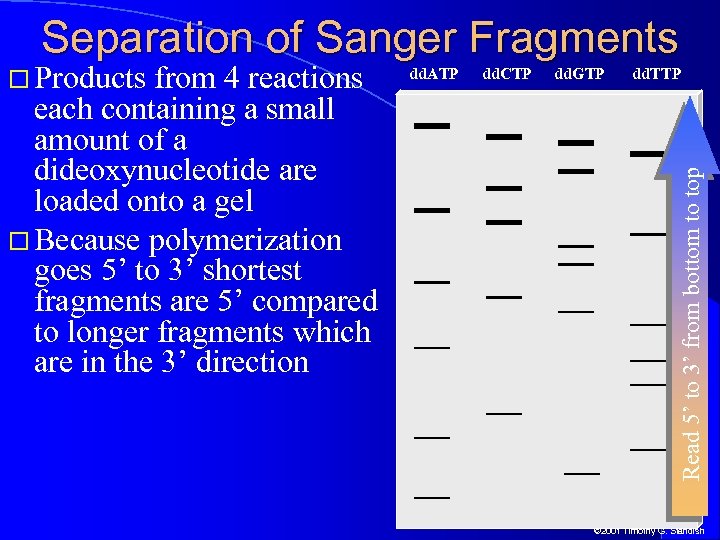 Separation of Sanger Fragments from 4 reactions each containing a small amount of a dideoxynucleotide are loaded onto a gel Because polymerization goes 5’ to 3’ shortest fragments are 5’ compared to longer fragments which are in the 3’ direction dd. ATP dd. CTP dd. GTP dd. TTP Read 5’ to 3’ from bottom to top Products © 2001 Timothy G. Standish
Separation of Sanger Fragments from 4 reactions each containing a small amount of a dideoxynucleotide are loaded onto a gel Because polymerization goes 5’ to 3’ shortest fragments are 5’ compared to longer fragments which are in the 3’ direction dd. ATP dd. CTP dd. GTP dd. TTP Read 5’ to 3’ from bottom to top Products © 2001 Timothy G. Standish
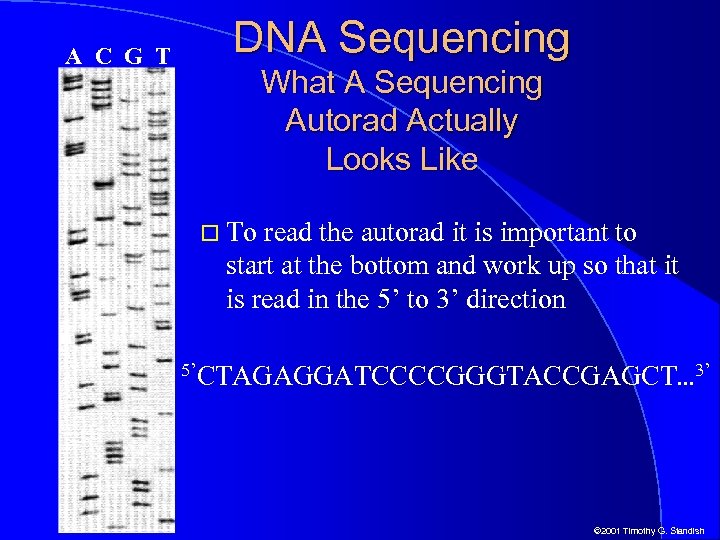 A C G T DNA Sequencing What A Sequencing Autorad Actually Looks Like To read the autorad it is important to start at the bottom and work up so that it is read in the 5’ to 3’ direction 5’CTAGAGGATCCCCGGGTACCGAGCT. . . 3’ © 2001 Timothy G. Standish
A C G T DNA Sequencing What A Sequencing Autorad Actually Looks Like To read the autorad it is important to start at the bottom and work up so that it is read in the 5’ to 3’ direction 5’CTAGAGGATCCCCGGGTACCGAGCT. . . 3’ © 2001 Timothy G. Standish
 Sequencing Method Refinements Because of difficulties intrinsic to the Maxam. Gilbert chemical sequencing strategy, efforts at improvement have been concentrated on the Sanger method Major improvements in the following areas have been achieved Labeling and detection Fragment separation DNA Polymerases used in sequencing and resulting strategies for generation of fragments Automation © 2001 Timothy G. Standish
Sequencing Method Refinements Because of difficulties intrinsic to the Maxam. Gilbert chemical sequencing strategy, efforts at improvement have been concentrated on the Sanger method Major improvements in the following areas have been achieved Labeling and detection Fragment separation DNA Polymerases used in sequencing and resulting strategies for generation of fragments Automation © 2001 Timothy G. Standish
 It Pros and Cons of the Sanger Method is more amenable to automation than Maxam. Gilbert Fewer dangerous chemicals are used, but acrylamide and P 32 or S 35 are still a problem Gels or autorads are generally cleaner looking and the reading of bases is a lot easier than Maxam. Gilbert data The bottom line: Without improvements in automation, detection and separation technologies Sanger sequencing is still very labor intensive © 2001 Timothy G. Standish
It Pros and Cons of the Sanger Method is more amenable to automation than Maxam. Gilbert Fewer dangerous chemicals are used, but acrylamide and P 32 or S 35 are still a problem Gels or autorads are generally cleaner looking and the reading of bases is a lot easier than Maxam. Gilbert data The bottom line: Without improvements in automation, detection and separation technologies Sanger sequencing is still very labor intensive © 2001 Timothy G. Standish
