8e439ebe4429deefd79cb07569d8fb1f.ppt
- Количество слайдов: 41
 Mass Spectrometry in Life Science: Technology and Data-Evaluation H. Thiele Bruker Daltonik, Germany
Mass Spectrometry in Life Science: Technology and Data-Evaluation H. Thiele Bruker Daltonik, Germany
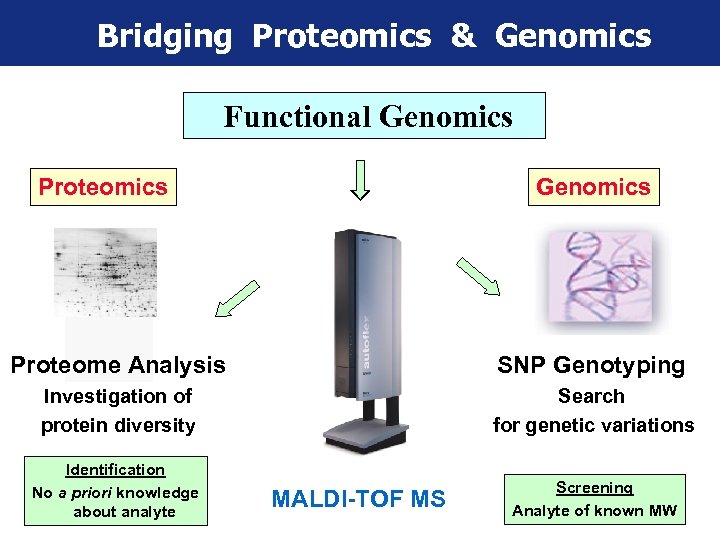 Bridging Proteomics & Genomics Functional Genomics Proteomics Genomics MALDI-TOF Mass Spectrometry Proteome Analysis SNP Genotyping Investigation of protein diversity Search for genetic variations Identification No a priori knowledge about analyte MALDI-TOF MS Screening Analyte of known MW
Bridging Proteomics & Genomics Functional Genomics Proteomics Genomics MALDI-TOF Mass Spectrometry Proteome Analysis SNP Genotyping Investigation of protein diversity Search for genetic variations Identification No a priori knowledge about analyte MALDI-TOF MS Screening Analyte of known MW
 The Technology Mass Spectrometer for Biopolymer Research
The Technology Mass Spectrometer for Biopolymer Research
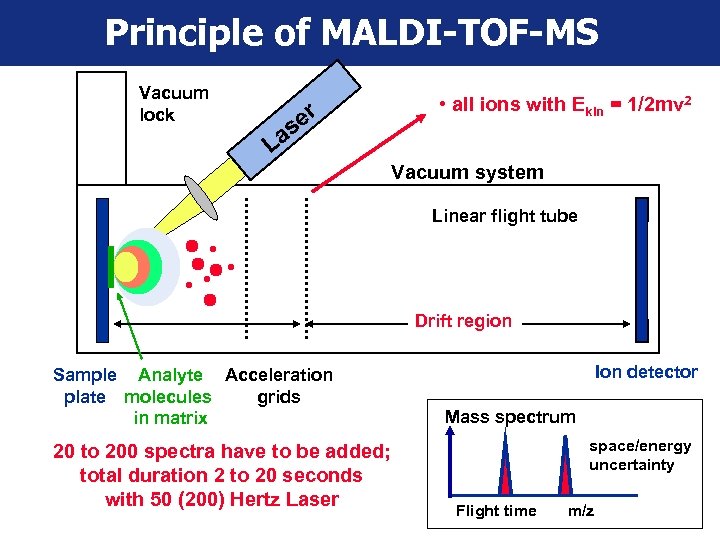 Principle of MALDI-TOF-MS Vacuum lock er as L • all ions with Ekin = 1/2 mv 2 Vacuum system Linear flight tube Drift region Sample Analyte Acceleration plate molecules grids in matrix 20 to 200 spectra have to be added; total duration 2 to 20 seconds with 50 (200) Hertz Laser Ion detector Mass spectrum space/energy uncertainty Flight time m/z
Principle of MALDI-TOF-MS Vacuum lock er as L • all ions with Ekin = 1/2 mv 2 Vacuum system Linear flight tube Drift region Sample Analyte Acceleration plate molecules grids in matrix 20 to 200 spectra have to be added; total duration 2 to 20 seconds with 50 (200) Hertz Laser Ion detector Mass spectrum space/energy uncertainty Flight time m/z
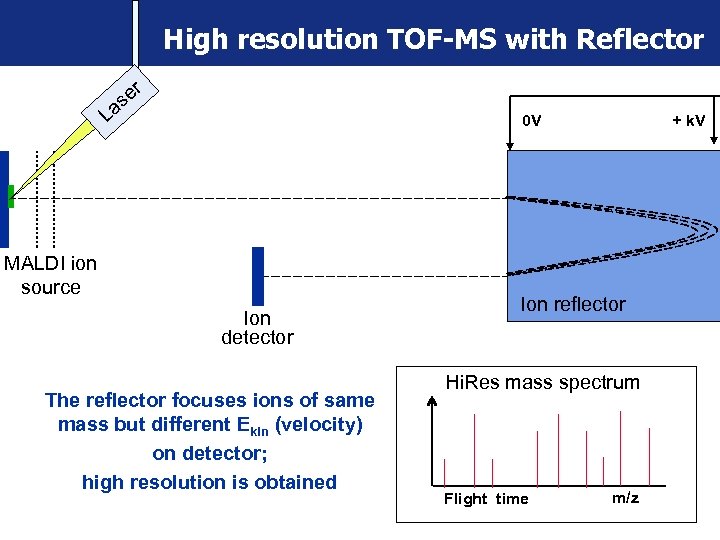 High resolution TOF-MS with Reflector er s La 0 V MALDI ion source Ion detector The reflector focuses ions of same mass but different Ekin (velocity) on detector; high resolution is obtained + k. V Ion reflector Hi. Res mass spectrum Flight time m/z
High resolution TOF-MS with Reflector er s La 0 V MALDI ion source Ion detector The reflector focuses ions of same mass but different Ekin (velocity) on detector; high resolution is obtained + k. V Ion reflector Hi. Res mass spectrum Flight time m/z
 MS/MS by PSD MS/MS = fragment ion or tandem mass spectromentry PSD = Post Source Decay
MS/MS by PSD MS/MS = fragment ion or tandem mass spectromentry PSD = Post Source Decay
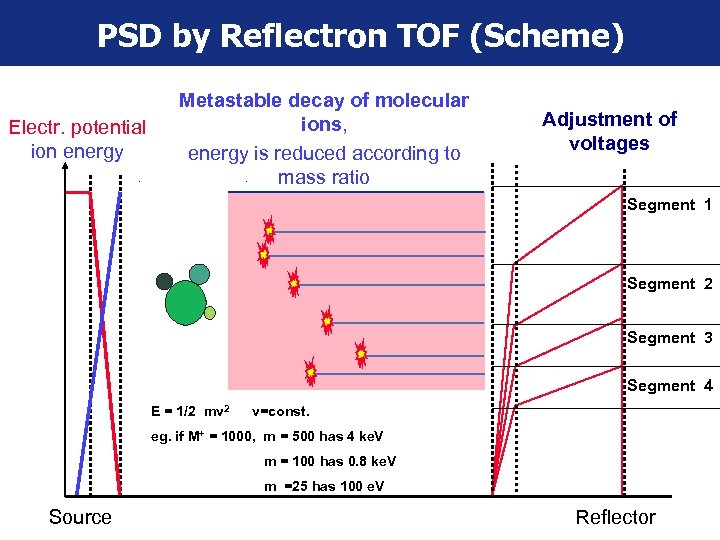 PSD by Reflectron TOF (Scheme) Electr. potential ion energy Metastable decay of molecular ions, energy is reduced according to mass ratio Adjustment of voltages Segment 1 Segment 2 Segment 3 Segment 4 E = 1/2 mv 2 v=const. eg. if M+ = 1000, m = 500 has 4 ke. V m = 100 has 0. 8 ke. V m =25 has 100 e. V Source Reflector