 Labeling and Detection Labeling using radioactive isotopes is difficult, dangerous and expensive Using biotin labeled primers has allowed conjugation of enzymes to fragments and their subsequent detection using substrates that change color in the presence of the enzyme This technique is clumsy, expensive, time consuming and unreliable It also may require transfer of fragments to membranes thus increasing labor and generally has not caught on © 2001 Timothy G. Standish
Labeling and Detection Labeling using radioactive isotopes is difficult, dangerous and expensive Using biotin labeled primers has allowed conjugation of enzymes to fragments and their subsequent detection using substrates that change color in the presence of the enzyme This technique is clumsy, expensive, time consuming and unreliable It also may require transfer of fragments to membranes thus increasing labor and generally has not caught on © 2001 Timothy G. Standish
 Labeling and Detection Another approach has involved development of very sensitive silver staining technologies I have tried this one, it is miserable and unreliable Read length on gels is typically short and creation of a permanent copy of the gel requires expensive additional equipment and supplies It may not involve isotopes, but it is such a hassle and the data is of such low quality that it is not worth the effort © 2001 Timothy G. Standish
Labeling and Detection Another approach has involved development of very sensitive silver staining technologies I have tried this one, it is miserable and unreliable Read length on gels is typically short and creation of a permanent copy of the gel requires expensive additional equipment and supplies It may not involve isotopes, but it is such a hassle and the data is of such low quality that it is not worth the effort © 2001 Timothy G. Standish
 Labeling and Detection The most significant advance in labeling has been the production of electrophoretically neutral dyes that fluoresce at specific wavelengths when excited by laser produced light over a very narrow range of wavelengths These dyes, when attached to primers allow detection down to 15 attomoles (10 -18) That’s less than 107 molecules! © 2001 Timothy G. Standish
Labeling and Detection The most significant advance in labeling has been the production of electrophoretically neutral dyes that fluoresce at specific wavelengths when excited by laser produced light over a very narrow range of wavelengths These dyes, when attached to primers allow detection down to 15 attomoles (10 -18) That’s less than 107 molecules! © 2001 Timothy G. Standish
 The Li-Cor System l l l Li-Cor of Lincoln, Nebraska was one of the first to implement fluorescent dyes as part of an automated sequencing system The Li-Cor system uses infrared lasers scanning a fixed line toward the bottom of an acrylamide slab gel Fluorescence of dyes attached to DNA fragments are detected as they pass the lasers and detectors Data in digital form is fed directly into a computer system where automated base calling is done A graphic representation of the data resembles a traditional autorad with bands appearing in 4 lanes © 2001 Timothy G. Standish