PSD by Reflectron TOF (Scheme) Electr. potential ion energy Metastable decay of molecular ions, energy is reduced according to mass ratio Adjustment of voltages Segment 1 Segment 2 Segment 3 Segment 4 E = 1/2 mv 2 v=const. eg. if M+ = 1000, m = 500 has 4 ke. V m = 100 has 0. 8 ke. V m =25 has 100 e. V Source Reflector
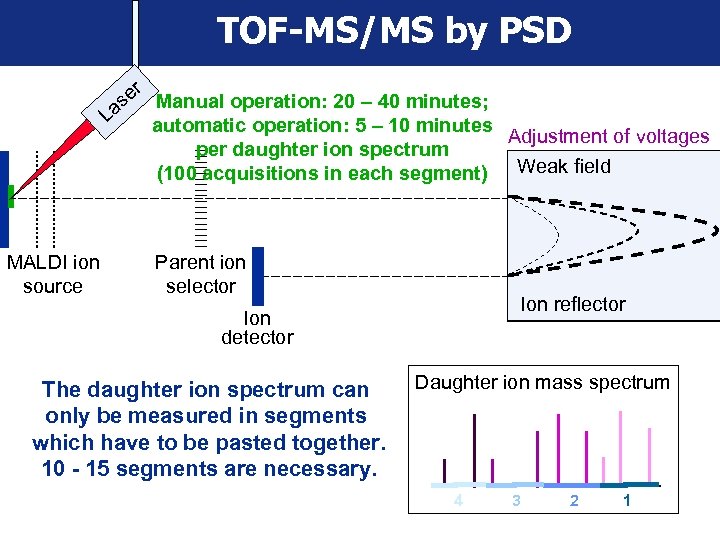 TOF-MS/MS by PSD er Manual operation: 20 – 40 minutes; s La automatic operation: 5 – 10 minutes Adjustment of voltages per daughter ion spectrum Weaker field (100 acquisitions in each segment) Strongfield MALDI ion source Parent ion selector Ion reflector Ion detector The daughter ion spectrum can only be measured in segments which have to be pasted together. 10 - 15 segments are necessary. Daughter ion mass spectrum 4 3 2 1
TOF-MS/MS by PSD er Manual operation: 20 – 40 minutes; s La automatic operation: 5 – 10 minutes Adjustment of voltages per daughter ion spectrum Weaker field (100 acquisitions in each segment) Strongfield MALDI ion source Parent ion selector Ion reflector Ion detector The daughter ion spectrum can only be measured in segments which have to be pasted together. 10 - 15 segments are necessary. Daughter ion mass spectrum 4 3 2 1
 In proteomics, many proteins have to be separated analysed fast to avoid degradation Regarding structure information, MALDI MS/MS appears to be optimal, but PSD is much too slow ! Consequence: Development of a fast MALDI MS/MS instrument !
In proteomics, many proteins have to be separated analysed fast to avoid degradation Regarding structure information, MALDI MS/MS appears to be optimal, but PSD is much too slow ! Consequence: Development of a fast MALDI MS/MS instrument !
 MALDI TOF/TOF with post-acceleration by potential LIFT
MALDI TOF/TOF with post-acceleration by potential LIFT
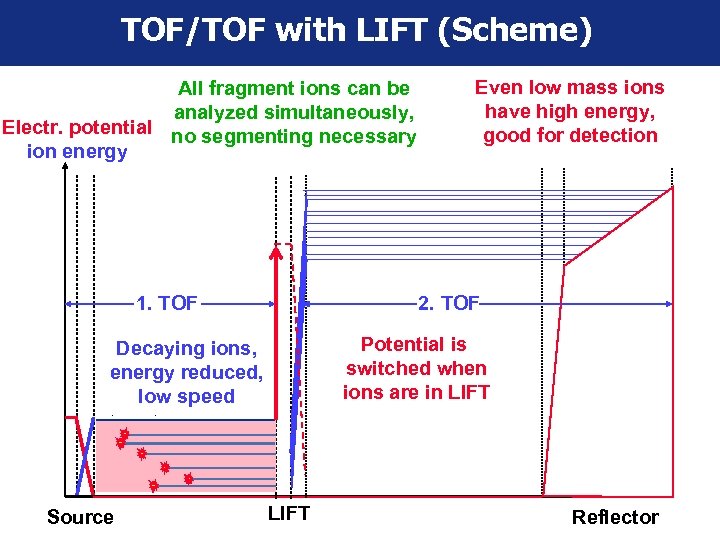 TOF/TOF with LIFT (Scheme) All fragment ions can be analyzed simultaneously, Electr. potential no segmenting necessary ion energy 1. TOF 2. TOF Potential is switched when ions are in LIFT Decaying ions, energy reduced, low speed Source Even low mass ions have high energy, good for detection LIFT Reflector
TOF/TOF with LIFT (Scheme) All fragment ions can be analyzed simultaneously, Electr. potential no segmenting necessary ion energy 1. TOF 2. TOF Potential is switched when ions are in LIFT Decaying ions, energy reduced, low speed Source Even low mass ions have high energy, good for detection LIFT Reflector
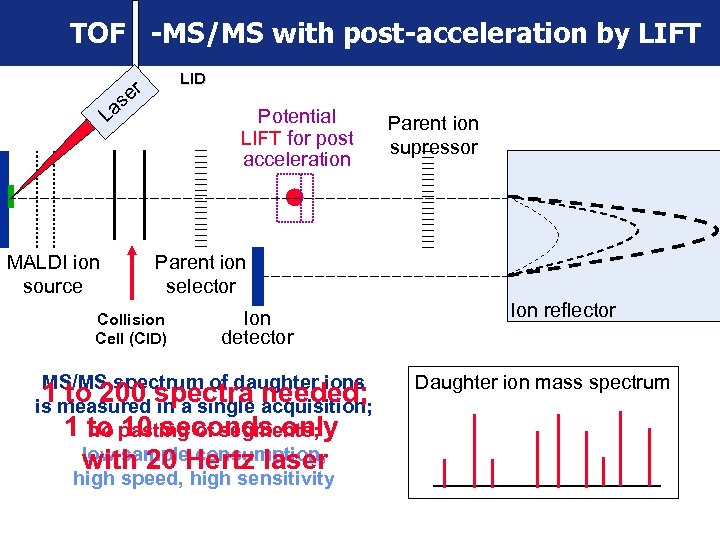 TOF -MS/MS with post-acceleration by LIFT LID er s La MALDI ion source Potential LIFT for post acceleration Parent ion supressor Parent ion selector Collision Cell (CID) Ion detector MS/MS spectrum of daughter ions 1 measured spectra acquisition; to 200 in a single needed; is 1 to pasting of segments; no 10 seconds only low sample consumption, with 20 Hertz laser high speed, high sensitivity Ion reflector Daughter ion mass spectrum
TOF -MS/MS with post-acceleration by LIFT LID er s La MALDI ion source Potential LIFT for post acceleration Parent ion supressor Parent ion selector Collision Cell (CID) Ion detector MS/MS spectrum of daughter ions 1 measured spectra acquisition; to 200 in a single needed; is 1 to pasting of segments; no 10 seconds only low sample consumption, with 20 Hertz laser high speed, high sensitivity Ion reflector Daughter ion mass spectrum
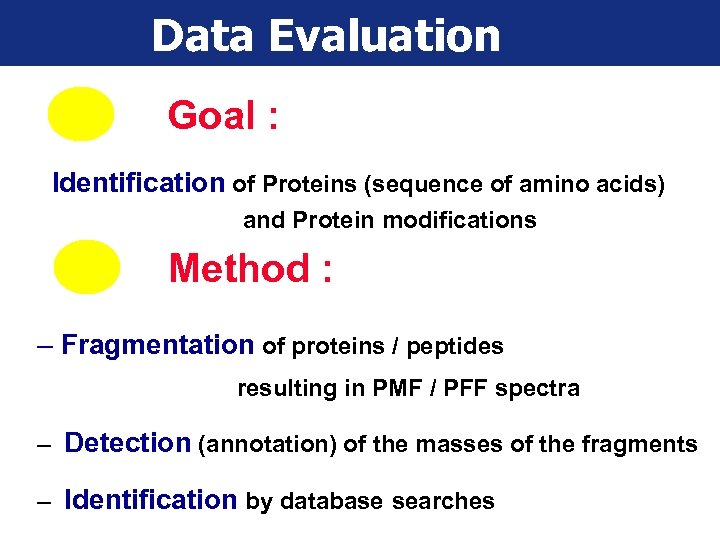 Data Evaluation Goal : Identification of Proteins (sequence of amino acids) and Protein modifications Method : – Fragmentation of proteins / peptides resulting in PMF / PFF spectra – Detection (annotation) of the masses of the fragments – Identification by database searches
Data Evaluation Goal : Identification of Proteins (sequence of amino acids) and Protein modifications Method : – Fragmentation of proteins / peptides resulting in PMF / PFF spectra – Detection (annotation) of the masses of the fragments – Identification by database searches
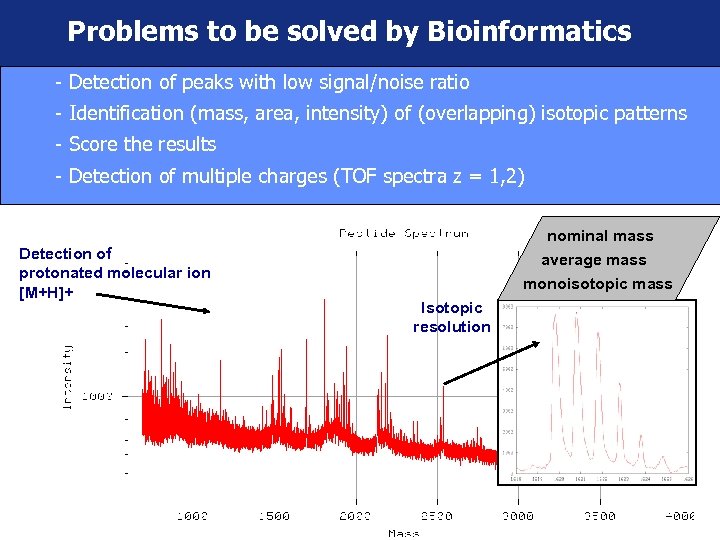 Problems to be solved by Bioinformatics - Detection of peaks with low signal/noise ratio - Identification (mass, area, intensity) of (overlapping) isotopic patterns - Score the results - Detection of multiple charges (TOF spectra z = 1, 2) Detection of protonated molecular ion [M+H]+ nominal mass average mass monoisotopic mass Isotopic resolution
Problems to be solved by Bioinformatics - Detection of peaks with low signal/noise ratio - Identification (mass, area, intensity) of (overlapping) isotopic patterns - Score the results - Detection of multiple charges (TOF spectra z = 1, 2) Detection of protonated molecular ion [M+H]+ nominal mass average mass monoisotopic mass Isotopic resolution
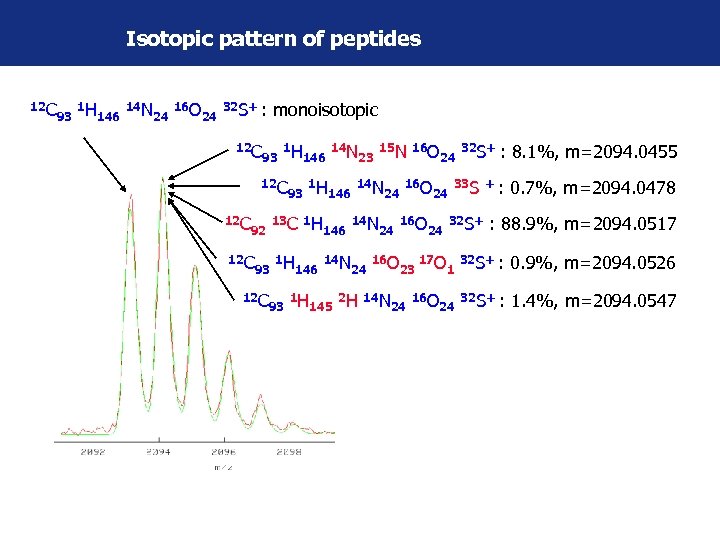 Isotopic pattern of peptides 12 C 93 1 H 146 14 N 24 16 O 24 32 S+ : 12 C monoisotopic 93 1 H 12 C 93 12 C 1 H 13 C 1 H 92 12 C 93 146 1 H 14 N 146 23 14 N 146 14 N 24 145 15 N 16 O 24 24 16 O 2 H 14 N 23 24 24 24 17 O 16 O 32 S+ : 8. 1%, m=2094. 0455 33 S + : 0. 7%, m=2094. 0478 24 32 S+ : 88. 9%, m=2094. 0517 1 32 S+ : 0. 9%, m=2094. 0526 24 32 S+ : 1. 4%, m=2094. 0547
Isotopic pattern of peptides 12 C 93 1 H 146 14 N 24 16 O 24 32 S+ : 12 C monoisotopic 93 1 H 12 C 93 12 C 1 H 13 C 1 H 92 12 C 93 146 1 H 14 N 146 23 14 N 146 14 N 24 145 15 N 16 O 24 24 16 O 2 H 14 N 23 24 24 24 17 O 16 O 32 S+ : 8. 1%, m=2094. 0455 33 S + : 0. 7%, m=2094. 0478 24 32 S+ : 88. 9%, m=2094. 0517 1 32 S+ : 0. 9%, m=2094. 0526 24 32 S+ : 1. 4%, m=2094. 0547