The Li-Cor System l l l Li-Cor of Lincoln, Nebraska was one of the first to implement fluorescent dyes as part of an automated sequencing system The Li-Cor system uses infrared lasers scanning a fixed line toward the bottom of an acrylamide slab gel Fluorescence of dyes attached to DNA fragments are detected as they pass the lasers and detectors Data in digital form is fed directly into a computer system where automated base calling is done A graphic representation of the data resembles a traditional autorad with bands appearing in 4 lanes © 2001 Timothy G. Standish
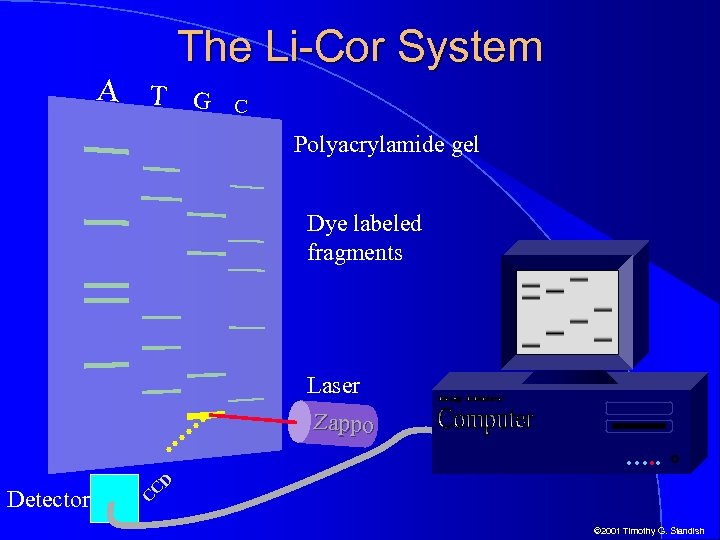 A T The Li-Cor System G C Polyacrylamide gel Dye labeled fragments Laser Zappo …. . Detector CD C © 2001 Timothy G. Standish
A T The Li-Cor System G C Polyacrylamide gel Dye labeled fragments Laser Zappo …. . Detector CD C © 2001 Timothy G. Standish
 The Pros and Cons Li-Cor systems major advantage is the lengths of its DNA reads – Because all fragments travel through the entire gel, resolution is sufficient to read over 1, 000 bases in a single run with over 99 % accuracy – This is better than just about any single run manual sequencing method Elimination of manual reading of autorads also eliminates human error and removes a labor intensive step P 32 or S 35 not used - another major advantage Tricky acrylamide gels still must be cast and loaded manually © 2001 Timothy G. Standish
The Pros and Cons Li-Cor systems major advantage is the lengths of its DNA reads – Because all fragments travel through the entire gel, resolution is sufficient to read over 1, 000 bases in a single run with over 99 % accuracy – This is better than just about any single run manual sequencing method Elimination of manual reading of autorads also eliminates human error and removes a labor intensive step P 32 or S 35 not used - another major advantage Tricky acrylamide gels still must be cast and loaded manually © 2001 Timothy G. Standish
 Applied Biosystems (ABI) has developed fluorescent dye systems further and improved methods for loading and electrophoresis Four dyes each of which fluoresce at a different wavelength, but having about the same impact on electrophoritic mobility can be used to label either primers or the nucleotides that terminate a reaction If terminator dyes are used, the entire sequencing reaction is reduced to one tube from 4 in conventional Sanger sequencing Instead of polyacrylamide slab gels, a single capillary can be used with a liquid polymer that is replaced after each individual run © 2001 Timothy G. Standish