 Deisotoping: Assigning monoisotopic masses SNAP approach: • Peak selection - Damping of chemical noise using FFT filtering Baseline correction noise calculation peak search • Iterative search for isotopic patterns – Analysing the largest peaks first – Alignment of patterns using peak list heuristic and FFT deconvolution – Nonlinear fit using asymmetric line shape – Subtraction of analysed patterns • Reevaluation – Fit of intensities of overlapping patterns, optional addition of ICAT masses – Calculation of Quality Factor
Deisotoping: Assigning monoisotopic masses SNAP approach: • Peak selection - Damping of chemical noise using FFT filtering Baseline correction noise calculation peak search • Iterative search for isotopic patterns – Analysing the largest peaks first – Alignment of patterns using peak list heuristic and FFT deconvolution – Nonlinear fit using asymmetric line shape – Subtraction of analysed patterns • Reevaluation – Fit of intensities of overlapping patterns, optional addition of ICAT masses – Calculation of Quality Factor
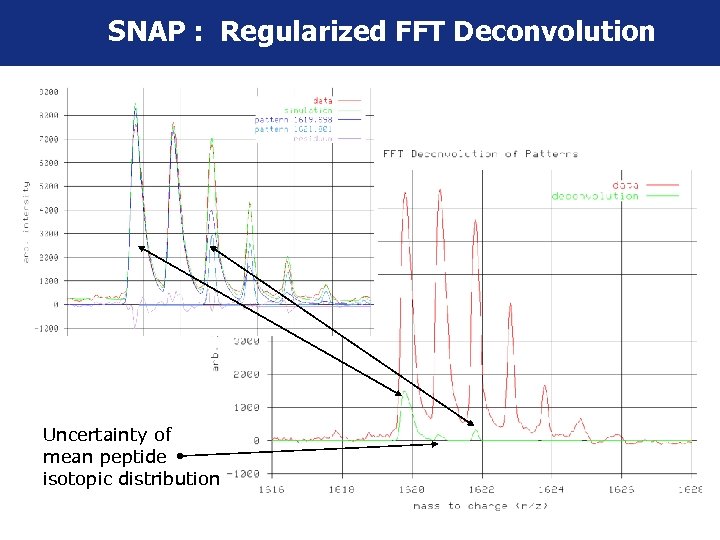 SNAP : Regularized FFT Deconvolution Uncertainty of mean peptide isotopic distribution
SNAP : Regularized FFT Deconvolution Uncertainty of mean peptide isotopic distribution
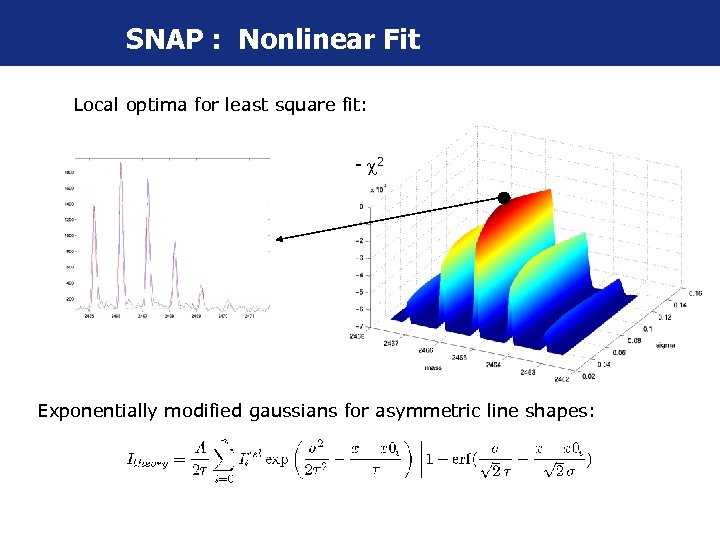 SNAP : Nonlinear Fit Local optima for least square fit: - 2 Exponentially modified gaussians for asymmetric line shapes:
SNAP : Nonlinear Fit Local optima for least square fit: - 2 Exponentially modified gaussians for asymmetric line shapes:
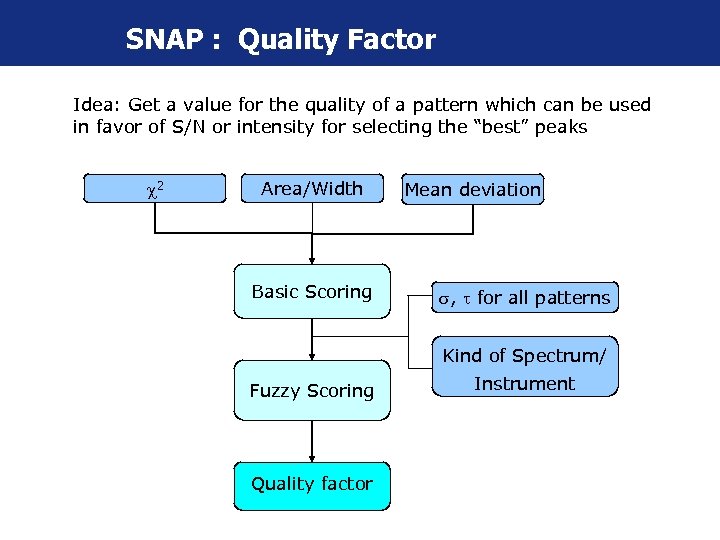 SNAP : Quality Factor Idea: Get a value for the quality of a pattern which can be used in favor of S/N or intensity for selecting the “best” peaks 2 Area/Width Basic Scoring Mean deviation , for all patterns Kind of Spectrum/ Fuzzy Scoring Quality factor Instrument
SNAP : Quality Factor Idea: Get a value for the quality of a pattern which can be used in favor of S/N or intensity for selecting the “best” peaks 2 Area/Width Basic Scoring Mean deviation , for all patterns Kind of Spectrum/ Fuzzy Scoring Quality factor Instrument
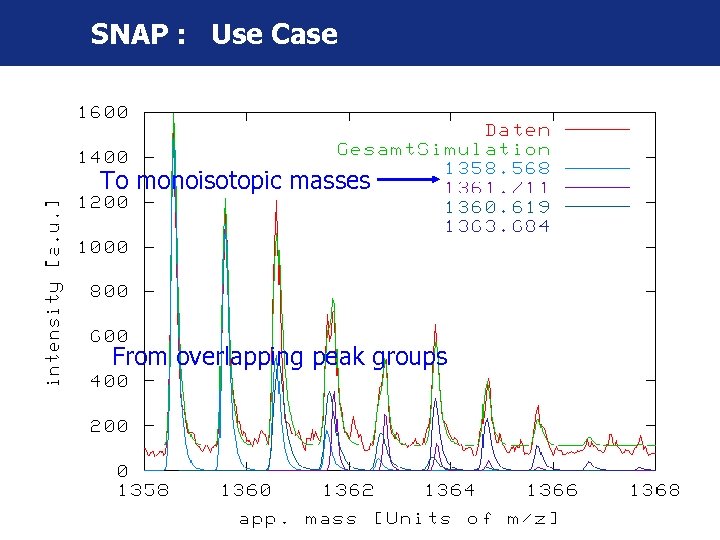 SNAP : Use Case To monoisotopic masses From overlapping peak groups
SNAP : Use Case To monoisotopic masses From overlapping peak groups
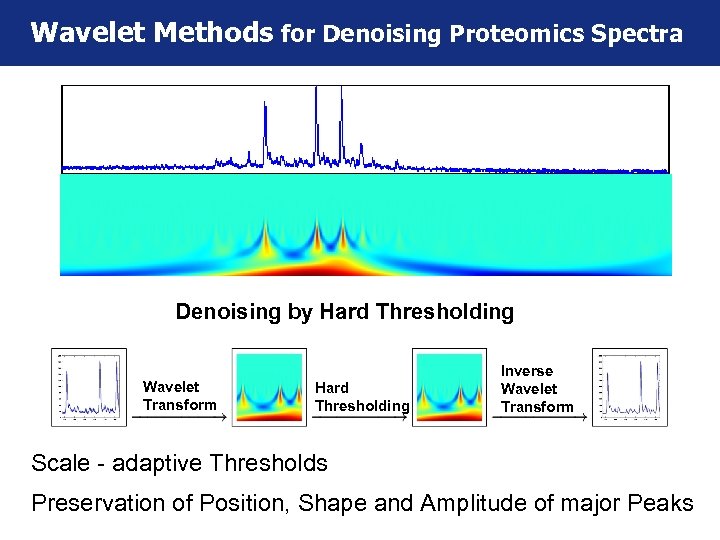 Wavelet Methods for Denoising Proteomics Spectra Denoising by Hard Thresholding Wavelet Transform Hard Thresholding Inverse Wavelet Transform Scale - adaptive Thresholds Preservation of Position, Shape and Amplitude of major Peaks
Wavelet Methods for Denoising Proteomics Spectra Denoising by Hard Thresholding Wavelet Transform Hard Thresholding Inverse Wavelet Transform Scale - adaptive Thresholds Preservation of Position, Shape and Amplitude of major Peaks
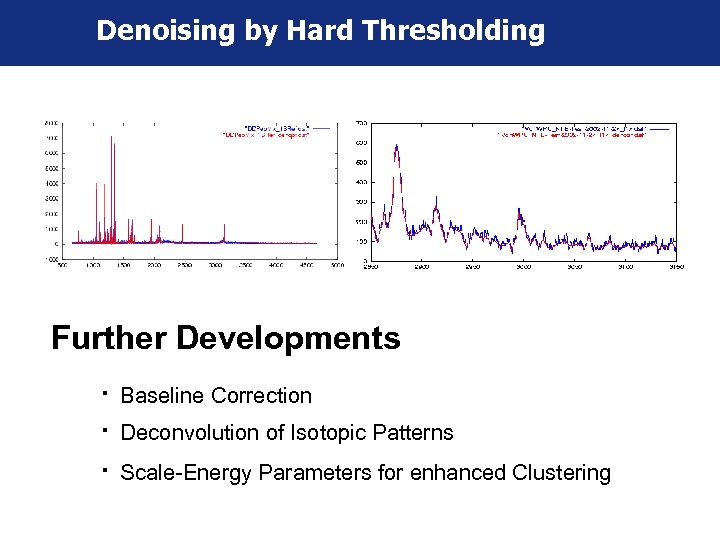 Denoising by Hard Thresholding Further Developments " Baseline Correction " Deconvolution of Isotopic Patterns " Scale-Energy Parameters for enhanced Clustering
Denoising by Hard Thresholding Further Developments " Baseline Correction " Deconvolution of Isotopic Patterns " Scale-Energy Parameters for enhanced Clustering
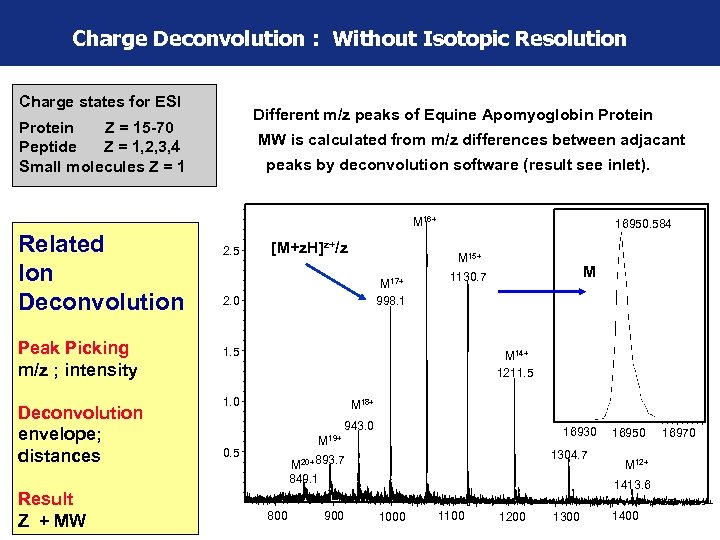 Charge Deconvolution : Without Isotopic Resolution Charge states for ESI Different m/z peaks of Equine Apomyoglobin Protein Z = 15 -70 Peptide Z = 1, 2, 3, 4 Small molecules Z = 1 MW is calculated from m/z differences between adjacant peaks by deconvolution software (result see inlet). M 16+ Related Ion Deconvolution Peak Picking m/z ; intensity Deconvolution envelope; distances Result Z + MW 2. 5 [M+z. H]z+/z 16950. 584 M 15+ M 17+ 998. 1 2. 0 M 1130. 7 1. 5 M 14+ 1211. 5 1. 0 M 18+ 943. 0 16930 M 19+ 0. 5 1304. 7 M 20+ 893. 7 849. 1 16950 16970 M 12+ 1413. 6 m/z 800 900 1000 1100 1200 1300 1400
Charge Deconvolution : Without Isotopic Resolution Charge states for ESI Different m/z peaks of Equine Apomyoglobin Protein Z = 15 -70 Peptide Z = 1, 2, 3, 4 Small molecules Z = 1 MW is calculated from m/z differences between adjacant peaks by deconvolution software (result see inlet). M 16+ Related Ion Deconvolution Peak Picking m/z ; intensity Deconvolution envelope; distances Result Z + MW 2. 5 [M+z. H]z+/z 16950. 584 M 15+ M 17+ 998. 1 2. 0 M 1130. 7 1. 5 M 14+ 1211. 5 1. 0 M 18+ 943. 0 16930 M 19+ 0. 5 1304. 7 M 20+ 893. 7 849. 1 16950 16970 M 12+ 1413. 6 m/z 800 900 1000 1100 1200 1300 1400
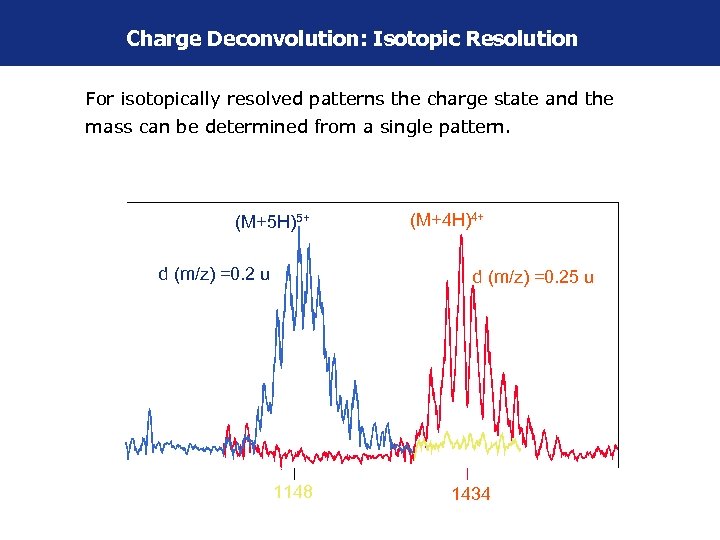 Charge Deconvolution: Isotopic Resolution For isotopically resolved patterns the charge state and the mass can be determined from a single pattern. (M+5 H)5+ d (m/z) =0. 2 u (M+4 H)4+ d (m/z) =0. 25 u 1148 1434
Charge Deconvolution: Isotopic Resolution For isotopically resolved patterns the charge state and the mass can be determined from a single pattern. (M+5 H)5+ d (m/z) =0. 2 u (M+4 H)4+ d (m/z) =0. 25 u 1148 1434
 Problems to be solved by Bioinformatics Get more accurate data Calibration
Problems to be solved by Bioinformatics Get more accurate data Calibration
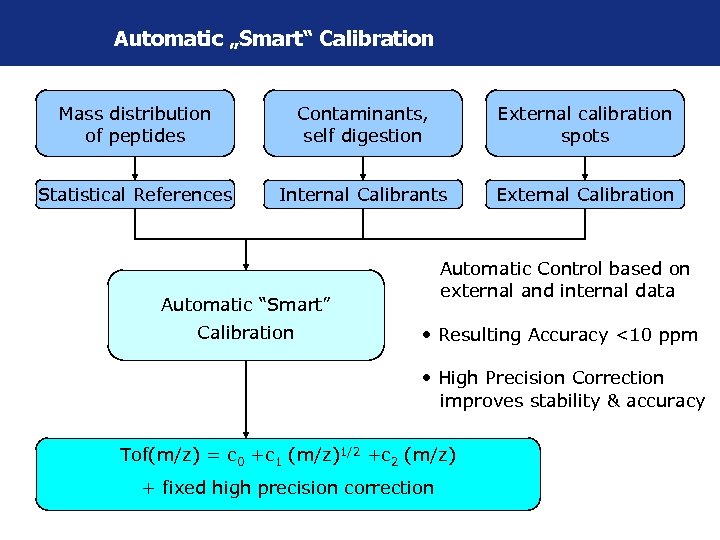 Automatic „Smart“ Calibration Mass distribution of peptides Contaminants, self digestion External calibration spots Statistical References Internal Calibrants External Calibration • Automatic Control based on external and internal data Automatic “Smart” Calibration • Resulting Accuracy <10 ppm • High Precision Correction improves stability & accuracy Tof(m/z) = c 0 +c 1 (m/z)1/2 +c 2 (m/z) + fixed high precision correction
Automatic „Smart“ Calibration Mass distribution of peptides Contaminants, self digestion External calibration spots Statistical References Internal Calibrants External Calibration • Automatic Control based on external and internal data Automatic “Smart” Calibration • Resulting Accuracy <10 ppm • High Precision Correction improves stability & accuracy Tof(m/z) = c 0 +c 1 (m/z)1/2 +c 2 (m/z) + fixed high precision correction