Applied Biosystems (ABI) has developed fluorescent dye systems further and improved methods for loading and electrophoresis Four dyes each of which fluoresce at a different wavelength, but having about the same impact on electrophoritic mobility can be used to label either primers or the nucleotides that terminate a reaction If terminator dyes are used, the entire sequencing reaction is reduced to one tube from 4 in conventional Sanger sequencing Instead of polyacrylamide slab gels, a single capillary can be used with a liquid polymer that is replaced after each individual run © 2001 Timothy G. Standish
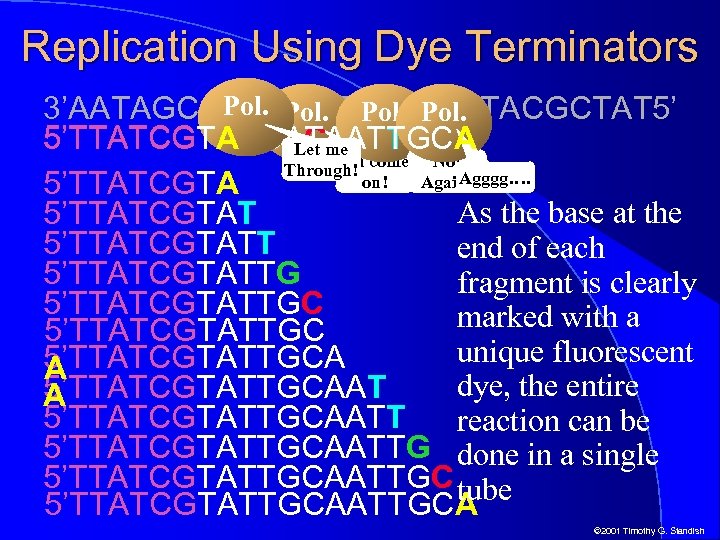 Replication Using Dye Terminators Pol. 3’AATAGCATAACGTTACGCTAT 5’ 5’TTATCGTACCATAATTGCA 5’TTATCGTACCATAATT 5’TTATCGTACCACme 5’TTATCGTA Let Oh come Not Through! Agggg…. on! Again! 5’TTATCGTA As the base at the 5’TTATCGTATT end of each 5’TTATCGTATTG fragment is clearly 5’TTATCGTATTGC marked with a 5’TTATCGTATTGC unique fluorescent 5’TTATCGTATTGCA A dye, the entire 5’TTATCGTATTGCAAT A 5’TTATCGTATTGCAATT reaction can be 5’TTATCGTATTGCAATTG done in a single 5’TTATCGTATTGCAATTGC tube 5’TTATCGTATTGCA © 2001 Timothy G. Standish
Replication Using Dye Terminators Pol. 3’AATAGCATAACGTTACGCTAT 5’ 5’TTATCGTACCATAATTGCA 5’TTATCGTACCATAATT 5’TTATCGTACCACme 5’TTATCGTA Let Oh come Not Through! Agggg…. on! Again! 5’TTATCGTA As the base at the 5’TTATCGTATT end of each 5’TTATCGTATTG fragment is clearly 5’TTATCGTATTGC marked with a 5’TTATCGTATTGC unique fluorescent 5’TTATCGTATTGCA A dye, the entire 5’TTATCGTATTGCAAT A 5’TTATCGTATTGCAATT reaction can be 5’TTATCGTATTGCAATTG done in a single 5’TTATCGTATTGCAATTGC tube 5’TTATCGTATTGCA © 2001 Timothy G. Standish
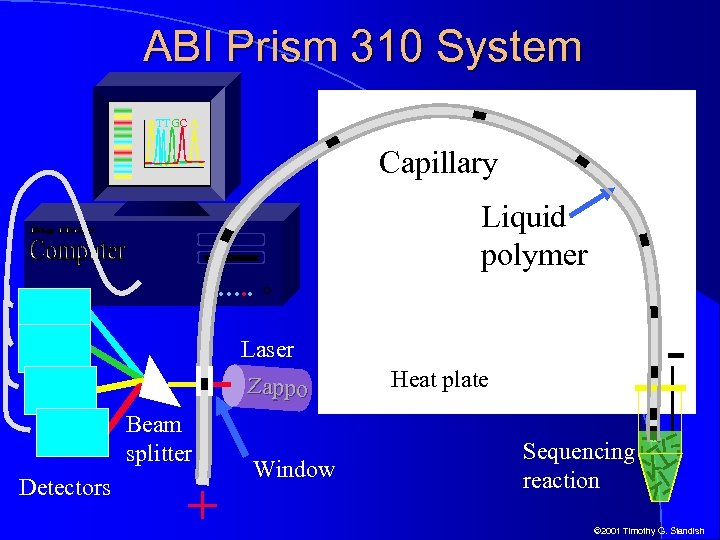 ABI Prism 310 System ATTGC A Capillary Liquid polymer …. . - Laser Zappo Beam splitter Detectors + Window Heat plate Sequencing reaction © 2001 Timothy G. Standish
ABI Prism 310 System ATTGC A Capillary Liquid polymer …. . - Laser Zappo Beam splitter Detectors + Window Heat plate Sequencing reaction © 2001 Timothy G. Standish
 The State of the Art ABI Prism 310 (1 capillary), 3100 (16 capillaries) and 3700 (96 capillaries) represent the current state of the art in automated sequencing machines A single ABI Prism 377 slab gel sequencer can run 115, 000 bases per day! The 3100 can run up to 184, 000 bases per day The 3700 can run up to 1, 104, 000 bases per day Large sequencing facilities, like Celera, have factories full of these machines which can run 24 hours a day with very little down time for routine maintenance © 2001 Timothy G. Standish
The State of the Art ABI Prism 310 (1 capillary), 3100 (16 capillaries) and 3700 (96 capillaries) represent the current state of the art in automated sequencing machines A single ABI Prism 377 slab gel sequencer can run 115, 000 bases per day! The 3100 can run up to 184, 000 bases per day The 3700 can run up to 1, 104, 000 bases per day Large sequencing facilities, like Celera, have factories full of these machines which can run 24 hours a day with very little down time for routine maintenance © 2001 Timothy G. Standish
 The State of the Art ABI Prism 3700 © 2001 Timothy G. Standish
The State of the Art ABI Prism 3700 © 2001 Timothy G. Standish
 © 2001 Timothy G. Standish
© 2001 Timothy G. Standish