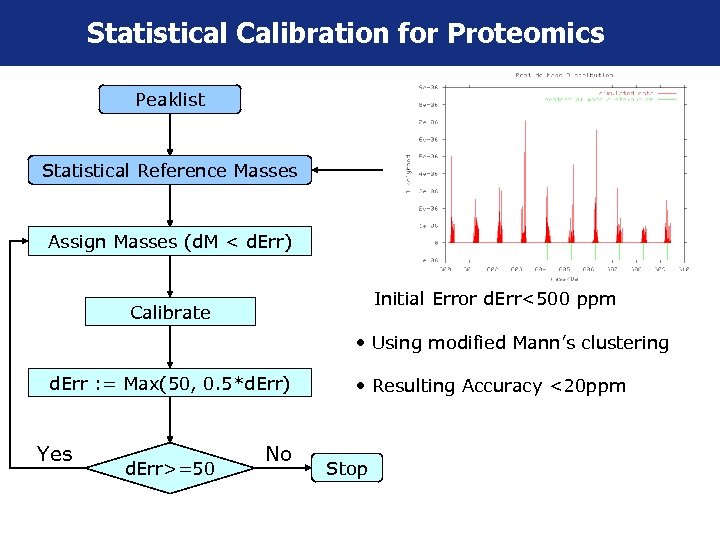 Statistical Calibration for Proteomics Peaklist Statistical Reference Masses Assign Masses (d. M < d. Err) • Initial Error d. Err<500 ppm Calibrate • Using modified Mann’s clustering d. Err : = Max(50, 0. 5*d. Err) Yes d. Err>=50 No • Resulting Accuracy <20 ppm Stop
Statistical Calibration for Proteomics Peaklist Statistical Reference Masses Assign Masses (d. M < d. Err) • Initial Error d. Err<500 ppm Calibrate • Using modified Mann’s clustering d. Err : = Max(50, 0. 5*d. Err) Yes d. Err>=50 No • Resulting Accuracy <20 ppm Stop
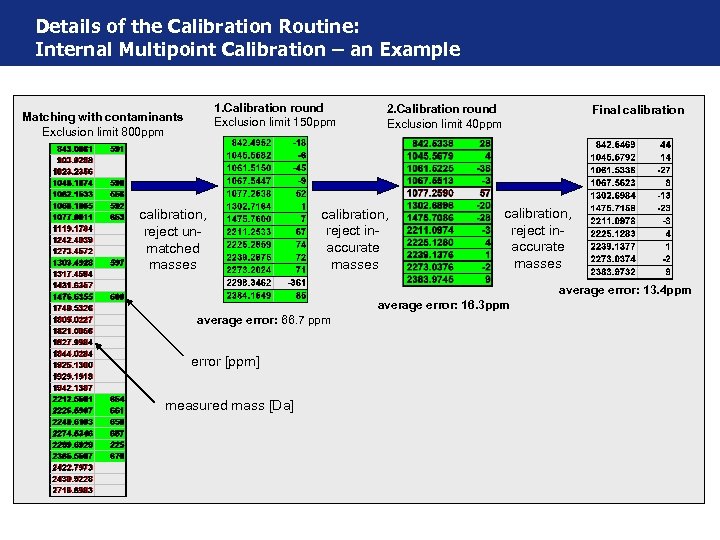 Details of the Calibration Routine: Internal Multipoint Calibration – an Example 1. Calibration round Exclusion limit 150 ppm Matching with contaminants Exclusion limit 800 ppm calibration, reject unmatched masses 2. Calibration round Exclusion limit 40 ppm calibration, reject inaccurate masses Final calibration, reject inaccurate masses average error: 13. 4 ppm average error: 16. 3 ppm average error: 66. 7 ppm error [ppm] measured mass [Da]
Details of the Calibration Routine: Internal Multipoint Calibration – an Example 1. Calibration round Exclusion limit 150 ppm Matching with contaminants Exclusion limit 800 ppm calibration, reject unmatched masses 2. Calibration round Exclusion limit 40 ppm calibration, reject inaccurate masses Final calibration, reject inaccurate masses average error: 13. 4 ppm average error: 16. 3 ppm average error: 66. 7 ppm error [ppm] measured mass [Da]
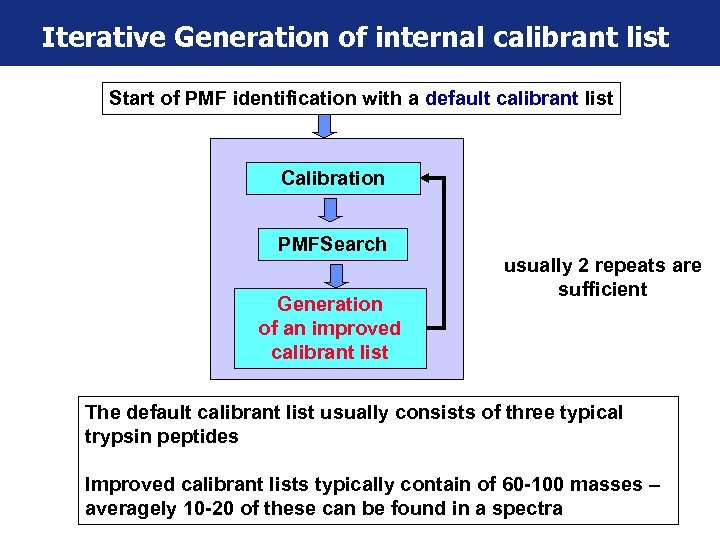 Iterative Generation of internal calibrant list Start of PMF identification with a default calibrant list Calibration PMFSearch Generation of an improved calibrant list usually 2 repeats are sufficient The default calibrant list usually consists of three typical trypsin peptides Improved calibrant lists typically contain of 60 -100 masses – averagely 10 -20 of these can be found in a spectra
Iterative Generation of internal calibrant list Start of PMF identification with a default calibrant list Calibration PMFSearch Generation of an improved calibrant list usually 2 repeats are sufficient The default calibrant list usually consists of three typical trypsin peptides Improved calibrant lists typically contain of 60 -100 masses – averagely 10 -20 of these can be found in a spectra
 Problems to be solved by Bioinformatics Search Engines MS based Identity Search
Problems to be solved by Bioinformatics Search Engines MS based Identity Search
 MS Protein Identification is Probability based How closely is a given protein or peptide sequence matching to the measured masses ? There are several strategies for a matching “ score“ : For example: -Probability based MOWSE score (Mascot) -Bayesian probability (Pro. Found) -Cross correlation (MS-Fit) Masses determined by MS are not unique Identification is probability based Problem of assigning true probabilities to a given identification
MS Protein Identification is Probability based How closely is a given protein or peptide sequence matching to the measured masses ? There are several strategies for a matching “ score“ : For example: -Probability based MOWSE score (Mascot) -Bayesian probability (Pro. Found) -Cross correlation (MS-Fit) Masses determined by MS are not unique Identification is probability based Problem of assigning true probabilities to a given identification
 Evaluation of PMF and Search Engines Part 1 Comparison of the performance of the search engines using a typical set of search parameters. Part 2 Successively changing various search parameters to test their influence. Optimisation of search parameters. Dataset: 168 MALDI PMF spectra the data was acquired in the environment of a typical proteome project About 10, 000 searches have been performed to establish a statistical basis
Evaluation of PMF and Search Engines Part 1 Comparison of the performance of the search engines using a typical set of search parameters. Part 2 Successively changing various search parameters to test their influence. Optimisation of search parameters. Dataset: 168 MALDI PMF spectra the data was acquired in the environment of a typical proteome project About 10, 000 searches have been performed to establish a statistical basis
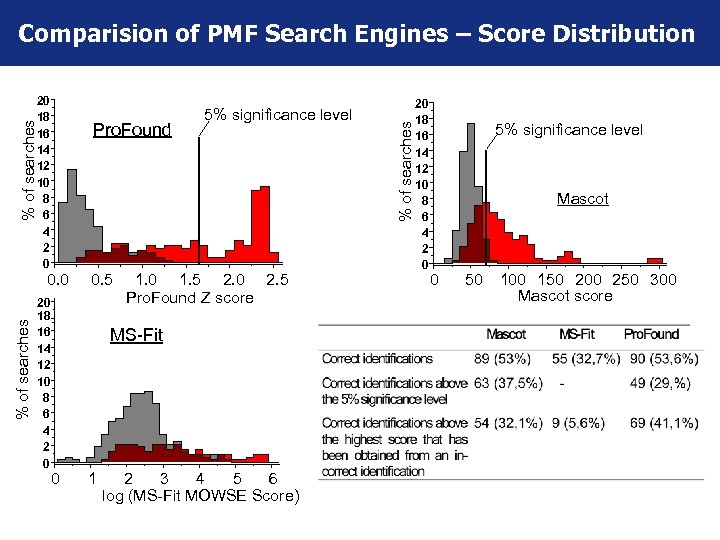 20 18 16 14 12 10 8 6 4 2 0 % of searches Pro. Found % of searches 0. 0 20 18 16 14 12 10 8 6 4 2 0 0. 5 5% significance level 1. 0 1. 5 2. 0 2. 5 Pro. Found Z score MS-Fit 0 1 2 3 4 5 6 log (MS-Fit MOWSE Score) % of searches Comparision of PMF Search Engines – Score Distribution 20 18 16 14 12 10 8 6 4 2 0 5% significance level Mascot 0 50 100 150 200 250 300 Mascot score
20 18 16 14 12 10 8 6 4 2 0 % of searches Pro. Found % of searches 0. 0 20 18 16 14 12 10 8 6 4 2 0 0. 5 5% significance level 1. 0 1. 5 2. 0 2. 5 Pro. Found Z score MS-Fit 0 1 2 3 4 5 6 log (MS-Fit MOWSE Score) % of searches Comparision of PMF Search Engines – Score Distribution 20 18 16 14 12 10 8 6 4 2 0 5% significance level Mascot 0 50 100 150 200 250 300 Mascot score
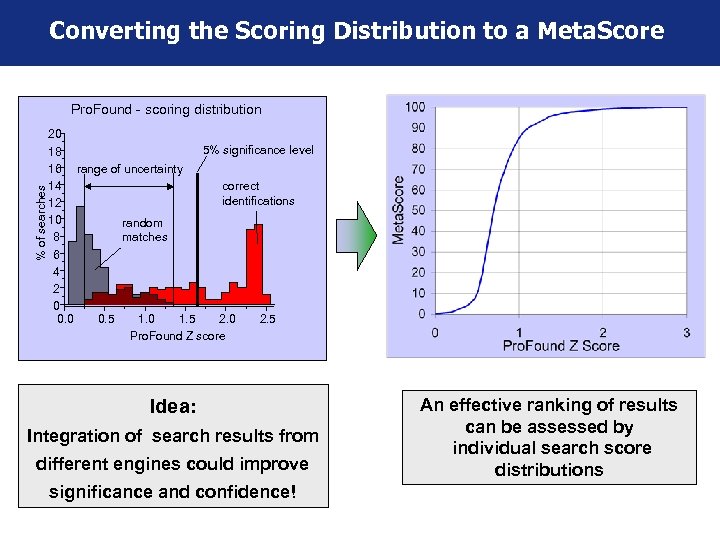 Converting the Scoring Distribution to a Meta. Score % of searches Pro. Found - scoring distribution 20 5% significance level 18 16 range of uncertainty 14 correct identifications 12 10 random 8 matches 6 4 2 0 0. 5 1. 0 1. 5 2. 0 2. 5 Pro. Found Z score Idea: Integration of search results from different engines could improve significance and confidence! An effective ranking of results can be assessed by individual search score distributions
Converting the Scoring Distribution to a Meta. Score % of searches Pro. Found - scoring distribution 20 5% significance level 18 16 range of uncertainty 14 correct identifications 12 10 random 8 matches 6 4 2 0 0. 5 1. 0 1. 5 2. 0 2. 5 Pro. Found Z score Idea: Integration of search results from different engines could improve significance and confidence! An effective ranking of results can be assessed by individual search score distributions
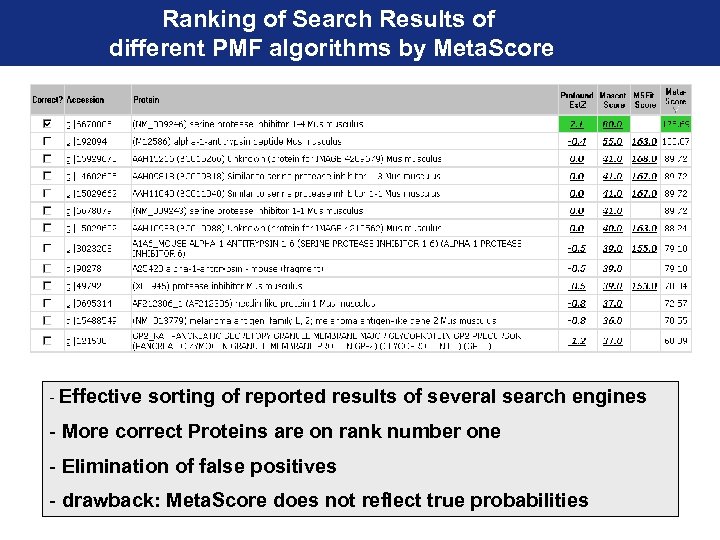 Ranking of Search Results of different PMF algorithms by Meta. Score - Effective sorting of reported results of several search engines - More correct Proteins are on rank number one - Elimination of false positives - drawback: Meta. Score does not reflect true probabilities
Ranking of Search Results of different PMF algorithms by Meta. Score - Effective sorting of reported results of several search engines - More correct Proteins are on rank number one - Elimination of false positives - drawback: Meta. Score does not reflect true probabilities
 Problems to be solved by Bioinformatics Search Engines Automated validation of Search Results
Problems to be solved by Bioinformatics Search Engines Automated validation of Search Results
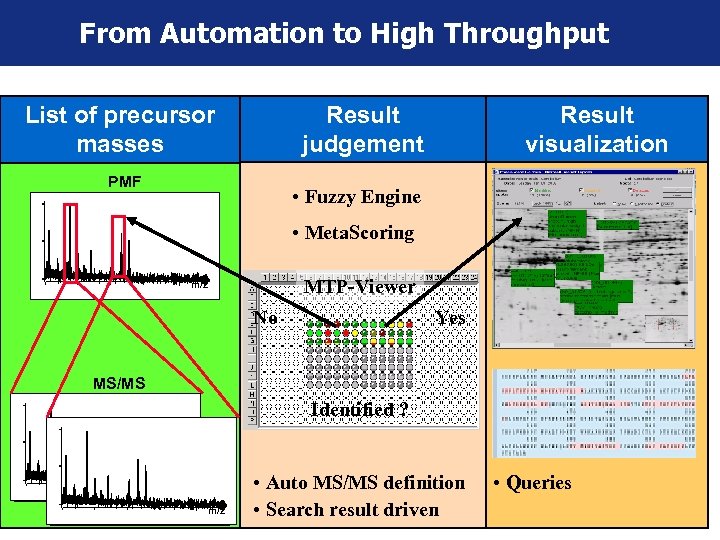 From Automation to High Throughput List of precursor masses Result judgement PMF Result visualization • Fuzzy Engine • Meta. Scoring MTP-Viewer m/z No Yes MS/MS Identified ? m/z • Auto MS/MS definition • Search result driven • Queries
From Automation to High Throughput List of precursor masses Result judgement PMF Result visualization • Fuzzy Engine • Meta. Scoring MTP-Viewer m/z No Yes MS/MS Identified ? m/z • Auto MS/MS definition • Search result driven • Queries
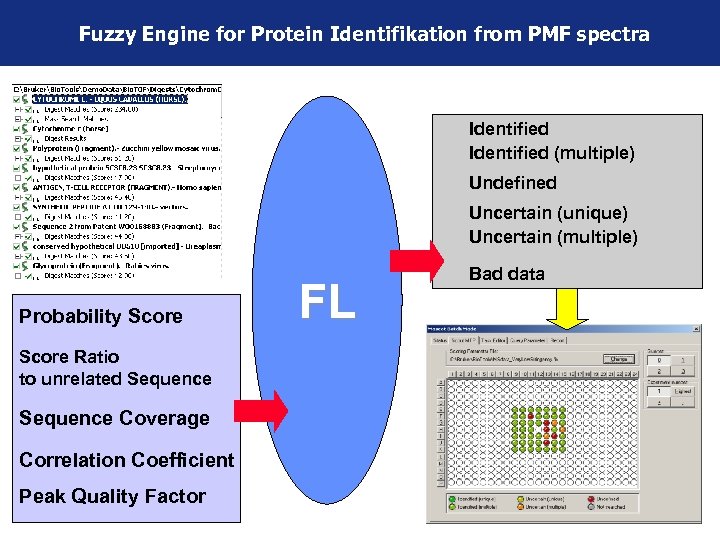 Fuzzy Engine for Protein Identifikation from PMF spectra Identified (multiple) Probability Score Undefined Uncertain (unique) Uncertain (multiple) Probability Score Ratio to unrelated Sequence Coverage Correlation Coefficient Peak Quality Factor FL Bad data
Fuzzy Engine for Protein Identifikation from PMF spectra Identified (multiple) Probability Score Undefined Uncertain (unique) Uncertain (multiple) Probability Score Ratio to unrelated Sequence Coverage Correlation Coefficient Peak Quality Factor FL Bad data
 Problems to be solved by Bioinformatics Automation & High Throughput Automated MS/MS Precursor Ion Selection
Problems to be solved by Bioinformatics Automation & High Throughput Automated MS/MS Precursor Ion Selection
 Strategies for automated MS/MS acquisition
Strategies for automated MS/MS acquisition
 Acknowledgement Bruker Daltonik Jens Decker , Michael Kuhn Martin Blüggel , Daniel Chamrad Peter Maaß Kristian Bredies
Acknowledgement Bruker Daltonik Jens Decker , Michael Kuhn Martin Blüggel , Daniel Chamrad Peter Maaß Kristian Bredies


